Abstract
The functions ascribed to the mammalian GTFs (general transcription factors) during the various stages of the RNAPII (RNA polymerase II) transcription reaction are based largely on in vitro studies. To gain insight as to the functions of the GTFs in living cells, we have analysed the genomic location of several human GTF and RNAPII subunits carrying a TAP (tandem-affinity purification) tag. ChIP (chromatin immunoprecipitation) experiments using anti-tag beads (TAP-ChIP) allowed the systematic localization of the tagged factors. Enrichment of regions located close to the TIS (transcriptional initiation site) versus further downstream TRs (transcribed regions) of nine human genes, selected for the minimal divergence of their alternative TIS, were analysed by QPCR (quantitative PCR). We show that, in contrast with reports using the yeast system, human TFIIF (transcription factor IIF) associates both with regions proximal to the TIS and with further downstream TRs, indicating an in vivo function in elongation for this GTF. Unexpectedly, we found that the Rpb7 subunit of RNAPII, known to be required only for the initiation phase of transcription, remains associated with the polymerase during early elongation. Moreover, ChIP experiments conducted under stress conditions suggest that Rpb7 is involved in the stabilization of transcribing polymerase molecules, from initiation to late elongation stages. Together, our results provide for the first time a general picture of GTF function during the RNAPII transcription reaction in live mammalian cells and show that TFIIF and Rpb7 are involved in both early and late transcriptional stages.
Keywords: chromatin immunoprecipitation, general transcription factor, genomic location, RNA polymerase II, tandem-affinity purification, transcription factor IIF (TFIIF)
INTRODUCTION
Since their first purification two decades ago, the set of mammalian factors required for transcription by RNAPII (RNA polymerase II) have drawn a great deal of attention. First believed to be required for the transcription of all class II genes, it is now generally accepted that the so-called GTFs (general transcription factors) TFII (transcription factor II) A, TFIIB, TFIID [also called TBP (TATA-box-binding protein)], TFIIE, TFIIF, TFIIH and TFIIS/TCEA1 [transcription elongation factor A (SII) 1] form the core of the machinery that participates in the transcription of a large fraction of class II genes (reviewed in [1]). Using this wide definition, the TFIID–STAGA (SPT3–TAF9–GCN5 acetyltransferase)–TFTC [TATA-box-binding protein-free TAF (TBP-associated transcription factor)-containing]–PCAF [p300/CREB (cAMP-response-element-binding protein)-binding protein-associated factor] and Mediator complexes can also be classified as GTFs because they also participate in the transcription of a large proportion of mammalian genes [2,3]. However, studies in yeast have revealed that some GTFs are not required for the transcription of a significant number of genes [4,5]. Similar studies in mammalian cells are still lacking.
Many reports have focused on defining the specific function of the GTFs during transcription. Biochemical analyses have revealed that the transcription reaction involves a number of successive steps that lead to the formation of a pre-mRNA [1]. In the first step, RNAPII positions its catalytic centre near the TIS (transcriptional initiation site) of promoter DNA. This first step is preceded by the formation of a pre-initiation complex that contains, in addition to RNAPII, a number of general initiation factors including TFIID, TFIIB, TFIIF, TFIIE and TFIIH. Assembly of the pre-initiation complex requires specific binding of some general initiation factors to core promoter elements such as binding of TBP to the TATA box, TFIIB to the BRE (TFIIB-recognition element) and TAFs of TFIID to the Inr (initiator element) and DPE (downstream promoter element). Both TFIIA and the Mediator complex have been shown to stimulate transcriptional initiation in reconstituted systems. Formation of the pre-initiation complex is accompanied by topological changes including bending and wrapping of promoter DNA around the protein core of the complex [6]. In the second step, promoter DNA is melted between nucleotides −9 and +2 in such a way that the template strand becomes accessible for NTP polymerization. Both TFIIF and TFIIE were shown to participate in promoter melting [7]. Two distinct single-stranded DNA helicases, XPB (xeroderma pigmentosum group B)/ERCC (excision repair cross-complementing rodent repair deficiency, complementation group) 3 and XPD (xeroderma pigmentosum group D)/ERCC2, have been identified as components of TFIIH. XPB/ERCC3 was attributed a major role in promoter DNA melting prior to initiation [8]. During the third step, RNAPII enters a cycle of abortive initiation events wherein the enzyme synthesizes many short 2–10 nt transcripts. TFIIH is responsible for the melting of the template DNA during promoter escape and for the phosphorylation of the CTD (C-terminal domain) of the Rpb1 subunit of RNAPII [9]. Crystallographic structures of RNAPII suggest that the dissociable heterodimer formed by Rpb4 and Rpb7 stabilizes the RNAPII complex in a clamp-closed conformation, which is believed to favour efficient initiation [10]. This view is consistent with the hypothesis that the Rpb4–Rpb7 heterodimer is dispensable for transcript elongation [11], as RNAPII adopts the same conformation when bound to DNA even in the absence of Rpb4–Rpb7. Formation of the mRNA is completed through transcript elongation and termination of the transcription reaction. While some reports have proposed a role for both TFIIE and TFIIH in transcript elongation [12,13], a large body of information has accumulated to support a role for TFIIF and TFIIS/TCEA1 after promoter clearance (reviewed in [14]). TFIIS/TCEA1 can rescue paused or arrested elongation complexes through stimulation of an intrinsic endoribonuclease activity that allows transcript cleavage by backtracked RNAPII [15]. TFIIF was shown to stimulate the rate of NTP addition by RNAPII and to modulate the activity of TFIIS/TCEA1 [16]. However, most of these studies have been performed using in vitro systems and the information on the roles of the various GTFs in vivo remains fragmentary.
To gain insight into the function of the GTFs in live mammalian cells and determine whether specific GTFs act at early versus late stages of the transcription reaction in vivo, we have affinity-tagged many components of the RNAPII transcription machinery, expressed the tagged polypeptides at physiological levels in human cells using an inducible system and analysed their genomic location by ChIP (chromatin immunoprecipitation) coupled with QPCR (quantitative PCR) detection of the enriched regions. The location of RNAPII, TFIIA, TFIIB, TFIIE, TFIIF, TFIID–STAGA–TFTC–PCAF, TFIIH, TFIIS/TCEA1 and the Mediator was analysed both in the region proximal to TIS and further downstream in TRs (transcribed regions) of the nine human genes that were selected according to (i) their active transcriptional status in HEK-293 cells (human embryonic kidney cells), (ii) the minimal divergence of their TIS and (iii) their belonging to various functional classes according to GO (gene ontology) terms. Our results shed light on the function of the GTFs in living cells by showing that although all the factors, including TFIIS/ TCEA1, can associate with regions proximal to the TIS, only TFIIF, TFIIS/TCEA1, the Mediator and STAGA–TFTC–PCAF complexes and the heterodimer Rpb4–Rpb7 are detected with RNAPII in TRs situated further downstream along actively transcribed genes.
EXPERIMENTAL
Generation of cell lines expressing tagged polypeptides
TAP (tandem-affinity purification)-tagged GTF and RNAPII subunits were cloned and expressed in human EcR 293 cells (HEK-293 cells stably transfected with the vector pVgRXR, expressing upon induction the heterodimeric ecdysone receptor) as previously described [17]. The following C-terminal TAP-tagged polypeptides were analysed: RNAPII (Rpb2, Rpb4, Rpb7 and Rpb11), TFIIF [RAP (RNAPII-associated protein) 30 and RAP74], TFIIB, TFIIA (the γ-subunit and the αβ precursor), TFIIE (TFIIE34 and TFIIE56), TFIIH [XPB/ERCC3 and CDK7 (cyclin-dependent kinase 7)], TFIID–STAGA–TFTC–PCAF (TAF10 and TAF13), the Mediator [SRB7 (suppressor of RNA polymerase B 7) and TRFP (TBP-related factor proximal homologue)] and TFIIS/TCEA1. Near physiological expression levels were obtained by inducing the cells for 24–48 h with 3–6 μM Ponasterone A (Invitrogen). In the case of TFIIS/TCEA1, two conditions were used in parallel: the cells were either exposed to a total dose of 12.5 J/m2 of UVC radiations and allowed to recover for 20 min at 37 °C or left unexposed, with a total of 48 h of culture in both cases. For the heat shock, the cells were incubated for 1 h at 42 °C in parallel with matched controls.
Affinity purification and identification of proteins by MS
Protein-affinity purification was performed as we have previously described [17] (detailed protocols are available on our website at http://www.ircm.qc.ca/microsites/hupi/en/733.html). The TAP eluates were run on SDS gels, stained with silver or SYPRO Ruby (Bio-Rad) and gel slices were excised and digested with trypsin as previously described [17]. The resulting tryptic peptides were purified and identified by either MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS) [18] or LC (liquid chromatography)-MS/MS (tandem MS) with microcapillary reversed-phase HPLC coupled with an LCQ DecaXP (Thermo Finnigan) or LTQ (Thermo Electron) quadrupole ion-trap mass spectrometer with a nanospray interface. Resulting peptide MS/MS spectra were interpreted using the MASCOT (Matrix Science) software and searched against proteins in either the NCBI (National Center for Biotechnology Information) non-redundant protein database or the Uniref protein database [19].
Western blots and antibodies
Proteins from TAP eluates were separated by SDS/PAGE and transferred on to a PVDF membrane and probed with the appropriate antibodies. The horseradish peroxidase-conjugated secondary antibodies were purchased from GE Healthcare. Signals were detected using the ECL® (enhanced chemiluminescence) system (GE Healthcare).
Transcription assay
In vitro transcription reactions were performed as previously described [17] using highly purified calf thymus RNAPII and recombinant TBP, TFIIB, TFIIE, TFIIF and TFIIH. RNAPII, TBP, TFIIE and TFIIH were independently omitted from the reconstituted system and replaced by the eluate of affinity-purified Rpb11–TAP, TFIIAαβ–TAP, TFIIE56–TAP and XPB–TAP. Control reactions in which an eluate from non-induced cells is added were included in each case.
ChIP
TAP-ChIP experiments were performed as previously described [17]. Briefly, 108 cells were cross-linked with formaldehyde at a final concentration of 1 %. A cross-link time of 10 min at room temperature (21 °C) was generally allowed before adding glycine to a final concentration of 125 mM. A reduced cross-link time of 1–2 min was necessary in the case of the cells expressing the TAP-tagged TFIIE56, TAF10, TAF13 and SRB7. Chromatin was sonicated to obtain fragments of an average length of 450 bp. After immunoprecipitation with IgG beads, washing and elution, the cross-links were reversed at 65 °C, and the DNA was purified. The enrichment of regions proximal to the TIS and further downstream TRs for the nine genes was evaluated by QPCR as previously described [17], using two control regions: an internal sequence of an expressed sequence tag located at chr17:49926700–49927999 and a sequence located in a 1.330 kbp gene-less region of the 13q21.33 band [20]. To ensure that the enrichment obtained was specific to the TAP-tagged factors, the immunoprecipitated DNA from the untagged EcR 293 parental cells was systematically analysed in parallel. These control experiments allowed us to estimate the background level, which ranged between 0.8- and 1.4-fold enrichment over control regions. In pilot experiments (results not shown), we found that treating the samples with proteinase K at a final concentration of 0.2 μg/μl during the cross-link reversal greatly decreased non-specific enrichment of TRs, which can result at this step, as reported previously [21].
The genes to be analysed were selected using the databases DBTSS (DataBase of Transcription Start Site; April 2003 version, http://dbtss.hgc.jp/samp_home.html) and MGC (Mammalian Gene Collection; March 2003 version, http://mgc.nci.nih.gov/). The selection criteria was the minimal divergence of their alternative TIS, empirically defined as the number of full-length cDNAs situated at a distance of less than 40 bp from the 5′-ends provided by RefSeq. Primers were designed by using the Primer3 algorithm (see Table 1 for the sequence of the primers and Supplementary Figure 2 at http://www.BiochemJ.org/bj/409/bj4090139add.htm for the genomic location of the PCR products, determined on the UCSC Genome Browser [22]). The fragments to be analysed have been chosen based on three criteria: (i) to include the TIS or a position situated at a distance of at least 1000 bp from the TIS; (ii) to rank between those with the best score generated by the primer design algorithm; and (iii) to be unique. All PCR-amplified fragments are unique as verified by BLAT on the April 2003 assembly version of the UCSC Genome Browser (http://genome.ucsc.edu/), by QPCR melting-curve analysis and by agarose-gel electrophoresis (results not shown).
Table 1. Sequences of the primers used to analyse the location of various polypeptides.
The position of the PCR products relative to the assigned TIS is indicated for each primer set. Fw, forward; Re, reverse; FTL, ferritin, light polypeptide; GNB2L1, guanine-nucleotide-binding protein (G-protein), β-polypeptide 2-like 1; GTF2F2, general transcription factor IIF, polypeptide 2, 30 kDa; POLA2, polymerase (DNA-directed), α2; POLR2E, polymerase (RNA) II (DNA-directed) polypeptide E; TIMM17A, translocase of inner mitochondrial membrane 17 homologue A (yeast).
| Official gene symbol | Gene length (bp) | Primer | Sequence | PCR product position
|
|
|---|---|---|---|---|---|
| Start | End | ||||
| POLA2 | 35 000 | TIS-Fw | 5′-aaagcaaggggaaggttt-3′ | − 61 | + 123 |
| TIS-Re | 5′-gaatggagggagcagaaat-3′ | ||||
| TR-Fw | 5′-ggtctgtgcaaatatgaatc-3′ | + 988 | + 1199 | ||
| TR-Re | 5′-atgactccttacactgctctac-3′ | ||||
| GTF2F2 | 163 566 | TIS-Fw | 5′-tttcttcagttatgctgacc-3′ | − 238 | 0 |
| TIS-Re | 5′-ttacctgccagaacactg-3′ | ||||
| TR-Fw | 5′-ctaagaggtctttcttgtcg-3′ | + 978 | + 1186 | ||
| TR-Re | 5′-actattctgggtatgacagg-3′ | ||||
| FTL | 1559 | TIS-Fw | 5′-gctgagactcctatgtgct-3′ | − 157 | + 33 |
| TIS-Re | 5′-acactgttgaagcaagagac-3′ | ||||
| TR-Fw | 5′-tatagaagccagctgaagat-3′ | + 957 | + 1176 | ||
| TR-Re | 5′-gtgaaatgaggctctgaa-3′ | ||||
| TIMM17A | 15 141 | TIS-Fw | 5′-aaccaatgctcatagacctt-3′ | − 80 | + 84 |
| TIS-Re | 5′-gcaagagaaatgcaaagac-3′ | ||||
| TR-Fw | 5′-caggaccttaaattactactctgg-3′ | + 910 | + 1094 | ||
| TR-Re | 5′-tatttacagtgtgctgagtcctac-3′ | ||||
| GNB2L1 | 6964 | TIS-Fw | 5′-gaatgtgcttgtttcagagt-3′ | − 200 | + 55 |
| TIS-Re | 5′-atggcttagagaaactagca-3′ | ||||
| TR-Fw | 5′-aaagggtgtctgtatttctg-3′ | + 998 | + 1165 | ||
| TR-Re | 5′-ttaactagagatgcggtttc-3′ | ||||
| SFRS2 | 3215 | TIS-Fw | 5′-gccaatcagaaggtttcat-3′ | − 86 | + 17 |
| TIS-Re | 5′-gcacctgagtaacaactgg-3′ | ||||
| TR-Fw | 5′-gaaggtccaagtccaagt-3′ | + 1005 | + 1194 | ||
| TR-Re | 5′-gattcccagacattaccat-3′ | ||||
| POLR2E | 7170 | TIS-Fw | 5′-aactgccgctctcgtaag-3′ | − 161 | + 90 |
| TIS-Re | 5′-atgatggtcttgcggatt-3′ | ||||
| TR-Fw | 5′-gagatagggtttcattctgtc-3′ | + 812 | + 1093 | ||
| TR-Re | 5′-gaccagtatgatcctgagag-3′ | ||||
| ENO1 | 17 717 | TIS-Fw | 5′-ggtgagggaatgagtgac-3′ | − 3 | + 151 |
| TIS-Re | 5′-accgaggtgaacgtaaag-3′ | ||||
| TR 1-Fw | 5′-taggccaagaaggatgtat-3′ | + 1140 | + 1289 | ||
| TR 1-Re | 5′-gaattagggacacggtaaat-3′ | ||||
| TR 2-Fw | 5′-gaacaagacctcaattgcta-3′ | + 3428 | + 3702 | ||
| TR 2-Re | 5′-agcactggactaaatactgg-3′ | ||||
| TR 3-Fw | 5′-gagaattgtgaaactccttc-3′ | + 5373 | + 5558 | ||
| TR 3-Re | 5′-gtgactcacagatggtgac-3′ ′ | ||||
| TR 4-Fw | 5′-gcacaagtttagagggttta-3′ | + 17106 | + 17321 | ||
| TR 4-Re | 5′-cagctcctcttcaattctt-3′ | ||||
| HSPA8 | 4643 | TIS-Fw | 5′-cttgtgattgggtcttgta-3′ | − 141 | + 88 |
| TIS-Re | 5′-aactcttgagcagaggttt-3′ | ||||
| TR 1-Fw | 5′-acctggagtccattgtagta-3′ | + 1175 | + 1463 | ||
| TR 1-Re | 5′-aataccattatccctgtcaa-3′ | ||||
| TR 2-Fw | 5′-ctgaaatctggataacgtaggag-3′ | + 2026 | + 2324 | ||
| TR 2-Re | 5′-ctctcccttgtattctacttggac-3′ | ||||
| TR 3-Fw | 5′-gtaccatttgtgatgcaagttc-3′ | + 3265 | + 3549 | ||
| TR 3-Re | 5′-tacagctctcttgttctcactgat-3′ | ||||
| TR 4-Fw | 5′-gtcagggagaaagaagggttatta-3′ | + 4152 | + 4439 | ||
| TR 4-Re | 5′-atgtgtggaacaatgctacatctac-3′ | ||||
In the present study, we consider that a region is occupied by a given factor when: (i) reproducible and statistically significant higher enrichment values are obtained for that region compared with control regions and (ii) low background signals are obtained for the same region in a parallel control ChIP experiments conducted with the parental cells in which the tagged factor is not expressed.
RESULTS AND DISCUSSION
TAP-tagged components of the RNAPII machinery are functional
The specific function of the various GTFs that assist mammalian RNAPII during the successive stages of the transcription reaction is largely based on in vitro studies (reviewed in [1]). Although these biochemical data have proved invaluable to understand transcription at a fine molecular level, the function of the various GTFs in living mammalian cells remains mainly unaddressed. We reasoned that a systematic analysis of the genomic location of many individual components of the general transcription machinery would help us to understand the roles of the GTFs in vivo. We took advantage of our expanding collection of cell lines programmed to express, upon induction, physiological levels of polypeptides carrying a TAP tag, including four subunits of RNAPII, two subunits of each TFIIA, TFIIE, TFIIF, TFIID–STAGA–TFTC–PCAF, TFIIH and the Mediator as well as TFIIB and TFIIS/TCEA1.
To ensure that the tagged polypeptides expressed at physiological levels in human cells behave properly (e.g. the tag does not impair their activity), we performed two lines of experiments. First, we affinity-purified all the tagged proteins in native conditions, analysed the eluates using MS and defined their network of interactions (see Figure 1A for an example and see Supplementary Figure 1 at http://www.BiochemJ.org/bj/409/bj4090139add.htm for details). Consistent with our previously published results [17], RNAPII is linked to TFIIB, TFIIF, FCP1 (TFIIF-interacting CTD phosphatase subunit 1)/CTDP1 (RNAPII CTD polypeptide A phosphatase subunit 1) and RPAP1 (RNAPII-associated protein 1) in vivo. Both RNAPII and TFIIF are connected to the Mediator complex, which itself is linked to TFIID–STAGA–TFTC–PCAF subunits. TFIID–STAGA–TFTC–PCAF associates with TFIIA and TRFs (TBP-related factors) in vivo. Tagged TFIIS/TCEA1 co-purifies with RNAPII subunits, as previously described [23]. Surprisingly, however, the RNAPII molecules that interact with TFIIS/TCEA1 appear to be mainly in the hypophosphorylated IIa form (Figure 1B); purifications from whole-cell lysates prepared in the presence of phosphatase inhibitors indicated that this finding is not merely the consequence of CTD dephosphorylation during the preparation of the cell extracts (results not shown). Because RNAPII with a hypophosphorylated Rpb1 CTD represents the form of the enzyme that is recruited to promoter DNA prior to transcription initiation, our results support the notion that TFIIS/TCEA1 can associate with RNAPII at promoters in vivo (see below). Also surprising is the finding that tagged Rpb11 co-purifies with TFIIS.1/TCEA2 (see Supplementary Figure 1). Comparison of the amino acid sequence of TFIIS/TCEA1 with TFIIS.1/TCEA2 shows that the C-terminal region, the portion of the polypeptide known to be necessary and sufficient for activity in vitro [24], is highly conserved (Figure 1C). A previous study has shown that the N-terminal region of human TFIIS/TCEA1 interacts with RNAPII holoenzyme, whereas its C-terminal portion binds free RNAPII [25]. To our knowledge, this is the first time that the protein encoded by the TFIIS.1/TCEA2 gene is purified and shown to associate with RNAPII. Notably, both TFIIE and TFIIH are only weakly linked to each other and to other GTFs in our network (Supplementary Figure 1). Both CDK7 and XPB/ERCC3 co-purify with the DNA repair factor ERCC5/XPG [xeroderma pigmentosum group G (Cockayne syndrome)]; XPB/ERCC3 co-purifies with BCR (breakpoint cluster region) (Figure 1A), the polypeptide forming the chimaeric BCR-ABL (Abelson tyrosine kinase fusion protein) protein in the Philadelphia chromosome. ERCC5/XPG connects TFIIH to TFIID–STAGA–TFTC–PCAF (Supplementary Figure 1). Together, these results indicate that the tagged factors have the ability to associate with their endogenous interaction partners in vivo and reveal new aspects of the TFIIS–RNAPII interaction in live mammalian cells.
Figure 1. TAP of the RNAPII general transcription machinery.
(A) SYPRO-stained SDS gels showing the CDK7- and XPB/ERCC3-TAP eluates. Although the identity of most visible bands is known, only interaction partners that were validated through examination of the literature are shown. Gene symbols and common names are provided. Tagged polypeptides are indicated by an asterisk. Western blots validating the presence of the TFIIH subunits as well as XPG and BCR in both CDK7- and XPB/ERCC3-TAP eluates are shown. (B) TAP-tagged TFIIS/TCEA1 co-purifies with Rpb1 carrying a hypophosphorylated CTD. The N-20 antibody, which detects both the hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of RNAPII in a whole cell extract (WCE) by being directed to the N-terminal part of Rpb1, revealed the presence of hypophosphorylated RNAPII in both the Rpb11- and TFIIS/TCEA1-TAP eluates. (C) Amino acid sequence alignment of the TFIIS/TCEA1 and TFIIS.1/TCEA2 proteins. The peptides sequences obtained by MS/MS that identify the TFIIS.1/TCEA2 protein in the Rpb11–TAP eluate are boxed. (D) In vitro transcription reactions were reconstituted using calf thymus RNAPII in the presence of the classically purified GTFs TBP, TFIIB, TFIIF and TFIIH. The linearized DNA template carries the AdML promoter and directs the synthesis of a 391-nt transcript. The TAP-tagged GTFs (TFIIAαβ–TAP, TFIIE56–TAP and XPB–TAP) and RNAPII (Rpb11–TAP) were used to replace their cognate highly purified counterparts after their omission from the reconstituted system (All). A control reaction performed with an eluate obtained from non-induced cells is included.
Secondly, the ability of the purified GTFs and RNAPII to support transcription was assessed. The GTFs and RNAPII affinity-purified using their tagged subunits were able to replace their highly purified counterparts in in vitro transcription assays (Figure 1D). Together, the results of the proteomic analysis and the biochemical assays support the notion that the tagged polypeptides associate with their cognate partners to form functionally active transcription factors in mammalian cells.
TAP-ChIP as a systematic method for the localization of proteins along mammalian genomic DNA
The genomic location analysis of transcription factors by ChIP in a systematic fashion is known to be difficult mainly because of the requirement of antibodies able to efficiently immunoprecipitate proteins after a cross-linking step. TAP-ChIP experiments used IgG beads, which target the Protein A moiety of the TAP tag, to pull down the TAP-tagged polypeptides covalently cross-linked to chromatin fragments. This method allowed us to systematically localize many components of the RNAPII general transcription machinery. Pilot experiments have revealed that the second affinity purification step (e.g. calmodulin beads) does not significantly improve the enrichment values of DNA fragments under our conditions (results not shown). After purification, enriched DNA sequences were analysed using QPCR with primer sets designed for detecting both regions proximal to the TIS and further downstream TRs of nine genes actively transcribed in HEK-293 cells.
The ability to discriminate between the location of a protein either in regions close to the TIS versus further downstream TRs of a gene demands that: (i) the TIS be known with accuracy and (ii) alternative, distant TIS not be used. To meet these requirements, we developed a procedure that allowed us to select genes having well-characterized TIS. Two databases reporting on experimentally determined human TISs using various methodologies were compared for the minimal divergence of alternative TISs. Among the selected genes, nine were chosen for (i) their belonging to various GO classes, (ii) their activity in HEK-293 cells as verified by quantitative RT (reverse transcriptase)–PCR (results not shown) and (iii) our ability to design primer sets that are efficient in QPCR detection.
In the present study, the regions to be analysed by QPCR were selected in such a way that early versus late transcription events can be monitored. A distinction between early steps of the reaction (e.g. promoter binding, first-bond formation and promoter clearance) is difficult to address principally because of the limitation imposed by the resolution of the ChIP technique, which depends on the size of the sonicated fragments (approx. 450 bp in our case). In addition, it is conceivable that the efficiency of bringing down a promoter fragment would be much higher when a protein is associated with a tightly bound, engaged polymerase as compared with a loosely bound polymerase in a pre-initiation complex.
The results of our TAP-ChIP experiments (Figure 2) indicate that, as expected, TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH and the Mediator assemble with RNAPII in regions proximal to the TIS of transcribed genes in live mammalian cells. The analysis of a second subunit confirmed the results obtained for these factors (see Supplementary Figure 3 at http://www.BiochemJ.org/bj/409/bj4090139add.htm). Moreover, when N-tagged factors or specific antibodies were used in validation experiments, we obtained similar location profiles as when using the C-tagged version (results not shown). The lack of detection of initiation factors in some regions close to the TIS may reflect a low rate of initiation events (e.g. under the detection limit of the ChIP procedure used here).
Figure 2. Occupancy of regions proximal to the TIS and further downstream TRs of selected human genes by the GTF and RNAPII subunits obtained by ChIP.
Fold enrichment of regions located close to the TIS (left-hand matrix) and further downstream TRs (right-hand matrix) over control regions are represented for each TAP-tagged factor, including subunits of RNAPII (Rpb11) and the GTFs TFIIA (TFIIAαβ), TFIIB, TFIIE (TFIIE56), TFIIH (XPB), TFIID/STAGA (TAF10 and TAF13), the Mediator (SRB7) and TFIIS/TCEA1. Colour code: black:> 5-fold enrichment; grey: significantly enriched with less than 5-fold enrichment values; white: not significantly enriched; X: not determined. The number of regions situated close to the TIS and further downstream TRs occupied by each factor is indicated as a ratio (observed/total tested). The results shown for TFIIS/TCEA1 were obtained by the analysis of cells cross-linked after an irradiation with a total dose of 12.5 J/m2 of UVC (TFIISexp). Student’s t tests were performed to determine the significance of the enrichment values obtained in four experiments, including at least two independent ChIP assays.
Our results also indicate that the transcription elongation factor TFIIS/TCEA1 is not stably recruited along the tested genes under normal growth conditions (results not shown). Notably, TFIIS/TCEA1 was detected at a number of regions close to the TIS and in further downstream TRs after cell exposure to UVC radiations (Figure 2), suggesting a role for TFIIS/TCEA1 both at early and late stages of transcription. Cyclobutane pyrimidine dimers, the major DNA lesions induced by UVC radiations, are known to arrest the polymerase and to induce the recruitment of TFIIS/TCEA1 in vitro [26]. Whether the association of TFIIS/TCEA1 with genomic DNA is simply a consequence of an increased number of stalled elongating RNAPII molecules along damaged DNA or the signature of an active role for this factor in TCR (transcription-coupled DNA repair), as suggested by genetic studies in yeast [27], is not clear. A recent study in human cells showed that TFIIS is recruited to lesion-arrested RNAPII via the CSA and CSB (Cockayne syndrome type A and type B) proteins, suggesting a non-specific role of TFIIS in TCR [28]. Although the location of TFIIS/TCEA1 at regions close to the TIS does not necessarily imply that it has a function in transcription initiation, the finding that TFIIS/TCEA1 also associates with the hypophosphorylated form of RNAPII (see Figure 1B) fully supports the conclusion that it plays a role in very early stages of transcription, as recently proposed in yeast TFIIS/TCEA1 [29].
Two TAFs, TAF10 and TAF13, were detected along our tested genomic regions with very similar occupancy profiles of regions close to the TIS but distinct profiles in further downstream TRs (Figure 2). Both TAFs were detected at half of the regions close to the TIS analysed, indicating that the complex recruited is TFIID, TAF13 being a TFIID-specific subunit [30]. Of note, at some regions close to the TIS (three out of eight), neither TAF10 nor TAF13 was detected, which is in agreement with a previous report showing that these TAFs are required for the expression of only a fraction of the yeast genes [31]. Our results indicate that TAF10, a component of both TFIID and the STAGA–TFTC–PCAF complexes (reviewed in [30]), is recruited to TRs (see Figure 2 and results not shown) and suggest a role for this TAF during transcriptional elongation of specific genes. The observation that TAF13 is not recruited to these regions suggests that the TAF10-containing complex present at these locations is STAGA, TFTC or PCAF. While the function of SAGA (SPT–ADA–GCN5 acetyltransferase) at different stages of the transcription reaction is supported by a number of reports (see [32] for a recent review), its human homologue complexes STAGA–TFTC–PCAF are only poorly characterized. In support of our observation, the STAGA complex has been implicated in diverse transcription-coupled processes such as chromatin modification, pre-mRNA splicing and DNA repair [33].
The Mediator subunit SRB7 was detected at a fraction of the regions close to the TIS that we have tested (Figure 2), which is in agreement with data reported by Gromoller and Lehming [34] showing that Srb7p is essential for the activation of a subset of genes and with recent observations in yeast that Mediator subunits are recruited to the promoter of highly transcribed genes [35,36]. Unexpectedly, SRB7 was also localized in one TR, suggesting a role in late stages of transcription for this co-activator.
TFIIF is involved in post-initiation stages of transcription in vivo
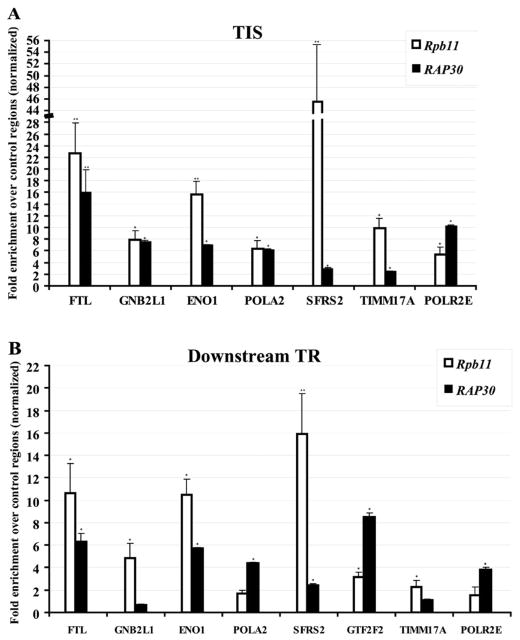
As shown in Figure 3, our results indicate that the TFIIF subunit RAP30 co-localizes with RNAPII in many regions located close to the TIS but also in further downstream TRs, demonstrating for the first time a role for this factor in transcriptional elongation in vivo. The analysis of the other TFIIF subunit RAP74 confirmed this observation (see Supplementary Figure 3). This result contrasts with in vivo results in yeast [18], but fully supports previous in vitro results (reviewed in [1,37–39]) showing that TFIIF acts at both the initiation and elongation stages of the transcription reaction and those of genetic screens, indicating a complex interdependence between TFIIF and the elongation factor TFIIS/TCEA1 [40].
Figure 3. Occupancy of regions proximal to the TIS and further downstream TRs of selected human genes by the TFIIF subunit RAP30 and the RNAPII subunit Rpb11 as determined by ChIP.
Fold enrichment of regions situated close to the TIS (A) and further downstream TRs (B) over control regions are represented for nine selected genes. Student’s t tests were performed to determine the significance of the enrichment values obtained in four experiments, including at least two independent ChIP assays (*P < 0.05, **P ≤ 0.001).
It is interesting to note the gene-specific differences in the relative occupancy of TFIIF and RNAPII (Figure 3): while the RNAPII subunit Rpb11 showed higher occupancy values than the TFIIF subunit RAP30 at most of the regions located close to the TIS, the relative occupancy ratio is inverted in the case of the POLR2E gene where RAP30 showed a higher signal than Rpb11. Furthermore, RNAPII/TFIIF occupancy ratios significantly differ between regions close to the TIS and those situated further downstream in TRs as, for half of the downstream TRs occupied by RAP30, this polypeptide showed higher occupancy signals than Rpb11. This finding may reflect the presence of transcription complexes with different polypeptide composition at these positions, affecting the accessibility of the tag and/or the cross-linking efficiency. This is supported by a recent paper showing that RNAPII stalled close to the promoter in vitro cannot respond to TFIIF and can enter in productive elongation only after the P-TEFb (positive transcription elongation factor b) has functioned, at which point it begins to elongate and move rapidly away from the promoter [41]. Another possible explanation is that TFIIF is preferentially recruited on certain sequences in order to support elongation and to suppress pausing of the polymerase as previously suggested [16].
Rpb7 associates with both the regions proximal to the TIS and with further downstream TRs of active genes
Previous reports showed that Rpb7 is required for the initiation but not the elongation stage of transcription in vitro [42,43]. In vivo studies provided evidence suggesting that human Rpb7 can interact with transcriptional activators such as the oncogenic fusion protein EAD-Fli1 [44] and regulate transcription of genes such as VEGF (vascular endothelial growth factor) [45]. Nevertheless, no previous study addressed the direct implication of this RNAPII subunit at distinct stages of transcription in vivo.
ChIP experiments using a TAP-tagged version of Rpb7 show that it associates with regions close to the TIS and, surprisingly, with further downstream TRs as well (Figure 4). As Rpb4 and Rpb7 are known to form a heterodimer that can dissociate from the polymerase (reviewed in [46]), we generated and used cell lines expressing the TAP-tagged version of Rpb4 in order to confirm the Rpb7 results. Indeed, Rpb4 was also detected in regions close to the TIS and in further downstream TRs (see Supplementary Figure 3). Neither Rpb7 nor Rpb4 was shown to have a direct function in transcription elongation to date (see [46] for a recent review).
Figure 4. Occupancy of regions proximal to the TIS and further downstream TRs of selected human genes by Rpb7 as determined by ChIP.
(A, B) Fold enrichment of regions situated close to the TIS (A) and further downstream TRs (B) over control regions are represented for Rpb7 and Rpb11 when cells are cultured under normal conditions. (C, D) Fold enrichment of regions situated close to the TIS (C) and further downstream TRs (D) after a heat shock (1 h at 42 °C) (HS) are represented for Rpb7 and Rpb11. (E, F) The distribution along the genes ENO1 (E) and HSPA8 (F) is represented for Rpb7 and Rpb11 cultured under normal conditions or after a heat shock (HS). Colour code: black: > 10-fold enrichment; dark grey: between 5- and 10-fold enrichment; light grey: significantly enriched with less than 5-fold enrichment values; white: not significantly enriched. Student’s t tests were performed to determine the significance of the enrichment values obtained in four experiments, including at least two independent ChIP assays (*P < 0.05, **P ≤ 0.001).
Because the Rpb4–Rpb7 dimer was previously suggested to have a role in the transcriptional response to stress, we next addressed the possible implication of Rpb7 in heat shock. As shown in Figure 4(C), and similarly to the core RNAPII subunit Rpb11, Rpb7 occupies all the regions close to the TIS after a heat shock of 1 h at 42 °C. We noted a significant increase (40 % or higher) in the enrichment of most regions close to the TIS (e.g. seven out of eight) by tagged Rpb7 when comparing heat shock with non-heat-shock conditions. Similar results were obtained with the tagged version of Rpb4, for which we observed a significant increase (> 50 %) in the enrichment of regions close to the TIS after a heat shock (results not shown). Notably, even at regions close to the TIS showing a decreased Rpb11 occupancy after heat shock [Figure 4C, for example SFRS2 (splicing factor, arginine/serine-rich 2)], a higher Rpb7 occupancy is observed. Conversely, in the case of POLR2E, a marked decrease in Rpb11 occupancy is accompanied by a lower Rpb7 occupancy after heat shock. This observation argues in favour of a function for the Rpb4–Rpb7 heterodimer in the stabilization of RNAPII during the early stages of transcription under stress conditions.
The same TRs were occupied by Rpb7 before and after heat shock, with similar or increased enrichments, suggesting that the requirement for this subunit during the elongation stage of transcription is maintained under stress conditions (Figure 4D). Of note, Rpb7 and Rpb4 were both detected in one TR which was not occupied by these polypeptides under normal conditions (Figures 4B and 4D and results not shown). In order to address in more detail the requirement of these subunits for transcriptional elongation under stress conditions, we designed primer sets along the entire TR of two constitutive heat-shock genes, known to present increased expression levels in response to a heat shock: ENO1 (enolase 1) and the HSPA8 (heat-shock 70 kDa protein 8). The change in the occupancy pattern observed for the RNAPII core subunit Rpb11 is in agreement with an increased expression of these genes in response to a heat shock, presenting higher occupancy values compared with normal conditions (Figure 4) and becoming detectable in downstream TRs which are not occupied under normal condition (Figures 4E and 4F). In contrast with the core subunit Rpb11, Rpb7 occupied only the TRs proximal to the TIS under normal conditions (up to approx. + 1200) of both genes, while Rpb11 is detected much further downstream under the same conditions (Figures 4E and 4F). This does not seem to be related to a detection limitation in the case of Rpb7, since occupancy values for these two RNAPII subunits are either similar or Rpb7 presents even higher occupancy values than Rpb11 in the TRs proximal to the TIS of these two genes. Interestingly, after heat shock, both Rpb7 and Rpb11 are detected down to the end of both TRs. Moreover, Rpb7 is detected at the 3′ extremity of the HSPA8 gene, which is in agreement with the hypothesis of its involvement in the formation of 3′ termini of the mRNA [47]. We cannot rule out the possibility that Rpb7 remains associated with the elongating polymerase all along the genes under normal conditions, but if this is the case, its conformation or its position in the elongation complex must be different from those under heat shock. Our results showing that Rpb4 is recruited to TRs (Supplementary Figure 3) confirm that the Rpb4–Rpb7 heterodimer has a function during transcriptional elongation. It has been previously shown that Rpb4 genetically interacts with the elongation factor TFIIS/TCEA1 and with the RNAPII subunit Rpb9, which also plays a role in elongation [48,49]. Moreover, it has been shown that yeast Rpb4 is a regulator of transcription-coupled DNA repair pathways [50]. Taken together, our results suggest that the Rpb4–Rpb7 heterodimer is involved in both the initiation and elongation stages of transcription, having a specific role in stress conditions such as heat shock.
In conclusion, we used a systematic affinity tagging procedure coupled with ChIP experiments and gene-specific QPCR detection of the enriched genomic regions to determine the location of components of the general transcription machinery along active class II genes in human cells. Based on the location of the various polypeptides, our results help define their function during the transcription reaction in living mammalian cells.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory, François Robert and the members of his laboratory and Jacques Archambault for helpful discussions and comments on this paper. We are also grateful to Jean-Marc Egly (IGBMC, Strasbourg, France) for antibodies. We thank Diane Bourque for artwork and Julie Edwards for a critical reading of this paper. This work is supported by grants from the Canadian Institutes for Health Research, Genome Canada and Genome Québec. M. C. and C. J. hold studentships from the Canadian Institutes for Health Research and the Fonds de recherche en Santé du Québec.
Abbreviations used
- BCR
breakpoint cluster region
- CDK7
cyclin-dependent kinase 7
- ChIP
chromatin immunoprecipitation
- CTD
C-terminal domain
- ENO1
enolase 1
- ERCC
excision repair cross-complementing rodent repair deficiency, complementation group
- GO
gene ontology
- HEK-293 cell
human embryonic kidney cell
- EcR 293 cells
HEK-293 cells stably transfected with the vector pVgRXR, expressing upon induction the heterodimeric ecdysone receptor
- GTF
general transcription factor
- HSPA8
heat-shock 70 kDa protein 8
- MS/MS
tandem MS
- PCAF
p300/CREB (cAMP-response-element-binding protein)-binding protein]-associated factor
- QPCR
quantitative PCR
- RAP
RNA polymerase II-associated protein
- RNAPII
RNA polymerase II
- SFRS2
splicing factor, arginine/serine-rich 2
- SRB7
suppressor of RNA polymerase B 7
- STAGA
SPT3–TAF9–GCN5–acetyltransferase
- TAF
TBP-associated transcription factor
- TAP
tandem-affinity purification
- TBP
TATA-box-binding protein
- TCEA1
transcription elongation factor A (SII) 1
- TCR
transcription-coupled DNA repair
- TFII
transcription factor II
- TFTC
TATA-box-binding protein-free TAF-containing
- TIS
transcriptional initiation site
- TR
transcribed region
- XPB
xeroderma pigmentosum group B
- XPG
xeroderma pigmentosum group G (Cockayne syndrome)
References
- 1.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller F, Tora L. The multicoloured world of promoter recognition complexes. EMBO J. 2004;23:2–8. doi: 10.1038/sj.emboj.7600027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 5.Fan X, Chou DM, Struhl K. Activator-specific recruitment of mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 6.Forget D, Langelier MF, Therien C, Trinh V, Coulombe B. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol Cell Biol. 2004;24:1122–1131. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 8.Bradsher J, Coin F, Egly JM. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J Biol Chem. 2000;275:2532–2538. doi: 10.1074/jbc.275.4.2532. [DOI] [PubMed] [Google Scholar]
- 9.Zurita M, Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet. 2003;19:578–584. doi: 10.1016/j.tig.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc Natl Acad Sci USA. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 12.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Hawley DK. Identification of a 3′ –> 5′ exonuclease activity associated with human RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:843–847. doi: 10.1073/pnas.90.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Burton ZF. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J Mol Biol. 2004;342:1085–1099. doi: 10.1016/j.jmb.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 17.Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 21.Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci USA. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu F, Pringle TH, Kuhn RM, Karolchik D, Diekhans M, Haussler D, Kent WJ. The UCSC Proteome Browser. Nucleic Acids Res. 2005;33:D454–D458. doi: 10.1093/nar/gki100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi T, Shimoaraiso M, Kubo T, Natori S. Structure–function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J Biol Chem. 1995;270:8991–8995. doi: 10.1074/jbc.270.15.8991. [DOI] [PubMed] [Google Scholar]
- 25.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 26.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong JM, Ingles CJ. A compromised yeast RNA polymerase II enhances UV sensitivity in the absence of global genome nucleotide excision repair. Mol Gen Genet. 2001;264:842–851. doi: 10.1007/s004380000374. [DOI] [PubMed] [Google Scholar]
- 28.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 29.Prather DM, Larschan E, Winston F. Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 31.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 32.Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromoller A, Lehming N. Srb7p is essential for the activation of a subset of genes. FEBS Lett. 2000;484:48–54. doi: 10.1016/s0014-5793(00)02123-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the srb8-11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Woychik NA, Hampsey M. The RNA polymerase II machinery. Structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 38.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 39.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fish RN, Ammerman ML, Davie JK, Lu BF, Pham C, Howe L, Ponticelli AS, Kane CM. Genetic interactions between TFIIF and TFIIS. Genetics. 2006;173:1871–1884. doi: 10.1534/genetics.106.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 42.Orlicky SM, Tran PT, Sayre MH, Edwards AM. Dissociable Rpb4–Rpb7 subassembly of RNA polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J Biol Chem. 2001;276:10097–10102. doi: 10.1074/jbc.M003165200. [DOI] [PubMed] [Google Scholar]
- 43.Rosenheck S, Choder M. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J Bacteriol. 1998;180:6187–6192. doi: 10.1128/jb.180.23.6187-6192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H, Lee KA. An hsRPB4/7-dependent yeast assay for trans-activation by the EWS oncogene. Oncogene. 2001;20:1519–1524. doi: 10.1038/sj.onc.1204135. [DOI] [PubMed] [Google Scholar]
- 45.Na X, Duan HO, Messing EM, Schoen SR, Ryan CK, di Sant’Agnese PA, Golemis EA, Wu G. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel–Lindau protein. EMBO J. 2003;22:4249–4259. doi: 10.1093/emboj/cdg410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem Sci. 2004;29:674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Mitsuzawa H, Kanda E, Ishihama A. Rpb7 subunit of RNA polymerase II interacts with an RNA-binding protein involved in processing of transcripts. Nucleic Acids Res. 2003;31:4696–4701. doi: 10.1093/nar/gkg688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V. Members of the SAGA and mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 2004;23:4232–4242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 50.Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.