Abstract
Lim domain only 2 (LMO2) is a transcriptional co-factor required for angiogenesis and the specification of haematopoietic cells during development. LMO2 is widely expressed within haematopoiesis with the exception of T-cells. Failure to downregulate LMO2 during T-cell maturation leads to leukaemia, thus underlining the critical nature of context-dependent regulation of LMO2 expression. We previously identified a distal regulatory element of LMO2 (element -25) that cooperates with the proximal promoter in directing haematopoietic expression. Here we dissected the functional activity of element -25 and showed it to consist of two modules that conferred independent and cell-type specific activities: a 3’ myeloid enhancer and a 5’ T-cell repressor. The myeloid enhancer was bound by GATA2 in progenitors and its activity depended on a highly conserved GATA motif, whereas the T-cell repressor moiety of element -25 was bound by the Core Binding Factor in T-cells and its repressive activity depended on a highly conserved RUNT motif. Since the myeloid enhancer and nearby downstream region is recurrently involved in oncogenic translocations, our data suggest that the -25 enhancer region provides an open chromatin environment prone to translocations, which in turn cause aberrant LMO2expression in T-cells due to the removal of the adjacent T-cell repressor.
Introduction
Lim domain only 2 (LMO2) is a zinc-finger transcriptional co-factor required for angiogenesis and emergence of haematopoietic stem cells during ontogenesis [1–4]. LMO2 forms a complex with the LIM domain-binding protein 1 (LDB1) and the DNA-binding E-box and GATA transcription factors. This complex has been shown to be critical for the specification of haematopoietic stem cells and erythroid lineage [5–8]. LMO2 was originally identified through its involvement in recurrent chromosomal translocations [9, 10]. LMO2 is a major oncogene and its ectopic expression leads to T-cell lymphoproliferative disease and T-cell acute lymphoblastic leukaemia (T-ALL) [11–13]. Recently, gene-expression profiling studies revealed high LMO2 expression in different subtypes of B-cell lymphoproliferative disorders or acute myeloid leukaemia, thus, suggesting a broader oncogenic effect in different haematopoietic lineages caused by failure of LMO2 down-regulation [14–22]. Juxtaposition of TCR enhancers is thought to be the main driving mechanism for ectopic LMO2 expression [23, 24]. However, this notion has recently been challenged by a detailed break point analysis in TCRdelta-LMO2 rearranged T-ALL patients [25]. Therefore, investigation of context-dependent regulation of important developmental genes such as LMO2 remains instrumental for understanding oncogenic deregulation in leukaemogenesis.
The LMO2 gene is localised on the short arm of chromosome 11 within band 13 (11p13) and its expression is tightly regulated in the haematopoietic system. LMO2 expression is directed by a proximal and a distal promoter element that generate transcripts with distinct 5’ untranslated regions but an identical coding region derived from exons 3–6 [26]. Additionally, our group recently reported an intermediate promoter element (mdp) that mediates LMO2 expression in a subset of T-acute lymphoblastic leukaemia patients [27]. We previously showed that the proximal promoter element of LMO2 confers endothelial-specific activity [28], although additional distal regulatory elements are required for a comprehensive and context-dependent regulation of Lmo2-expression in haematopoiesis [29]. We identified a distal regulatory element located 25kb upstream of the proximal promoter (element -25) that cooperates with the proximal promoter in specifying cell-context dependent haematopoietic expression to foetal liver in transient transgenic mouse assays [29]. This element is highly enriched for acetylation of lysine 9 of histone H3 (H3K9ac), a histone mark associated with accessible regions of chromatin, in the haematopoietic progenitor cell line HPC7 and the myeloid progenitor cell line 416B [29]. Interestingly, medium to low levels of H3K9ac enrichment were found in endothelial (MS1), erythroid (MEL) and T- (BW5147) cell lines as well as whole adult murine thymus, suggesting dynamic occupancy of element -25 during differentiation. Moreover, when enhancer activity was tested in conjunction with the proximal promoter element in stable transfections, element -25 was the strongest from an array of 14 elements in the myeloid progenitor 416B cell line [29].
In the current study, we have dissected in detail the regulatory activity of element -25. We demonstrate that element -25 retains dual and cell-type specific activity, acting as a myeloid enhancer and a T-cell repressor. We defined a myeloid enhancer module in the 3’ region of element -25 that is dependent on a highly conserved GATA binding site and a T-cell repressor module localised in the 5’ region, which activity depends on a highly conserved RUNT binding site.
Materials and Methods
Cell lines and cell culture
The murine multipotent myeloid progenitor cell line 416B [30], erythroleukaemia cell line F4N [31], endothelial cell line MS1 [32] as well as the human T-cell lines Jurkat (DSMZ, ACC282) and Molt4 (DSMZ, ACC362) were cultured as previously described [27–29].
Sequence analysis
Homologous genomic LMO2 sequences of element -25 derived from human, mouse, cow, dog and cat were downloaded from Ensembl, aligned using multi-Lagan [33] and displayed using Genedoc [34]. Candidate transcription factor binding sites were identified using Transcription Factor Binding Sites Search (TFBSsearch) [35] and the Transcription Element Search Software (TESS: www.cbil.upenn.edu/tess ) programs [36].
Reporter constructs
The LMO2 luciferase reporter constructs were amplified from human genome using primers listed in S1 Table, cloned into pGL2-luciferase vectors (Promega Corporation, Madison, WI) and confirmed by sequencing. Deletion constructs were produced by restriction enzyme digestion and re-ligation. Mutation constructs were generated with QuickChange XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA) using primers listed in S2 Table. All constructs were confirmed by sequencing.
Stable transfection experiments
All cell lines were stably transfected by electroporation as previously described [37]. G418 was added 24 hours post transfection and resistant cells were assayed 14 days later. Transfection experiments were performed at least in triplicate and at least on two different occasions. Results are shown as mean and standard error of the mean (SEM). Comparison among two groups was performed by t-test (Fig 1B and 1C). Comparison among more than two groups was performed by one-way analysis of variance followed by post-hoc analysis with the Bonferroni test for selected pairs of columns (Fig 1A) or Dunnett's test (Figs 2, 3D and 4D) to evaluate the significance of the differences between two groups. Statistical significance was assumed when P < 0.01.
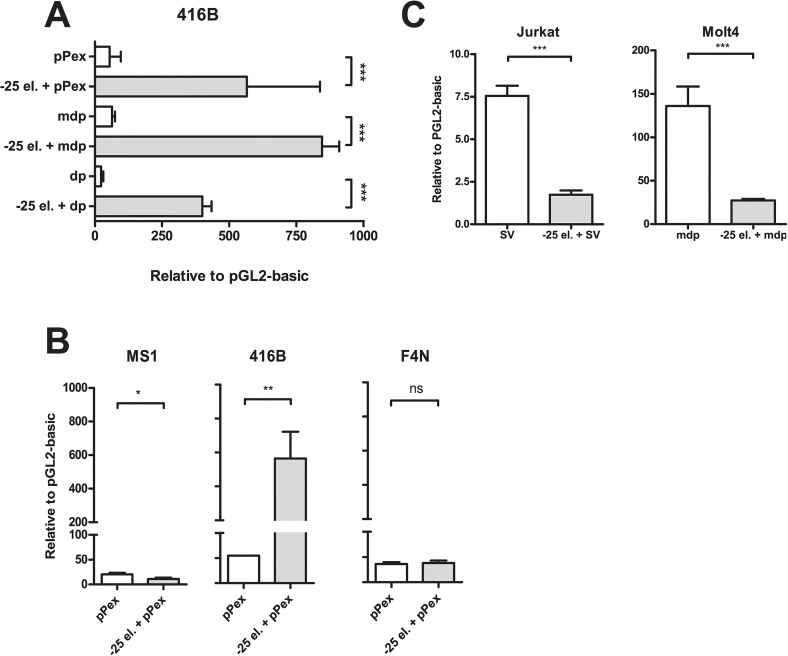
Fig 1. Cell-type specific activity of LMO2 element -25.
A) Promoter-independent enhancer activity of element -25 in multipotent myeloid progenitors 416B. Luciferase activity of LMO2 proximal (pPex), intermediate (md) or distal (dp) promoter elements in the presence and absence of element -25 (-25 el.) was measured in 416B cells. B) Cell-type specific activity of -25kb DRE. Luciferase experiments were performed in endothelial MS1 and erythroid F4N cells using LMO2 proximal (pPex) promoter element in the presence and absence of element -25 (-25 el.). For comparison purposes, 416B data from panel A is also shown. C) T-cell repressor activity of element -25. Luciferase experiments were performed in LMO2 expressing Molt4 and LMO2 non-expressing Jurkat cells, using, respectively, LMO2 intermediate (md) or minimal SV promoter elements in the presence and absence of element -25 (-25 el.). In all cases, mean and standard error of the means (SEM) for at least two independent stable transfections (each one performed at least in triplicate) are shown. Values are expressed relative to empty vector, pGL2 basic. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
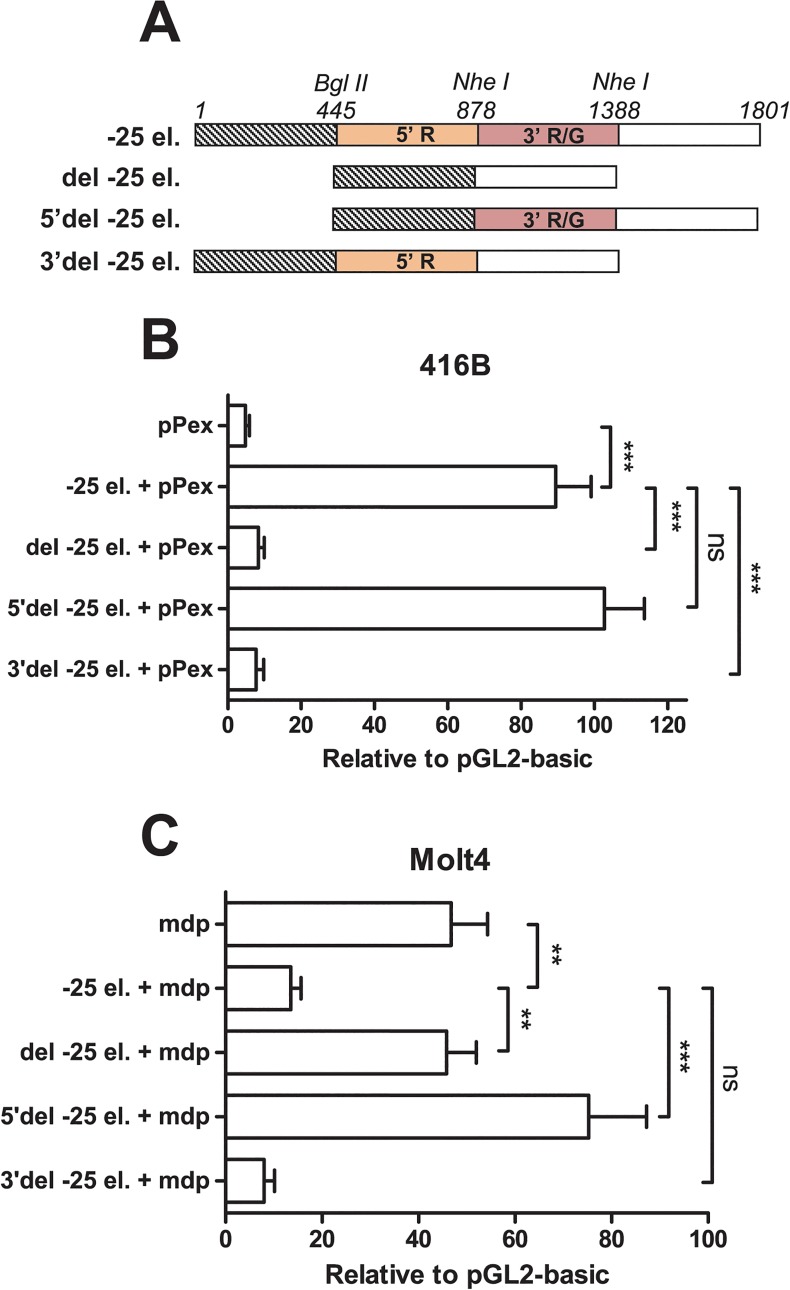
Fig 2. Dual myeloid enhancer and T-cell repressor activity of element -25.
A) Diagram of element -25 and deletion constructs. 5’ T-cell repressor and 3’ myeloid enhancer modules of element -25 (-25 el.) are coloured in orange and red, respectively. The presence of the conserved 5’ RUNT (5’ R), 3’ RUNT and GATA (3’ R/G) motifs is indicated. Three deletion constructs (del -25 el., 5’del -25 el. and 3’del -25 el.) were produced by restriction enzyme digestion using BglII (position 445) and NheI (positions 878 and 1388). B, C) Independent myeloid enhancer and T-cell repressor activities of 2 modules within element -25. Deletion constructs were stably transfected together with either LMO2 proximal (pPex) or intermediate (mdp) promoter elements in myeloid progenitors 416B (B) and T-cells Molt4 (C), respectively. In all cases, mean and standard error of the means (SEM) for at least two independent stable transfections (each one performed at least in triplicate) are shown. Values are expressed relative to empty vector, pGL2 basic. **P < 0.01; ***P < 0.001; ns, not significant.
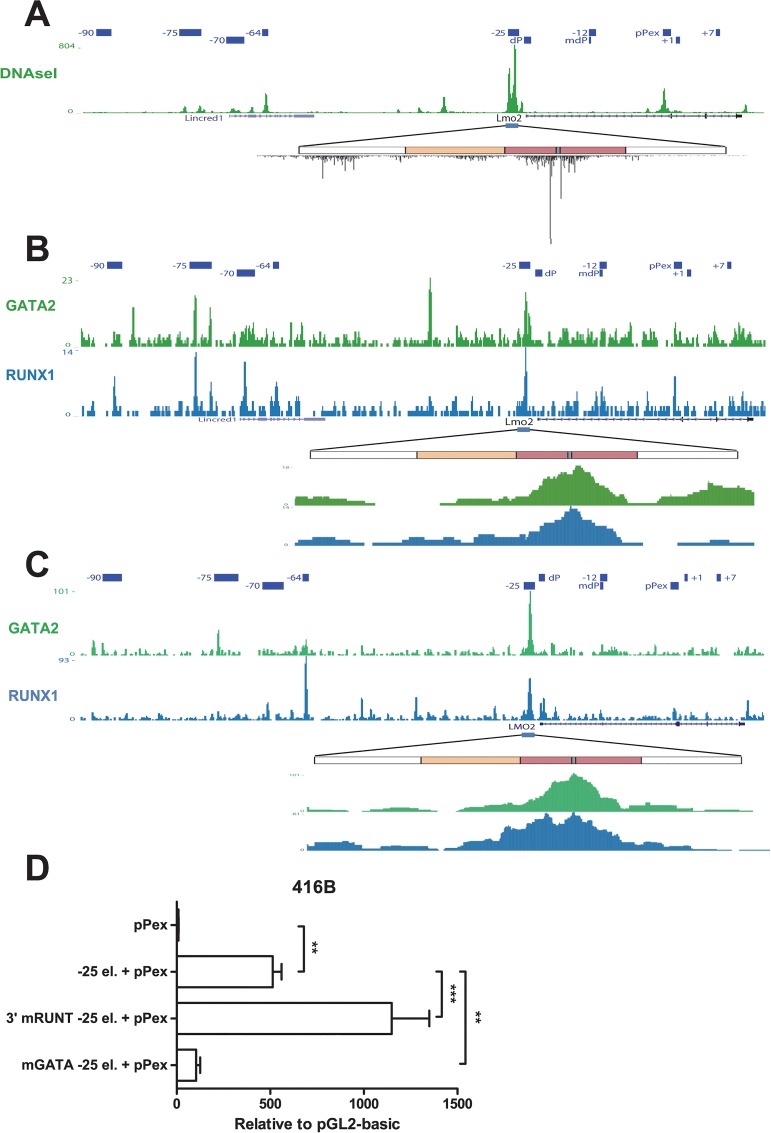
Fig 3. The 3’ myeloid enhancer activity of element -25 is dependent on GATA binding.
A) DNaseI-Seq profile of Lmo2 locus region in mouse myeloid progenitors 416B cells. Elements previously described to be involved in the regulation of Lmo2 expression are indicated. The strongest DHS in the Lmo2 locus co-localises with element -25. A magnification of the region corresponding to element -25 is shown. Diagram of element -25 as in Fig 2A. DNaseI profile shows a strong protection in the region corresponding to conserved GATA motif (location shown in grey). B, C) ChIP-Seq profile of RUNX1 and GATA2 in LMO2 locus region in murine haematopoietic progenitors HPC7 (B) and human CD34+ (C) cells. Elements previously described to be involved in the regulation of LMO2 expression are indicated. Strong binding for GATA2 and RUNX1 can be detected at element -25. A magnification of the region corresponding to element -25 is shown. Diagram of element -25 as previously. Binding of RUNX1 and GATA2 specifically takes place at the 3‘ myeloid enhancer module of element -25. D) Myeloid enhancer activity depends on a conserved GATA motif. Conserved RUNT and GATA motifs present in the myeloid enhancer module of element -25 were mutated (3’mRUNT -25 el. and mGATA -25 el., respectively). Mutated constructs were stably transfected together with LMO2 proximal promoter element (pPex) in myeloid progenitor 416B cells and luciferase activity was measured. Mean and standard error of the means (SEM) for at least two independent stable transfections (each one performed at least in triplicate) are shown. Values are expressed relative to empty vector, pGL2 basic. **P < 0.01; ***P < 0.001.
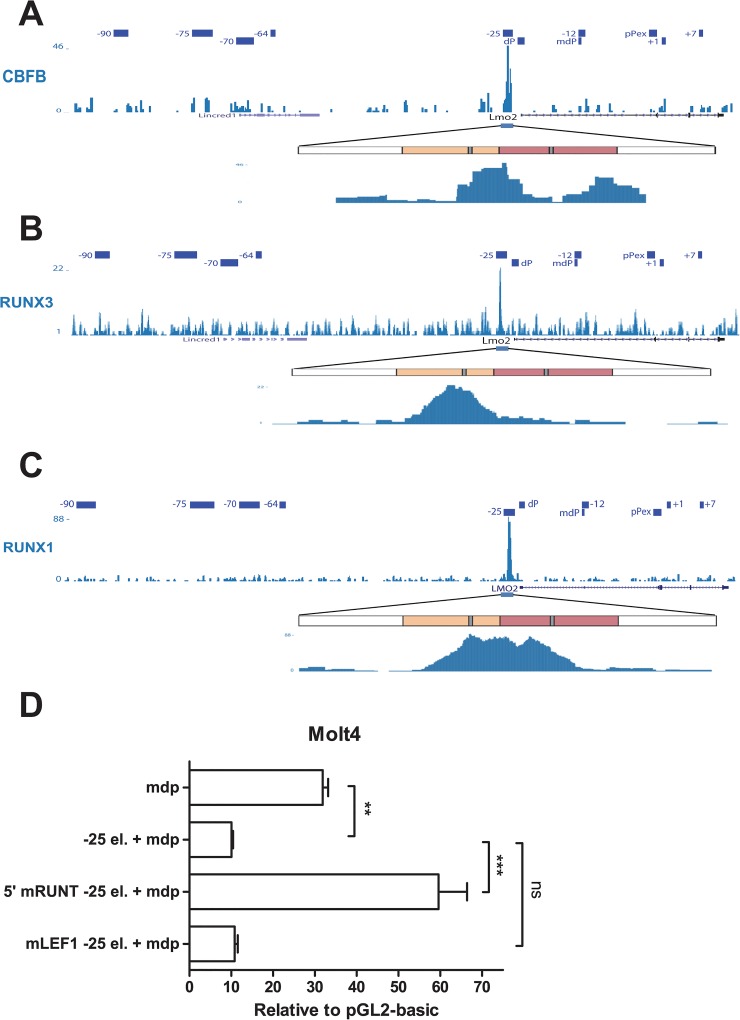
Fig 4. The 5’ T-cell repressor activity of element -25 is dependent on Runx binding.
A, B, C) ChIP-Seq profile of the LMO2 locus region for CBFB in murine thymocytes (A), RUNX3 in murine CD8+ SP T-cells (B) and RUNX1 in Jurkat cells (C). Elements previously described to be involved in the regulation of LMO2 expression are indicated. Binding for CBF can be detected at element -25. A magnification of the region corresponding to element -25 is shown. Diagram of element -25 as previously. While binding of CBFB can be detected at both modules of element -25, RUNX1 and RUNX3 are preferentially bound to the 5‘ T-cell repressor module of element -25 (location of conserved RUNT motifs is shown in grey). D) T-cell repressor activity depends on a conserved RUNT motif. Conserved RUNT and LEF-1 motifs present in the T-cell repressor module of element -25 were mutated (5’ mRUNT -25 el. and mLEF1–25 el., respectively). Mutated constructs together with LMO2 intermediate promoter element (mdp) were stably transfected in Molt4 T-cells and luciferase activity was measured. Mean and standard error of the means (SEM) for a representative experiment of at least two independent stable transfections (each one performed at least in triplicate) are shown. Values are expressed relative to empty vector, pGL2 basic. **P < 0.01; ***P < 0.001; ns, not significant.
Display of ChIP-Seq traces
ChIP-Sequencing traces were retrieved from the publicly available compendium of Next Generation Sequencing experiments Codex (http://codex.stemcells.cam.ac.uk/) [38] and displayed in UCSC genome browser (http://genome.ucsc.edu/).
Results
Element -25 acts as a cell-type-specific enhancer
Element -25 consistently directs staining to foetal liver cells in transgenics and shows strong enrichment for active chromatin marks in the multipotential hematopoietic progenitors HPC7 and myeloid progenitors 416B [29]. Element -25 also displays strong enhancer activity in the myeloid progenitor cell line 416B in stable transfections [29]. To investigate its regulatory function in detail, element -25 in conjunction with each promoter element of LMO2 (proximal, intermediate or distal) was inserted into the pGL2 basic luciferase vector and stably transfected into 416B cells. Of note, an extended version of the proximal promoter element (pPex) reported to provide stronger expression was used [29]. We found a consistent increase (over 10 fold) of luciferase activity of all 3 promoters in the presence of element -25 (Fig 1A), indicating that element -25 confers promoter-independent enhancer activity in 416B cells.
To investigate potential context-dependent activity of this enhancer, stable transfections were performed using the element -25 in collaboration with pPex (the most active promoter element, data not shown) in endothelial (MS1) and erythroleukaemia (F4N) cell lines, both of which express Lmo2. Element -25 did not exhibit any activity in either of these 2 cell lines (Fig 1B). These results suggest that element -25 acts as a cell-type-specific enhancer that controls the expression of LMO2 in multipotent myeloid progenitors.
Dual myeloid enhancer and T-cell repressor activity of element -25
Our data indicates that element -25 displays enhancer activity in specific cell types. Previous work from Hammond et al described a specific T-cell repressor region 2.5 kb upstream of the distal promoter element (dp) of LMO2 [39]. Interestingly, this specific T-cell repressor region is contained within element -25. LMO2 is not expressed in mature T cells hence we investigated the activity of element -25 in the T-ALL cell lines Jurkat and Molt4. Jurkat is a non-LMO2-expressing cell line while Molt4 expresses LMO2 in the absence of a translocation involving LMO2. We found that a minimal version of the SV40 promoter directed low reporter expression in Jurkat cells and we decided to use the intermediate promoter element (mdp) in the cell line Molt4 since our group previously showed high activity of this element in this cell line [27]. We then performed stable transfections of the element -25 together with minimal SV40 or intermediate promoters in Jurkat and Molt4 T-cells, respectively. In line with the report from Hammond et al. [39], we found a consistent 80% repression inferred by element -25 in both cell lines (Fig 1C). These results indicate that element -25 shows a promoter-independent repressor activity in T-cells. Promoter-independent activity seems critical for genes such as LMO2 where the gene locus contains three distinct promoter elements. Taken together, our results show that element -25 contains a cell-type-specific dual activity as both myeloid enhancer and T-cell repressor.
Element -25 contains myeloid enhancer and T-cell repressor modules
To further understand the dual function of element -25, we produced three deletion constructs (Fig 2A) and analysed the effect of these deletions on the activity of element -25. To this end, we performed stable transfections of the different versions of element -25 together with the proximal or intermediate promoter elements in 416B and Molt4 cell lines, respectively (Fig 2B and 2C). When the central part of element -25 (445–1388 bp) was removed (del -25), we observed that both myeloid enhancer and T-cell repressor activities were completely abolished. To further dissect the element, a 5’ fragment (445–878 bp) was deleted (5’del -25); of note, this fragment included the T-cell repressor region previously described [39]. This deletion did not have any effect on the enhancer activity in 416B myeloid cells but completely abolished the repressor effect of this element in Molt4 T-cells. Finally, a fragment located in the 3’ region of element -25 (878–1388 bp) was deleted (3’del -25). In this case, deletion completely abolished enhancer activity in myeloid cells but did not have an effect on the repressor activity in T-cells. Taken together, our results show that element -25 comprises 2 juxtaposed independent modules that act as repressor and enhancer elements in a cell-type-specific manner.
The 3’ myeloid enhancer activity of element -25 is dependent on GATA binding
To determine which transcription factors could be involved in the enhancer activity of element -25, comparative genomic analysis was performed. We produced a multi-specie sequence alignment for the 3’ region of element -25 (S1 Fig) and identified highly conserved transcription factor consensus binding sites in this region. Several potential transcription-factor binding sites that were highly conserved between species were identified using TFBSsearch [35] and TESS [36], comprising putative binding sites for Interferone Response Element 1 (IRF1) as well as E-twenty six (ETS), RUNT and GATA.
Our previous studies showed that ETS and GATA2 factors are bound to element -25 in 416B myeloid progenitors [29]; however, in order to gain insight into which binding motifs were actually occupied in this region, we interrogated a DNAseI genome-wide dataset performed in 416B cells [40] (Fig 3A). The most prominent DNaseI hypersensitive (DHS) region within the Lmo2 locus in 416B cells can be detected in the genomic region corresponding to element -25, specifically in the region corresponding to the 3’ enhancer module of the element. A more detailed look at this module shows that there is a protected region between 2 highly hypersensitive sites that colocalises with the putative RUNT and GATA motifs. This pattern of DNaseI cleavage represents the classical pattern seen for regions that are strongly bound by transcription factors [41]. Investigation of recently published ChIP-Seq datasets furthermore supported GATA2 and RUNX1 binding to the myeloid enhancer moiety of this region in haematopoietic progenitors (HPC7) [42] and human CD34+ peripheral blood cells [43] (Fig 3B and 3C).
In order to test the importance of the RUNT and GATA motifs for the myeloid enhancer activity of element -25, we mutated the highly conserved RUNT (3’ RUNT) and GATA motifs present in the myeloid module of element -25 and then analysed the effect on luciferase activity in myeloid progenitors 416B. While the mutation of the RUNT motif did not reduce the activity of the promoter, mutation of the GATA motif drastically reduced the myeloid enhancer activity of element -25 (Fig 3D). Taken together, these results show that although the 3’ module of element -25 is bound by RUNX1 and GATA2 in human and mouse progenitors, the specific enhancer activity in myeloid progenitors only depends on the GATA motif.
5’ T-cell repressor activity depends on RUNT consensus binding site
In a similar way, we performed multi-specie alignments for the 5’ T-cell repressor module of element -25 (S1 Fig). The 5’ T-cell repressor shows less homology than the 3’ enhancer region revealing only highly conserved motifs for Lymphoid enhancer-binding factor 1 (LEF1) and RUNT proteins. During normal T-cell development, LMO2 is expressed at the double-negative (DN) stage but is progressively down regulated during T-cell development. Although LMO2 is expressed at fairly high levels in DN1 cells, its expression is severely reduced at DN2 stage and is barely detectable at the stages DN3 and DN4, double-positive (DP) and single-positive (SP) CD4+. LMO2 cannot be detected in CD8+ SP cells [44].
RUNX1 and RUNX3 are important at different stages of T-cell differentiation. Both proteins can interact with the CBFb protein to constitute the so-called Core Binding Factor (CBF). We interrogated a previously published dataset for binding of CBFb in thymocytes [45]. Strong binding can be detected in the region corresponding to the repressor module for CBFb (Fig 4A). Of note, CBFb binding was also detected at the myeloid moiety of the enhancer, very likely due to the mixed population present in the sample. RUNX3 is known to act as a repressor of CD4 in CD8+ SP cells. We also processed and interrogated a dataset for RUNX3 in CD8+ SP cells [46] (Fig 4B) where strong binding for RUNX3 specifically at the 5’ repressor module can be detected. In a similar way, we interrogated a previously published dataset for RUNX1 in the Jurkat T-cell line [47], which does not express LMO2 (Fig 4C). Like in CD8+SP cells, strong binding for RUNX1 can be detected at 5’ repressor module of the element -25. These results strongly suggest that the highly conserved RUNT motif found in the repressive module could be responsible for the repressor activity of this element.
We then decided to explore this hypothesis. We generated a version of element -25 where the RUNT motif located in the 5’ region was mutated and then we evaluated the effect in luciferase assays in Molt4 cells. We found complete abrogation of T-cell repressor activity by removing the RUNT binding motif at position 735bp (5’ RUNT), whereas activity remained unaffected after mutation of the LEF binding motif at position 549bp (Fig 4D). Our mutational analysis suggests therefore that the 3’ myeloid enhancer relies mainly on the GATA binding motif at position 1091bp whereas the 5’ T-cell repressor activity requires the RUNT binding motif at position 735bp (5’ RUNT).
Discussion
We previously showed that the LMO2 proximal promoter element drives LMO2 expression in endothelial cells as well as in haematopoietic cells by transient transgenic mouse assays [28, 29]. However, cooperation with additional regulatory modules, such as element -25, was required for robust expression in haematopoietic cells [29]. In our previous study, we described that element -25 was important for LMO2 expression in foetal liver cells, which comprises both erythroid and myeloid progenitors. In this study, we performed a detailed functional characterisation of element -25. We identified conserved transcription factor regions and provided functional demonstration of the effect of their deletion and mutation using stable transfection of haematopoietic cell lines. Moreover, we took advantage of previously published next generation sequence experiments and mined publicly available whole-genome transcription factor binding datasets to further support our results. Our current study reveals a dual cell-context dependent function of element -25. We identified 2 juxtaposed modules within element -25, one located in the 5’ region that acted as a repressor in T-cells and one located in the 3’ region that conferred enhancer activity in myeloid progenitor cells.
The spatial clustering of a myeloid enhancer and T-cell repressor may facilitate rapid “on-off switching” during the transition from a haematopoietic stem-progenitor to a T-cell lymphoid regulatory program. One potential mode of action would be that the close proximity of the 3’ myeloid enhancer and 5’ T-cell repressor module may accelerate the deployment of enzymes associated with the epigenetic machinery to the adjacent region of opposite functionality. It has been shown before that chromatin states can spread from an initial nucleation event (reviewed in [48, 49]). It will be intriguing therefore to investigate whether similar clustering may constitute a general characteristic of critical regulatory elements of developmental genes required for a transition from stem/progenitor cell to a more differentiated program.
The activity of the 3’ myeloid enhancer is strongly dependent on a highly conserved GATA site that is bound by GATA2 in mouse and human haematopoietic progenitor cells. Surprisingly, even though there is a highly conserved RUNT motif in this part of element -25, the myeloid enhancer activity was not only independent on this motif but the activity increased following the mutation of this motif. However, strong protection within the DHS could be detected in the region corresponding to the RUNT motif in 416B myeloid progenitors which indicates that this site is occupied in these cells and RUNX1 is also strongly bound to this region in progenitors (Fig 3). These results suggest that RUNX1 may exist in different complexes in myeloid progenitor cells that can have either activating or repressing enhancer activity. Direct interaction between RUNX1 and GATA2 and recruitment of RUNX1 in the absence of RUNT motif by GATA2 has been previously reported [42]. It is then reasonable to assume that RUNX1 recruitment to the myeloid enhancer module of element -25 is still important, although this may be mediated by GATA2 when the RUNX site is mutated. RUNX1-activating complexes would then contain GATA2 and mainly depend on GATA2 binding. A RUNX1-fusion transcription factor complex has been recently described in the Kasumi-1 myeloid cells [50, 51]. In these cells, the RUNX1-fusion complex mainly acts as a repressor [51] and lacks GATA2 [50]. In Kasumi-1 cells, coexistence of RUNX1 active and repressive complexes ensures fine regulation of the expression of multiple genes [51]. Interestingly, the strongest binding event in the LMO2 locus for AML1-ETO containing-transcription factor complex (of which LMO2 is a key component) in Kasumi-1 cells (that express high levels of LMO2) is located at the 5’ enhancer region of element -25 [51]. Similarly to our results, treatment of Kasumi-1 cells with shRNA against the fusion protein resulted in a slight upregulation of the expression of LMO2 [50].
The 5’ T-cell repressor module of element -25 that we describe overlaps with a previously described repressor element in LMO2 [39]. In contrast to our results, the previous study reported that the RUNT motif is not important for the repressor activity. These discrepancies may be due to at least two reasons. Firstly, we used Molt4 cells as a model in contrast to the previous study which used Jurkat cells. The use of Molt4 cells is supported by our previous observation that Molt4 cells express LMO2 at higher levels than Jurkat cells [27] in the absence of any known translocation involving the LMO2 locus. Secondly, the manner in which luciferase assays were performed differed in both studies. In our study we performed stable transfections in contrast to that of Hammond et al. which used transient transfections. Only the use of stable transfection assays allows a comprehensive evaluation of any transcriptional effects associated with integration into chromatin, which is known to be pivotal for multiple aspects of transcriptional repressor activity.
Critical role of RUNX factors in down-regulation of LMO2 during T-cell development
LMO2 down-regulation is known to be critically required for terminal differentiation of T-cells. In murine models, enforced expression of Lmo2 in thymocytes blocks differentiation at the double negative stage and, eventually progression to a T-cell lymphoproliferative disease takes place [9–13]. On the other hand, up-regulation of Runx1 is essential at late double negative stages and, in Runx1 deficient mice, development of T-cell lymphoma has been reported [52]. The role of RUNX proteins in T-cell development has been extensively studied, especially in the context of the CD4/CD8 specification process. Thus, it is known that a 434 bp repressor located in the CD4 locus is responsible for the appropriate spatial and temporal expression of CD4 during T-cell development. This repressor is bound by RUNX1 at early stages of development (DN) and by RUNX3 in CD8+ SP cells and requirement for these factors in CD4 silencing was confirmed using transgenic mice deficient in Runx1 and Runx3 at the appropriate stages [53, 54]. We show in our study that CBF binds to the 5’ T-cell repressor region in thymocytes and CD8+ SP cells, similarly to the CD4 repressor. Our results therefore support a regulatory model, in which the up-regulation of RUNX proteins during T cell differentiation is required for the down-regulation of LMO2 mediated by CBF binding to the 5’ T-cell repressor module of element -25. According to this model, translocation or functional abrogation of the 5’ repressor within element -25 would lead to failure of LMO2 down-regulation in a lymphoid cell context and to ectopic LMO2 expression. Our results provide a molecular link between RUNX proteins and LMO2 and underline the importance of RUNX transcription factors not only in the specification of haematopoietic stem cells but also during maturation of T-cells [55].
Segregation of the RUNX-dependent 5’ T-cell repressor module as a main mechanism for ectopic LMO2 expression in TCR-LMO2 rearranged T-ALL
The mechanisms deregulating LMO2 expression are numerous and more varied than initially assumed. Nearly 50% of human T-ALLs that lack chromosomal lesions involving LMO genes present overexpression of LMO2 or its family member LMO1 [56]. Recently our group reported activation of a novel intermediate promoter element at –11.8 kb upstream of the proximal promoter in a number of cytogenetically normal T-ALL patients as a possible mechanism for ectopic LMO2 expression [27]. In the case of chromosomal abnormalities that involve LMO2, including translocations and deletions, the presence of activating elements in the vicinity of LMO2 as a result of the abnormality was believed to be the cause. However, it has been suggested that removal of the T-cell repressor region was the truly activating mechanism [25, 26, 39]. Most of the studied LMO2 break-point regions in rearranged T-ALL patients clustered in the region between the T-cell specific repressor and the proximal promoter element (~1.7 kb) and break-points contained within this region were very strongly associated with high LMO2 expression levels [25, 57–62]. The factors that govern the occurrence of recurrent translocations in this area are still unknown. Two potent cryptic recombination signal sequences that resemble TCR/Ig recombination signal sequence have been identified in this region. Interestingly, one is located within the 3’ enhancer here described and a second one is located 370 bp downstream of element -25 [25]. The existence of an enhancer element located immediately downstream of the repressor that is active at the early stages of T-cell development could potentially provide chromatin accessibility for RAG mistargeting which could explain the recurrent oncogenic translocations into this region [25]. It may be the dual nature of the -25 region therefore, containing both activating and repressing activities, that makes it a particularly vulnerable target for oncogenic translocations that cause ectopic expression of LMO2.
Supporting Information
Homologous genomic sequences of the element -25 were downloaded from Ensembl for human, mouse, cow, dog and cat, aligned using multi-Lagan and displayed using Genedoc. Regions corresponding to the 5’ T-cell repressor and 3’ myeloid enhancer are indicated. Highly conserved sequences are depicted in black and candidate transcription factor binding sites are indicated. Arrowheads indicate the previously described T-cell repressor-region [39].
(PDF)
(DOCX)
(DOCX)
Acknowledgments
We are grateful to Rebecca Hannah and Manuel Sánchez-Castillo for their help with displaying some of the tracks and to Nicola Wilson for helpful discussions about the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Leukaemia and Lymphoma Research (BG) (http://leukaemialymphomaresearch.org.uk/, grants number 07060 and 12029), a fellowship from the Swiss National Science Foundation (NB) (http://www.snf.ch/) and Wellcome Trust Infrastructure support funding for the Cambridge Institute for Medical Research ((http://www.wellcome.ac.uk/) grant number 100140/Z/12/Z) and the Wellcome Trust and MRC Cambridge Stem Cell Institute (http://www.mrc.ac.uk/, grant number 097922/Z/11/Z). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sanchez-Garcia I, Rabbitts TH. LIM domain proteins in leukaemia and development. Seminars in cancer biology. 1993;4(6):349–58. Epub 1993/12/01. . [PubMed] [Google Scholar]
- 2. Nam CH, Rabbitts TH. The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion. Molecular therapy: the journal of the American Society of Gene Therapy. 2006;13(1):15–25. Epub 2005/11/02. 10.1016/j.ymthe.2005.09.010 . [DOI] [PubMed] [Google Scholar]
- 3. Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78(1):45–57. Epub 1994/07/15. . [DOI] [PubMed] [Google Scholar]
- 4. Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3890–5. Epub 1998/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osada H, Grutz G, Axelson H, Forster A, Rabbitts TH. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(21):9585–9. Epub 1995/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valge-Archer VE, Osada H, Warren AJ, Forster A, Li J, Baer R, et al. The LIM protein RBTN2 and the basic helix-loop-helix protein TAL1 are present in a complex in erythroid cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(18):8617–21. Epub 1994/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. The EMBO journal. 1997;16(11):3145–57. Epub 1997/06/02. 10.1093/emboj/16.11.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valge-Archer V, Forster A, Rabbitts TH. The LMO1 and LDB1 proteins interact in human T cell acute leukaemia with the chromosomal translocation t(11;14)(p15;q11). Oncogene. 1998;17(24):3199–202. Epub 1999/01/01. 10.1038/sj.onc.1202353 . [DOI] [PubMed] [Google Scholar]
- 9. Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(10):4367–71. Epub 1991/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Royer-Pokora B, Loos U, Ludwig WD. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene. 1991;6(10):1887–93. Epub 1991/10/01. . [PubMed] [Google Scholar]
- 11. Larson RC, Lavenir I, Larson TA, Baer R, Warren AJ, Wadman I, et al. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. The EMBO journal. 1996;15(5):1021–7. Epub 1996/03/01. [PMC free article] [PubMed] [Google Scholar]
- 12. Larson RC, Osada H, Larson TA, Lavenir I, Rabbitts TH. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene. 1995;11(5):853–62. Epub 1995/09/07. . [PubMed] [Google Scholar]
- 13. McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327(5967):879–83. Epub 2010/01/23. 10.1126/science.1182378 . [DOI] [PubMed] [Google Scholar]
- 14. Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26(3):447–54. Epub 2007/12/19. 10.1200/JCO.2007.13.0690 . [DOI] [PubMed] [Google Scholar]
- 15.Natkunam Y. The biology of the germinal center. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2007:210–5. Epub 2007/11/21. 10.1182/asheducation-2007.1.210 . [DOI] [PubMed]
- 16. Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109(4):1636–42. Epub 2006/10/14. 10.1182/blood-2006-08-039024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blenk S, Engelmann J, Weniger M, Schultz J, Dittrich M, Rosenwald A, et al. Germinal center B cell-like (GCB) and activated B cell-like (ABC) type of diffuse large B cell lymphoma (DLBCL): analysis of molecular predictors, signatures, cell cycle state and patient survival. Cancer informatics. 2007;3:399–420. Epub 2007/01/01. [PMC free article] [PubMed] [Google Scholar]
- 18. Arribas AJ, Campos-Martin Y, Gomez-Abad C, Algara P, Sanchez-Beato M, Rodriguez-Pinilla MS, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119(3):e9–e21. Epub 2011/11/24. 10.1182/blood-2011-02-339556 . [DOI] [PubMed] [Google Scholar]
- 19. Younes SF, Beck AH, Ohgami RS, Lossos IS, Levy R, Warnke RA, et al. The efficacy of HGAL and LMO2 in the separation of lymphomas derived from small B cells in nodal and extranodal sites, including the bone marrow. American journal of clinical pathology. 2011;135(5):697–708. Epub 2011/04/20. 10.1309/AJCP7Z2BIBUNQPLZ . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cobanoglu U, Sonmez M, Ozbas HM, Erkut N, Can G. The expression of LMO2 protein in acute B-cell and myeloid leukemia. Hematology. 2010;15(3):132–4. Epub 2010/06/19. 10.1179/102453309X12583347113618 . [DOI] [PubMed] [Google Scholar]
- 21. Malumbres R, Fresquet V, Roman-Gomez J, Bobadilla M, Robles EF, Altobelli GG, et al. LMO2 expression reflects the different stages of blast maturation and genetic features in B-cell acute lymphoblastic leukemia and predicts clinical outcome. Haematologica. 2011;96(7):980–6. Epub 2011/04/05. 10.3324/haematol.2011.040568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Calero-Nieto FJ, Joshi A, Bonadies N, Kinston S, Chan WI, Gudgin E, et al. HOX-mediated LMO2 expression in embryonic mesoderm is recapitulated in acute leukaemias. Oncogene. 2013;32(48):5471–80. Epub 2013/05/28. 10.1038/onc.2013.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372(6502):143–9. Epub 1994/11/10. 10.1038/372143a0 . [DOI] [PubMed] [Google Scholar]
- 24. De Keersmaecker K, Marynen P, Cools J. Genetic insights in the pathogenesis of T-cell acute lymphoblastic leukemia. Haematologica. 2005;90(8):1116–27. Epub 2005/08/05. . [PubMed] [Google Scholar]
- 25. Dik WA, Nadel B, Przybylski GK, Asnafi V, Grabarczyk P, Navarro JM, et al. Different chromosomal breakpoints impact the level of LMO2 expression in T-ALL. Blood. 2007;110(1):388–92. Epub 2007/03/16. 10.1182/blood-2006-12-064816 . [DOI] [PubMed] [Google Scholar]
- 26. Royer-Pokora B, Rogers M, Zhu TH, Schneider S, Loos U, Bolitz U. The TTG-2/RBTN2 T cell oncogene encodes two alternative transcripts from two promoters: the distal promoter is removed by most 11p13 translocations in acute T cell leukaemia's (T-ALL). Oncogene. 1995;10(7):1353–60. Epub 1995/04/06. . [PubMed] [Google Scholar]
- 27. Oram SH, Thoms JA, Pridans C, Janes ME, Kinston SJ, Anand S, et al. A previously unrecognized promoter of LMO2 forms part of a transcriptional regulatory circuit mediating LMO2 expression in a subset of T-acute lymphoblastic leukaemia patients. Oncogene. 2010;29(43):5796–808. Epub 2010/08/03. 10.1038/onc.2010.320 . [DOI] [PubMed] [Google Scholar]
- 28. Landry JR, Kinston S, Knezevic K, Donaldson IJ, Green AR, Gottgens B. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106(8):2680–7. Epub 2005/07/05. 10.1182/blood-2004-12-4755 . [DOI] [PubMed] [Google Scholar]
- 29. Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113(23):5783–92. Epub 2009/01/28. 10.1182/blood-2008-11-187757 . [DOI] [PubMed] [Google Scholar]
- 30. Dexter TM, Allen TD, Scott D, Teich NM. Isolation and characterisation of a bipotential haematopoietic cell line. Nature. 1979;277(5696):471–4. Epub 1979/02/08. . [DOI] [PubMed] [Google Scholar]
- 31. Dube SK, Pragnell IB, Kluge N, Gaedicke G, Steinheider G, Ostertag W. Induction of endogenous and of spleen focus-forming viruses during dimethylsulfoxide-induced differentiation of mouse erythroleukemia cells transformed by spleen focus-forming virus. Proceedings of the National Academy of Sciences of the United States of America. 1975;72(5):1863–7. Epub 1975/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arbiser JL, Moses MA, Fernandez CA, Ghiso N, Cao Y, Klauber N, et al. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(3):861–6. Epub 1997/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, et al. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome research. 2003;13(4):721–31. Epub 2003/03/26. 10.1101/gr.926603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholas KB, Nicholas HBJ. GeneDoc: a tool for editing and annotating multiple sequence alignments 1997.
- 35. Chapman MA, Donaldson IJ, Gilbert J, Grafham D, Rogers J, Green AR, et al. Analysis of multiple genomic sequence alignments: a web resource, online tools, and lessons learned from analysis of mammalian SCL loci. Genome research. 2004;14(2):313–8. Epub 2004/01/14. 10.1101/gr.1759004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al]. 2008;Chapter 2:Unit 2 6. Epub 2008/04/23. 10.1002/0471250953.bi0206s21 . [DOI] [PubMed]
- 37. Gottgens B, McLaughlin F, Bockamp EO, Fordham JL, Begley CG, Kosmopoulos K, et al. Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene. 1997;15(20):2419–28. Epub 1997/12/12. 10.1038/sj.onc.1201426 . [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Castillo M, Ruau D, Wilkinson AC, Ng FS, Hannah R, Diamanti E, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic acids research. 2014. Epub 2014/10/02. 10.1093/nar/gku895 . [DOI] [PMC free article] [PubMed]
- 39. Hammond SM, Crable SC, Anderson KP. Negative regulatory elements are present in the human LMO2 oncogene and may contribute to its expression in leukemia. Leukemia research. 2005;29(1):89–97. Epub 2004/11/16. 10.1016/j.leukres.2004.05.013 . [DOI] [PubMed] [Google Scholar]
- 40. Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome biology. 2012;13(8):418 Epub 2012/08/15. 10.1186/gb-2012-13-8-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bowers SR, Mirabella F, Calero-Nieto FJ, Valeaux S, Hadjur S, Baxter EW, et al. A conserved insulator that recruits CTCF and cohesin exists between the closely related but divergently regulated interleukin-3 and granulocyte-macrophage colony-stimulating factor genes. Molecular and cellular biology. 2009;29(7):1682–93. Epub 2009/01/23. 10.1128/MCB.01411-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell stem cell. 2010;7(4):532–44. Epub 2010/10/05. 10.1016/j.stem.2010.07.016 . [DOI] [PubMed] [Google Scholar]
- 43. Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122(14):e12–22. Epub 2013/08/27. 10.1182/blood-2013-03-490425 . [DOI] [PubMed] [Google Scholar]
- 44. Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nature immunology. 2000;1(2):138–44. Epub 2001/03/15. 10.1038/77819 . [DOI] [PubMed] [Google Scholar]
- 45. Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Molecular cell. 2012;45(3):330–43. Epub 2012/02/14. 10.1016/j.molcel.2011.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, Groner Y. Runx3-mediated transcriptional program in cytotoxic lymphocytes. PloS one. 2013;8(11):e80467 Epub 2013/11/16. 10.1371/journal.pone.0080467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer cell. 2012;22(2):209–21. Epub 2012/08/18. 10.1016/j.ccr.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301(5634):798–802. Epub 2003/08/09. 10.1126/science.1086887 . [DOI] [PubMed] [Google Scholar]
- 49. Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nature reviews Genetics. 2006;7(10):793–803. Epub 2006/09/20. 10.1038/nrg1920 . [DOI] [PubMed] [Google Scholar]
- 50. Sun XJ, Wang Z, Wang L, Jiang Y, Kost N, Soong TD, et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature. 2013;500(7460):93–7. Epub 2013/07/03. 10.1038/nature12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell reports. 2014;8(6):1974–88. Epub 2014/09/23. 10.1016/j.celrep.2014.08.024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kundu M, Compton S, Garrett-Beal L, Stacy T, Starost MF, Eckhaus M, et al. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood. 2005;106(10):3621–4. Epub 2005/07/30. 10.1182/blood-2005-04-1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111(5):621–33. Epub 2002/12/05. . [DOI] [PubMed] [Google Scholar]
- 54. Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nature reviews Immunology. 2009;9(2):106–15. Epub 2009/01/24. 10.1038/nri2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunological reviews. 2006;209:191–211. Epub 2006/02/02. 10.1111/j.0105-2896.2006.00352.x . [DOI] [PubMed] [Google Scholar]
- 56. Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer cell. 2002;1(1):75–87. Epub 2002/06/28. . [DOI] [PubMed] [Google Scholar]
- 57. Fitzgerald TJ, Neale GA, Raimondi SC, Goorha RM. Rhom-2 expression does not always correlate with abnormalities on chromosome 11 at band p13 in T-cell acute lymphoblastic leukemia. Blood. 1992;80(12):3189–97. Epub 1992/12/15. . [PubMed] [Google Scholar]
- 58. Yoffe G, Schneider N, Van Dyk L, Yang CY, Siciliano M, Buchanan G, et al. The chromosome translocation (11;14)(p13;q11) associated with T-cell acute lymphocytic leukemia: an 11p13 breakpoint cluster region. Blood. 1989;74(1):374–9. Epub 1989/07/01. . [PubMed] [Google Scholar]
- 59. Boehm T, Baer R, Lavenir I, Forster A, Waters JJ, Nacheva E, et al. The mechanism of chromosomal translocation t(11;14) involving the T-cell receptor C delta locus on human chromosome 14q11 and a transcribed region of chromosome 11p15. The EMBO journal. 1988;7(2):385–94. Epub 1988/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cheng JT, Yang CY, Hernandez J, Embrey J, Baer R. The chromosome translocation (11;14)(p13;q11) associated with T cell acute leukemia. Asymmetric diversification of the translocational junctions. The Journal of experimental medicine. 1990;171(2):489–501. Epub 1990/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Garcia IS, Kaneko Y, Gonzalez-Sarmiento R, Campbell K, White L, Boehm T, et al. A study of chromosome 11p13 translocations involving TCR beta and TCR delta in human T cell leukaemia. Oncogene. 1991;6(4):577–82. Epub 1991/04/01. . [PubMed] [Google Scholar]
- 62. Boehm T, Mengle-Gaw L, Kees UR, Spurr N, Lavenir I, Forster A, et al. Alternating purine-pyrimidine tracts may promote chromosomal translocations seen in a variety of human lymphoid tumours. The EMBO journal. 1989;8(9):2621–31. Epub 1989/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Homologous genomic sequences of the element -25 were downloaded from Ensembl for human, mouse, cow, dog and cat, aligned using multi-Lagan and displayed using Genedoc. Regions corresponding to the 5’ T-cell repressor and 3’ myeloid enhancer are indicated. Highly conserved sequences are depicted in black and candidate transcription factor binding sites are indicated. Arrowheads indicate the previously described T-cell repressor-region [39].
(PDF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.