SUMMARY
We have performed a survey of soluble human protein complexes containing components of the transcription and RNA processing machineries using protein affinity purification coupled to mass spectrometry. Thirty-two tagged polypeptides yielded a network of 805 high-confidence interactions. Remarkably, the network is significantly enriched in proteins that regulate the formation of protein complexes, including a number of previously uncharacterized proteins for which we have inferred functions. The RNA polymerase II (RNAP II)-associated proteins (RPAPs) are physically and functionally associated with RNAP II, forming an interface between the enzyme and chaperone/scaffolding proteins. BCDIN3 is the 7SK snRNA methylphosphate capping enzyme (MePCE) present in an snRNP complex containing both RNA processing and transcription factors, including the elongation factor P-TEFb. Our results define a high-density protein interaction network for the mammalian transcription machinery and uncover multiple regulatory factors that target the transcription machinery.
INTRODUCTION
The information contained in mammalian genomes is decoded through the concerted action of a myriad of proteins that recognize and interpret specific signals at active genomic sites in such a way that mRNA precursors are synthesized and processed to form mature mRNA that will then serve for protein synthesis. The machinery required for gene transcription includes RNA polymerase II (RNAP II) and its general transcription factors TFIIA, TFIIB, TFIID/STAGA, TFIIE, TFIIF, TFIIH, TFIIS, and Mediator (Hampsey, 1998; Hahn, 2004; Coulombe and Burton, 1999). Many additional proteins regulating the activity of RNAP II at various stages of the transcription reaction have also been purified and characterized from various organisms. Among the regulatory proteins, many were found to modulate the structure of chromatin, the substrate of RNAP II in vivo (Smith and Peterson, 2005; Fischle et al., 2003).
Although information on the composition, function, and mechanisms of these protein complexes has accumulated in the literature, the protein-protein interactions that participate in their functional recruitment at their site of action along genomic DNA are often elusive. Emerging evidence shows that the RNAP II transcription apparatus is involved in this process, particularly through the CTD of RPB1, the largest subunit of RNAP II, which was shown to bind several regulatory complexes at various stages of transcription (Hampsey and Reinberg, 2003; Hirose and Manley, 2000). The CTD was found to interact with transcription elongation factors, chromatin-modifying complexes, pre-mRNA maturation enzymes, and other proteins, providing a molecular basis for the coupling between transcription and RNA processing. The pre-mRNA processing events, including 5′ end capping, splicing, and 3′ end cleavage/polyadenylation, were shown to occur cotranscriptionally (Proudfoot et al., 2002; Bentley, 2005; Aguilera, 2005). Indeed, during the transcription cycle, CTD phosphorylation stimulates efficient recruitment of 5′ and 3′ end processing factors to the pre-mRNA. The splicing machinery is comprised of small-nuclear ribonucleoprotein particles (snRNPs), each composed of various proteins and a specific snRNA (U1, U2, U4, U5, and U6 snRNAs), which together form the spliceosome responsible for performing the catalytic events leading to exon excision and intron ligation (Zhou et al., 2002; Jurica and Moore, 2003). Many proteins such as the hnRNPs and the SR proteins regulate splice site selection (Black, 2003; Shin and Manley, 2004).
The spliceosomal U1-U5 snRNA genes are transcribed by RNAP II while U6 is synthesized by RNA polymerase III (RNAP III) (reviewed in Kiss [2004]). In contrast to the RNAP II-synthesized snRNAs, which receive a 7-methyl guanosine (m7G) cap during transcription and are subsequently methylated posttranscriptionally to form the trimethylguanosine (TMG) cap, RNAP III-transcribed snRNAs acquire a methyl group on the gamma-phosphate of their 5′ end posttranscriptionally (Reddy et al., 1992). Another snRNA synthesized by RNAP III, 7SK, is an abundant 331 nucleotide transcript found in association with the positive transcription elongation factor b (P-TEFb) (Nguyen et al., 2001; Yang et al., 2001). P-TEFb, which is composed of CDK9 and CCNT1/cyclin T1 or CCNT2/cyclin T2, stimulates the elongation phase of transcription by phosphorylating the CTD of RNAP II and reversing the effects of negative elongation factors (for recent reviews, see Peterlin and Price [2006] and Zhou and Yik [2006]). P-TEFb not only plays an important role in the transcription of cellular genes but is also a key factor for the expression of the human immunodeficiency virus type 1 (HIV-1) genome (Zhu et al., 1997). Previous studies have shown that a complex formed of the 7SK snRNA and proteins termed HEXIMs can interact with P-TEFb and repress its stimulatory activity on transcriptional elongation by RNAP II (Barboric et al., 2005; Blazek et al., 2005; Byers et al., 2005; Li et al., 2005; Yik et al., 2005).
Although the protein complexes actively involved in transcription on genomic DNA have been extensively studied, very little is known about the organization of the transcription machinery in the soluble nuclear compartment, prior to or following its association with genomic DNA, where its formation and recycling likely take place. To determine the composition and organization of the soluble machinery for mRNA formation and obtain insight into its mechanisms of formation and/or recycling, we performed a survey of the soluble protein complexes comprising key components of the RNAP II machinery. Tagged proteins were purified and their associating partners identified by mass spectrometry. High-confidence interactions were selected computationally and used to draw a map of interactions connecting these various complexes. The composition and organization of this network revealed important features of the machinery involved in mRNA formation in eukaryotes.
RESULTS
A Network of Protein Complexes Involving the Human Transcription and RNA Processing Machineries
To survey soluble protein complexes that comprise the transcription machinery and define their interaction network, several subunits of RNAP II and basal transcription factors carrying a tandem affinity purification (TAP) tag were purified and their interaction partners identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (see Figure 1A for an overview of our procedure and see Figure S1A in the Supplemental Data available with this article online for a schematic representation of the expression system used). A number of identified interaction partners, including RNA processing factors, were also tagged and submitted to the same procedure, thereby enriching the interaction data set (see Figure S1B for examples). Of note, all the tagged transcription factors that we purified and tested were active in transcription reactions in vitro and were present in transcribed regions in chromatin immunoprecipitation (ChIP) experiments in vivo (M. Cojocaru, C.J., D. F., A.B., D.B., P. Côte, G.G.P., J.G., and B. Coulombe, unpublished data), indicating that bona fide, functional protein complexes were purified.
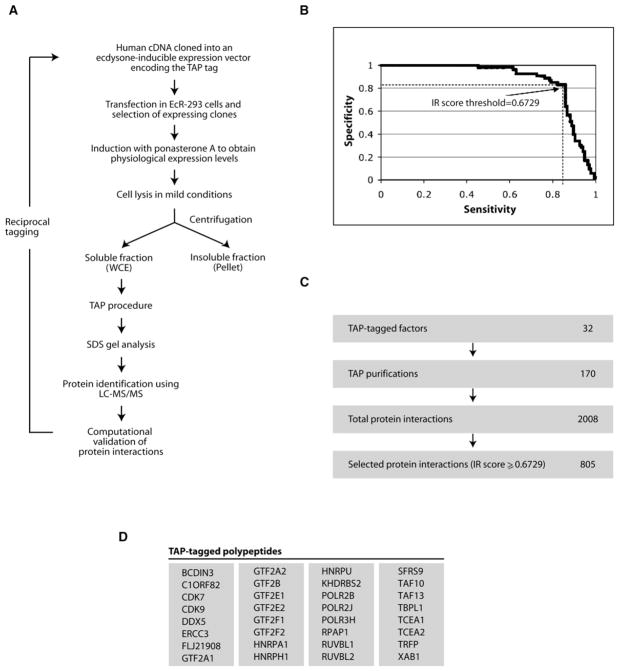
Figure 1. A Method for the Analysis of Protein Complexes in Human Cells.
(A) Flow chart of the procedure used for the affinity purification and mass spectrometry identification of protein complexes in HEK293 cells.
(B) Computational validation of protein interactions. Sensitivity/specificity ROC curve, parameterized by the IR score, for the discrimination between literature-supported interactions and dubious interactions (see Table S1). The chosen IR score threshold results in a sensitivity of 83% and a specificity of 83%. The IR was calculated based on the method described in the Supplemental Experimental Procedures.
(C) Summary of the overall screen for protein interactions.
(D) List of the 32 TAP-tagged polypeptides used in this study.
To select high-confidence interactions and minimize the number of false positives in our data set, we developed a computational method that, first, filters out spurious interactions caused by proteins that bind nonspecifically to our columns and very abundant cellular proteins that may have remained as contaminants after affinity purification and, second, attributes an interaction reliability (IR) score to each protein interaction detected by mass spectrometry (see the Supplemental Data for a description of the method; Figure S2 illustrates the overall procedure, and the Supplemental Experimental Procedures describes the computational algorithm used).
Figure 1B illustrates the ability of our IR score to separate true protein interactions from likely false positives. Of the 2008 putative protein interactions detected by mass spectrometry (see Table S1), 805 were judged highly reliable by our classifier (IR score above 0.6729) (Figure 1C), involving a total of 436 proteins. This set of highly reliable interactions contains 83% of the interactions supported by the literature, while excluding 83% of those that were judged likely false positives. As summarized in Figure 1C, 170 successful affinity purifications using 32 tagged proteins yielded 2008 putative protein interactions (see Figure 2 and Table S1 for a list of the tagged proteins). The 805 distinct interactions with an IR score above 0.6729 constitute a network of protein interactions in which we have high confidence.
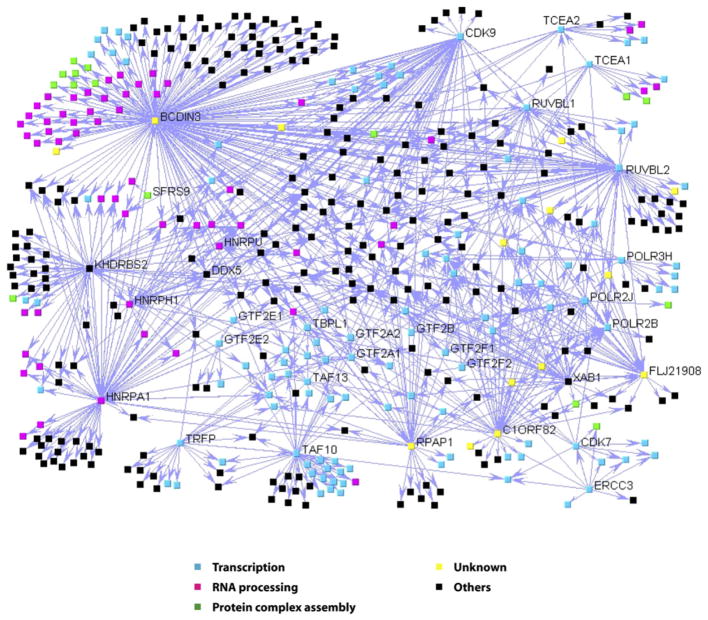
Figure 2. A Network of Protein Complexes Involving the RNAP II Basal Transcription Machinery.
Overview of the interaction map showing the 805 validated interactions obtained using 32 tagged polypeptides. The interactions are represented as directional edges extending from the tagged protein using the web-based software VisANT (http://visant.bu.edu). The thickness of the line that connects two proteins is proportional to the IR score obtained for this interaction (see Table S1). The nodes are colored according to their GO annotations (http://www.geneontology.org). The tagged proteins are indicated.
Figure 2 shows the interaction network for the protein complexes comprising all the tagged transcription and RNA processing factors used in our experiments and their interaction partners. Not surprisingly, gene ontology (GO) annotations (Gene-Ontology database, http://www.geneontology.org) revealed that many proteins of the network are either transcription factors (Figure 2; blue nodes) or RNA processing factors (Figure 2; pink nodes), which are sometimes interconnected as should be expected for two closely related cellular machineries. Unexpectedly, however, GO annotations also revealed that a significant number of network components are proteins involved in modulating the formation of protein complexes (P value = 1.8e-5) (see Figure 2; green nodes). Importantly, our protein network also contains several proteins that are in the databases but have not been previously characterized (Figure 2; yellow nodes).
To further assess the efficiency of our affinity-purification method for the discovery of functionally relevant protein interaction partners, five proteins occupying key positions in the overall interaction network, namely RNAP II-associated protein (RPAP)1, C1ORF82, FLJ21908, XAB1, and BCDIN3, were used in experiments aimed at validating their network connections both physically (reciprocal tagging and gel filtration) and functionally (bio-informatics, silencing of the yeast homolog, and biochemical assays). These five proteins were selected because our initial bioinformatic analysis revealed that they may represent unprecedented regulatory factors targeting two key components of the transcription machinery, RNAP II and P-TEFb. As shown in a previous study in yeast (Krogan et al., 2004), the affinity-purification procedure used is also efficient in identifying RNA components of the purified complexes.
The RPAPs-XAB1 Physically and Functionally Connect RNAP II to Regulators of Protein Complex Formation
Our protein interaction network reveals the existence of a group of four polypeptides, RPAP1 (Jeronimo et al., 2004), XAB1, C1ORF82, and FLJ21908, that are tightly connected to the enzyme RNAP II itself in the soluble nuclear compartment (Figure 3A). Gel filtration experiments using the tagged-RPB11/POLR2J eluate confirmed that RPAP1, FLJ21908, and XAB1 cofractionate with RNAP II (Figure 3B). Because of their physical association with RNAP II, which may be direct or indirect, the two previously uncharacterized proteins C1ORF82 and FLJ21908 were named RPAP2 and RPAP3, respectively. The RPAPs-XAB1 also interact with the regulatory complex Mediator, GRINL1A/Gdown1, a recently discovered RNAP II interaction partner that mediates Mediator responsiveness during transcriptional activation (Hu et al., 2006), and the Integrator, a complex involved in the processing of RNAP II-synthesized snRNAs (Baillat et al., 2005) (Figure 3A).
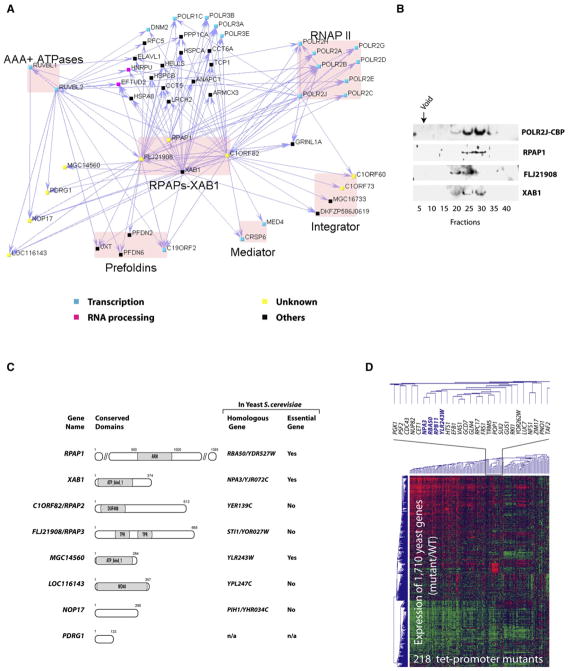
Figure 3. Previously Uncharacterized Protein Complexes Containing RNAP II in Association with Regulators of Protein Complex Assembly.
(A) Network highlighting the interactions of RPAPs-XAB1 with RNAP II, the regulatory complexes Integrator and Mediator, and a group of proteins with chaperone/scaffolding activity.
(B) Elution profile of RNAP II and the RPAPs-XAB1 in gel filtration.
(C) Previously uncharacterized proteins are associated with RNAP II. For each human protein, a representation with putative functional domains provided by the NCBI’s conserved domain database, the name of the closest yeast homologous gene as determined by a BLASTP search on the June 2005 assembly version of the UCSC genome browser, and gene essentiality in yeast provided by the SGD are shown. ARM, ARM repeat protein superfamily; ATP_bind_1, conserved hypothetical ATP-binding protein; DUF408, domain of unknown function; TPR, tetratricopeptide repeat domain; WD40, WD40 domain.
(D) Clustering analysis of mRNA expression profiles with tet-promoter alleles corresponding to RBA50, NPA3, YLR243W, and RPB11, in comparison to profiles from 214 other tet-promoter mutants representing diverse cellular functions (Mnaimneh et al., 2004).
Examination of the protein interaction network further reveals that the RPAPs-XAB1 are placed at the interface between RNAP II and a group of proteins able to assemble into molecular chaperone complexes, including the prefoldins (C19ORF2/URI, PFDN6, PFDN2, and UXT) (Gstaiger et al., 2003; Vainberg et al., 1998; Zhao et al., 2005) and the AAA+ chaperone-like ATPases RUVBL1 and RUVBL2 (Kanemaki et al., 1999; Ikura et al., 2000) (Figure 3A). Notably, a bioinformatics analysis revealed that motifs unique to chaperones and scaffolding proteins are also present in the RPAPs-XAB1 proteins themselves (Figure 3C). RPAP1 contains an ARM domain (Jeronimo et al., 2004), which is a structural element known to mediate protein-protein interactions (Huber et al., 1997). FLJ21908/RPAP3 contains helix-turn-helix tetratricopeptide repeat (TPR) motifs, which characterize cofactors of chaperones (Smith, 2004). XAB1 is part of a family of polypeptides containing an ATP-binding domain, termed “ATP_bind_1,” that are conserved among eukaryotes and archaea; of note, energy-driven conformational changes are central to the formation of many protein complexes. Although previous reports have shown that human XAB1 binds to the DNA repair factor XPA and participates in transcriptional regulation at methylated promoters (Nitta et al., 2000; Lembo et al., 2003), its mechanism of action remains undetermined. The RPAPs-XAB1 are also associated with a number of additional previously uncharacterized proteins (MGC14560, LOC116143, NOP17, and PDRG1) (Figure 3A). MGC14560 is another member of the ATP_bind_1 family, and LOC116143 contains a WD40 domain (Figure 3C), which is believed to coordinate multiprotein complex assemblies (Smith et al., 1999). Our results demonstrate that RNAP II is associated with a number of proteins known or predicted to regulate protein complex formation.
A number of interactions among the RPAPs-XAB1, RNAP II, and their other human interaction partners are also observed in yeast (Saccharomyces genome database, SGD). Interestingly, silencing the yeast homologs of XAB1 and MGC14560, namely NPA3 and YLR243W, which are both essential for cell growth and are known to physically interact in yeast (see Figure 3C and SGD), had global effects on yeast gene expression similar to those of silencing either the RNAP II subunit RPB11 or RBA50 (the closest homologous gene to RPAP1 in yeast) (Figure 3D). These results along with their position at the interface of RNAP II regulatory complexes and molecular chaperones/scaffolding proteins suggest a role for the RPAPs-XAB1 in the formation of multicomponent transcription complexes.
A 7SK snRNP Complex Is Comprised of RNA Processing and Transcription Factors, Including the Elongation Factor P-TEFb, and the Previously Uncharacterized Protein BCDIN3
Previous studies have shown that two forms of P-TEFb complexes, one being large and inactive and the other being small and active, can be purified from mammalian cells. Indeed, the cyclin-dependent kinase activity of P-TEFb is inhibited upon binding of proteins termed HEXIMs and the 7SK snRNA (Barboric et al., 2005; Blazek et al., 2005; Byers et al., 2005; Li et al., 2005; Yik et al., 2005). P-TEFb also binds to the bromodomain protein Brd4 to form the transcriptionally active P-TEFb complex (Jang et al., 2005; Yang et al., 2005). Our protein interaction network revealed that, in the soluble fraction, besides its positive (Brd4) and negative (HEXIMs and 7SK snRNA) regulators, P-TEFb is associated with many other proteins including RNA processing factors, SART3 (also referred to as Tip110), and the previously uncharacterized protein BCDIN3 (Bicoid-interacting 3, homolog [Drosophila]) (see Figure 4A and Table S1). Reciprocal tagging of BCDIN3 confirmed these interactions. Interestingly, SART3, a surface antigen in several cancers (Yang et al., 1999), has independently been described as (1) a coregulator of HIV-1 gene expression that directly associates with Tat (Liu et al., 2002), and (2) a negative regulator of transcriptional activation by the androgen receptor (Liu et al., 2004).
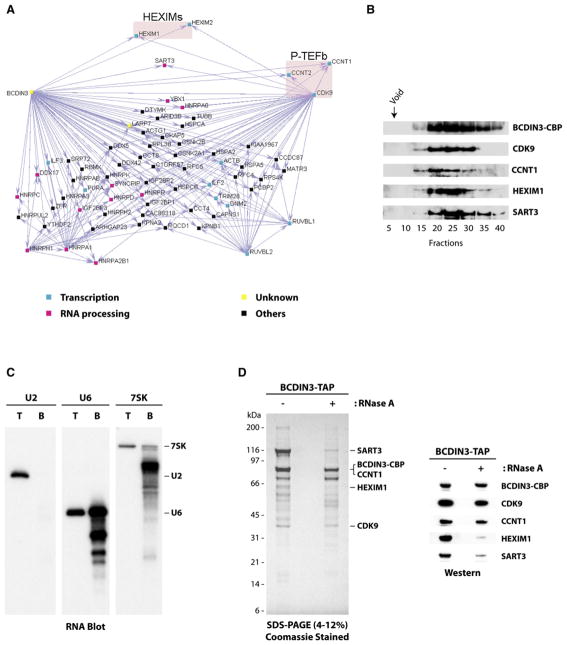
Figure 4. A Complex Comprising the Elongation Factor P-TEFb, the 7SK and U6 snRNAs, and the Previously Uncharacterized Protein BCDIN3.
(A) Network highlighting the interactions of BCDIN3 and CDK9 with various RNA processing factors, P-TEFb (cyclin T1/cyclin T2 subunits), and its regulatory factors HEXIM1 and HEXIM2.
(B) Gel filtration analysis showing that P-TEFb, HEXIM1, and the RNA processing factor SART3 cofractionate with BCDIN3.
(C) The 7SK and U6 snRNAs are present in the BCDIN3-TAP eluate. RNA blots were performed on total RNA extracted from HeLa cells (T; 900 ng) or RNA extracted from the BCDIN3-TAP eluate (B; 60 ng) and probed with RNA oligos specific for U2, U6, or 7SK snRNAs. Migration of the three intact RNA species is indicated.
(D) The 7SK snRNP complex is partly resistant to extensive digestion with RNase A. SDS gel showing BCDIN3 affinity eluates prepared from cell extracts treated or not with RNase A. Western blots confirming the presence of BCDIN3, P-TEFb, HEXIM1, and SART3 in both eluates are shown.
The affinity-purified complex was further characterized by running the BCDIN3 eluate on a gel filtration column (Figure 4B). This experiment confirmed that P-TEFb, HEXIM1, and SART3 cofractionate with BCDIN3. Because subunits of P-TEFb and the HEXIMs were found associated with BCDIN3, we reasoned that 7SK might also be present, because P-TEFb and HEXIM1 do not interact in the absence of 7SK (Li et al., 2005; Michels et al., 2003; Yik et al., 2003). RNA was isolated from a portion of a BCDIN3 eluate and probed for the presence of 7SK, U6, and U2 snRNAs. The probes specifically recognized their respective cellular RNAs from total HeLa cell RNA, but only 7SK and U6 were found in the BCDIN3-tagged eluate (Figure 4C). Both 7SK and U6 RNAs were partially degraded, presumably due to extensive incubations utilized in the affinity-purification procedure.
To assess the roles of protein-RNA and protein-protein interactions in maintaining the integrity of the 7SK snRNP, the complex was submitted to extensive digestion with RNase A prior to its purification on our affinity columns. This treatment led to a partial dissociation of SART3 and HEXIM1 (Figure 4D), suggesting that RNA-protein interactions contribute to, but are not essential for, the association of these proteins with the complex. This experiment also ruled out the possibility that our observed interactions are caused by contaminating RNA molecules associated nonspecifically with some protein components of the complex. The association of P-TEFb (and most other proteins of the eluate; data not shown) with tagged BCDIN3 is resistant to RNase A treatment (Figure 4D), indicating that P-TEFb and BCDIN3 are associated mainly through direct or indirect protein-protein interactions.
BCDIN3 Is a Conserved 7SK snRNA Methylphosphate Capping Enzyme
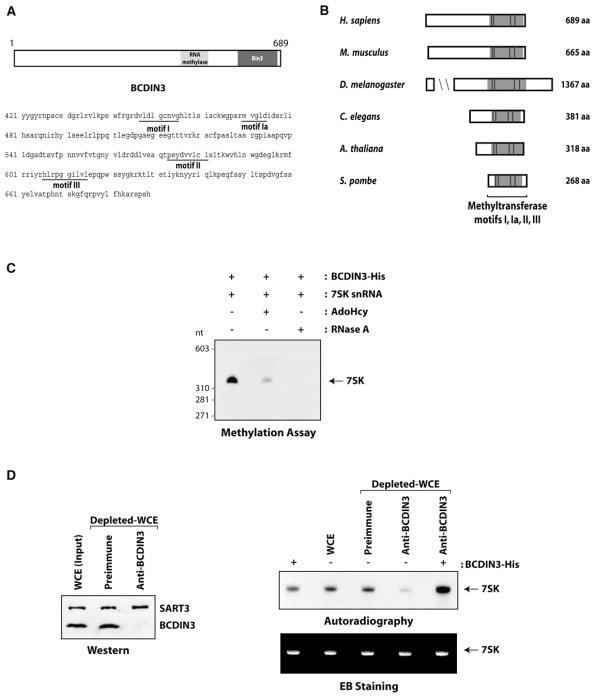
Bioinformatics analysis revealed that BCDIN3 contains a consensus S-adenosyl methionine (AdoMet)-binding domain (Figure 5A), AdoMet being the methyl donor used by methyltransferases (Lu, 2000). As in the other members of the major class (class I) of AdoMet-dependent methyltransferases, BCDIN3 has the four conserved amino acid sequence motifs (I, Ia, II, and III) that form a seven-strand twisted beta sheet (Kagan and Clarke, 1994). A database search with BCDIN3 revealed the existence of homologous proteins in metazoans (Mus musculus, Drosophila melanogaster, Caenorhabditis elegans), plants (Arabidopsis thaliana), and lower eukaryotes (Schizosaccharomyces pombe) (Figure 5B; see Experimental Procedures for homolog accession numbers). The Drosophila BCDIN3 homologous protein, named Bin3 (Bicoid-interacting protein 3), was previously identified in association with Bicoid, a DNA-binding factor that directs pattern formation in the early developing embryo (Zhu and Hanes, 2000). BCDIN3 and its homologous proteins show the highest homology in their C-terminal regions, which contain the conserved AdoMet-dependent methyltransferase motifs (Figure 5B), as well as the Bin3 domain (Figure 5A). Notably, a BCDIN3 homolog is absent in S. cerevisiae (Figure 5B).
Figure 5. BCDIN3 Is a Conserved Methyltransferase that Targets the 7SK snRNA.
(A) Linear representation of BCDIN3 with its RNA methylase and Bin3 domains provided by the NCBI’s conserved domain database. The amino acid sequence of the C-terminal region of BCDIN3 containing the putative S-adenosyl methionine (AdoMet)-binding domain is shown. The signature seven β strand methyltransferase motifs I, Ia, II, and III are indicated.
(B) Evolutionary conservation of human BCDIN3. A schematic representation of BCDIN3 and its protein homolog according to the NCBI’s Homolo-Gene system is provided. The conserved regions containing the methyltransferase motifs I, Ia, II, and III are indicated.
(C) The 7SK snRNA is methylated in vitro by BCDIN3. Recombinant 7SK snRNA was incubated with His-tagged BCDIN3 and 3H-AdoMet in the presence of the buffer alone, AdoHcy, or RNase A. The 3H-methylated RNA product was detected by autoradiography.
(D) Immunodepletion of BCDIN3 impairs the 7SK methylation activity of whole-cell extracts and addition of recombinant BCDIN3-His to the depleted extract rescues 7SK methylation. HEK293 whole-cell extracts (WCE) were incubated or not with protein A Sepharose beads coupled to either pre-immune or anti-BCDIN3 serum, and supernatants were immunoblotted with antibodies directed against BCDIN3 and SART3 (as a loading control). WCE incubated or not with beads coated with preimmune or anti-BCDIN3 serum were assayed for methylation of recombinant 7SK snRNA in the absence or presence of His-tagged BCDIN3. RNA was visualized by ethidium bromide (EB) staining (lower panel), and the 3H-methylated product was detected by autoradiography (upper panel).
To start addressing the function of this previously un-characterized methyltransferase, we searched for specific substrates within members of the 7SK snRNP complex. Because BCDIN3 harbors a putative RNA methylase domain (NCBI’s conserved domain database) (Figure 5A), we investigated whether the 7SK snRNA could serve as a substrate for BCDIN3 in an in vitro methylation assay. His-tagged BCDIN3 expressed and purified from bacteria was incubated with recombinant 7SK in the presence of radiolabeled AdoMet (3H-AdoMet). Figure 5C shows that the 7SK is methylated by BCDIN3-His. Moreover, competition with AdoHcy, a potent competitive inhibitor of a number of methyltransferases (Ueland, 1982), confirmed the enzymatic nature of the BCDIN3 methylation reaction.
To confirm that BCDIN3 is a bona fide 7SK methyltransferase in human cells, we depleted endogenous BCDIN3 from HEK293 whole-cell extracts using a specific antibody and assessed the effect on the ability of the extract to support 7SK methylation. As shown in Figure 5D, the depletion of BCDIN3 resulted in a marked decrease in the 7SK methylation activity of the extract. Addition of recombinant BCDIN3 to the depleted extract fully restored the methylation activity. These results indicate that BCDIN3 is an active 7SK methyltransferase in human cell extracts.
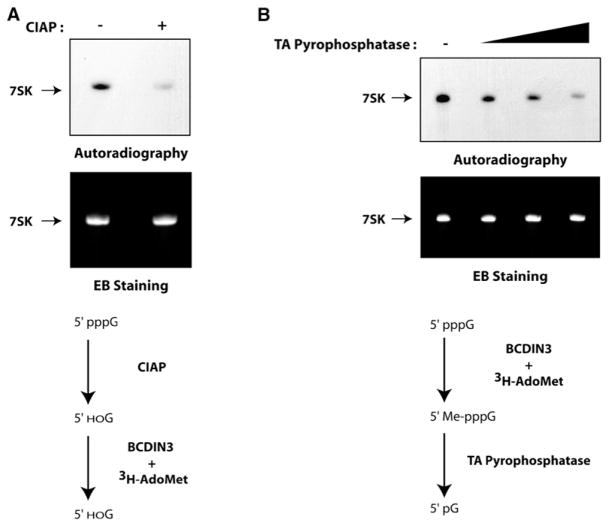
Previous studies have shown that RNAP III-synthesized snRNAs such as 7SK are capped posttranscriptionally through the addition of a methyl group directly on the gamma-phosphate of their first 5′ nucleotide (Gupta et al., 1990; Shumyatsky et al., 1990). However, up to now the enzyme responsible for this function had not been identified. Consequently, BCDIN3 is a candidate for being a methylphosphate capping enzyme. To test this hypothesis, we performed two independent experiments. First, recombinant 7SK was treated with calf intestinal alkaline phosphatase (CIAP), specifically removing the phosphate groups from the 5′ end of RNA, prior to its incubation with His-tagged BCDIN3 in the in vitro methylation assay. Treatment of 7SK with CIAP decreased the methylation signal (Figure 6A, upper panel), showing that the presence of the 5′ phosphates is required for efficient methylation by BCDIN3. Second, the 7SK snRNA was methylated in vitro by BCDIN3-His and the reaction product treated with tobacco acid (TA) pyrophosphatase, an enzyme that specifically removes the gamma and beta phosphates from the 5′ end of RNA (Shinshi et al., 1976). Figure 6B shows that treatment with this decapping enzyme removes the radiolabeled methyl from the 7SK. To ensure that equal amounts of RNA were used and that RNA was not degraded during the dephosphorylation and decapping assays, the gels containing the methylation reactions were stained with ethidium bromide prior to their autoradiography (Figures 6A and 6B, lower panels). Essentially the same results were observed using endogenous BCDIN3 (data not shown). Taken together, these results demonstrate that BCDIN3 is capable of acting as a 7SK methylphosphate capping enzyme, hereby renamed MePCE.
Figure 6. BCDIN3 Is the 7SK snRNA MePCE.
(A) Dephosphorylation of the 7SK snRNA impairs its methylation by BCDIN3. Recombinant 7SK snRNA was treated with (+) or without (−) calf intestinal alkaline phosphatase (CIAP) prior to its incubation with His-tagged BCDIN3 and 3H-AdoMet. The RNA was visualized by ethidium bromide (EB) staining (lower panel), and the methylated product was detected by autoradiography (upper panel). A schematic representation of the assay is shown.
(B) Enzymatic decapping of BCDIN3-methylated 7SK removes the radiolabeled methyl group. Recombinant 7SK was incubated with His-tagged BCDIN3 and 3H-AdoMet and then decapped or not (−) with different amounts of tobacco acid (TA) pyrophosphatase. The RNA was visualized by ethidium bromide (EB) staining (lower panel), and the methylated product was detected by autoradiography (upper panel). A schematic representation of the assay is shown.
BCDIN3/MePCE Silencing Decreases the Steady-State Level of Cellular 7SK
Previous studies have shown that 7SK capping through gamma-phosphate methylation is important for protecting the RNA from exonucleolytic degradation. Indeed, it has been shown that the cap structure enhances the stability of U6 and 7SK snRNAs (Shumyatsky et al., 1990) and that uncapped U6 snRNA is rapidly degraded (Hamm et al., 1990). Moreover, other known cap structures such as m7G and TMG also enhance the half-life of RNAs (Cougot et al., 2004). To determine whether BCDIN3/MePCE is involved in regulating 7SK steady-state levels, we used small interfering RNA (siRNA) to silence its expression in HEK293 cells and analyzed the effect on 7SK RNA levels. The results show that reducing the expression of BCDIN3/MePCE by about 70% in vivo caused a decrease in the steady-state level of cellular 7SK as determined by RNA blot (Figure 7). BCDIN3/MePCE knockdown had no effect on U2 and U6 snRNA levels (Figure 7). Together, our results indicate that BCDIN3/MePCE stabilizes the 7SK snRNA in human cells through the addition of a methyl-phosphate cap at its 5′ end.
Figure 7. BCDIN3 Silencing Decreases the Steady-State Level of Cellular 7SK.
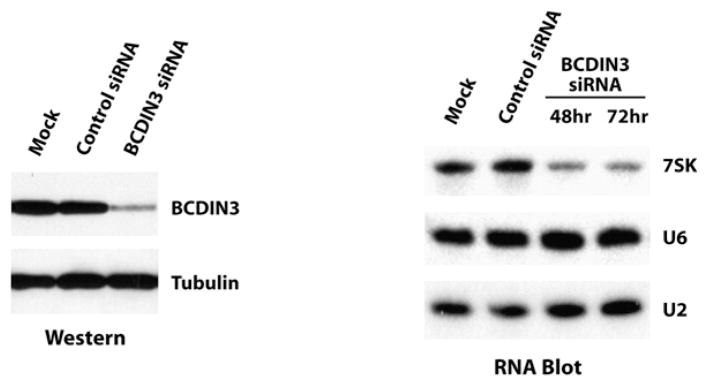
BCDIN3 silencing was monitored by western blotting using extracts from HEK293 cells mock transfected or transfected with control or BCDIN3 siRNA (tubulin was used as a loading control). 7SK steady-state levels were assessed by RNA blotting using total RNA extracted from HEK293 cells mock transfected or transfected with control or BCDIN3 siRNA (48 hr and 72 hr posttransfection) and probed with RNA oligonucleotides that specifically detect 7SK, U6, or U2 snRNAs.
DISCUSSION
In this article, we report the development of a general procedure allowing (1) the purification of affinity-tagged proteins in native conditions, (2) the identification of purified proteins using sensitive mass spectrometry, and (3) the computational analysis of the data for minimizing the rate of both false positives and false negatives, and its use to define a high-confidence protein interaction data set for transcription and RNA processing factors. The results were used to build a high-density interaction network connecting components of the transcription and RNA processing machineries in human cells. Although the transcription and RNA processing machineries have been studied for decades, our results define many additional proteins that interact with some of their key components. In particular, important transcription factors are associated with proteins that regulate the formation of other protein complexes. We believe that these proteins have not been identified in previous studies because they apparently do not regulate these machineries during active transcription, although this possibility remains to be tested. For example, the RPAPs-XAB1 are tightly associated both physically and functionally with RNAP II. Because they also interact with chaperones/scaffolding proteins and themselves contain motifs characteristic of chaperones/scaffolding proteins (see Figures 3A and 3C), our results suggest that we have identified and purified proteins involved in assembly of the protein complexes composing the cellular machinery for mRNA synthesis.
The composition of the 7SK snRNP, which was discovered more than two decades ago, has been elusive (Wassarman and Steitz, 1991; Gurney and Eliceiri, 1980). Association of the transcription elongation factor P-TEFb with a putative methyltransferase prompted us to use this tagged protein in reciprocal affinity-purification experiments. Our results confirmed that BCDIN3 associates with P-TEFb, its negative regulatory factors HEXIMs, and the RNA processing factor SART3. Our results also indicate that the 7SK and U6 snRNAs are part of the BCDIN3 affinity-purified complex. Indeed, previous studies have shown that HEXIM1 and HEXIM2 bind to the 7SK snRNA through their arginine-rich RNA-binding motifs (Byers et al., 2005; Yik et al., 2004, 2005). Of note, SART3 contains two RNA recognition motifs (RRMs) that bind directly to the U6 snRNA (Bell et al., 2002). Interestingly, it has been proposed that SART3 acts as an RNA chaperone. Indeed, SART3 interacts via its C-terminal domain with other components of the U6 snRNP, the LSM proteins, which assemble as heteroheptamers on the 3′ end of the U6 snRNA (Achsel et al., 1999). In addition, it has also been shown that SART3 interacts through its TPR motifs with the U4/U6-specific factor PRPF3, suggesting that it provides the scaffolding interface for the assembly of the U4/U6 snRNP (Medenbach et al., 2004). It will be interesting to investigate whether SART3 participates in the formation of the 7SK snRNP.
In this study, we show that BCDIN3, a conserved protein that is part of a family of AdoMet-dependent methyl-transferases, is a 7SK MePCE. To date, the methylated gamma-phosphate cap structure has been identified in three other RNAP III-transcribed small RNAs, namely U6, rodent B2, and plant U3 (Shumyatsky et al., 1990; Shimba et al., 1992; Liu et al., 1992). Our results indicate that siRNA-mediated silencing of MePCE expression reduces the steady-state level of cellular 7SK, suggesting that MePCE protects 7SK from exonucleolytic degradation. Interestingly, previous studies have shown that the activity of P-TEFb is highly modulated via alternative interactions with its positive (Brd4) and negative (HEXIMs and 7SK snRNA) regulators (Nguyen et al., 2001; Yang et al., 2001, 2005; Yik et al., 2003; Michels et al., 2003, 2004). Although the role of 7SK capping by MePCE remains to be detailed, this structure might be involved in regulating the formation (e.g., assembly and/or transport) of transcriptionally active P-TEFb to be recruited to gene promoters. Further analyses are necessary to unravel the mechanism of action of the capped 7SK and the pathway controlling the association of capped 7SK with P-TEFb.
Despite decades of detailed biochemical and genetic analysis of the RNAP II basal transcription machinery, we identified many previously uncharacterized factors, some being involved in regulating the network of interactions formed by the transcription and RNA processing machineries. Expanding this network promises to reveal numerous additional interactions of general or regulatory importance and may be similarly revealing for other cellular functions.
EXPERIMENTAL PROCEDURES
Human Cell Lines Carrying Inducible TAP-Tagged Proteins
Selected human polypeptides were cloned into the mammalian expression vector pMZI (Zeghouf et al., 2004) carrying a TAP tag at its C terminus (Rigaut et al., 1999). Stable human embryonic kidney cell lines EcR-293 (derived from HEK293) carrying these constructs were produced as previously described (Jeronimo et al., 2004). The conditions for expression, affinity purification, and mass spectrometry identification of proteins are detailed in the Supplemental Experimental Procedures.
DNA Microarray Analysis of Tet-Promoter Mutants
DNA microarray analyses of tet-promoter mutants were performed as described (Mnaimneh et al., 2004). See the Supplemental Experimental Procedures for further details.
siRNA Knockdown and RNA Blot
BCDIN3 (ON-TARGETplus SMART pool) and control (siCONTROL Non-targeting pool) siRNAs (Dharmacon) were transfected in 50% confluent HEK293 cells using Lipofectamine 2000 (Invitrogen) at an siRNA final concentration of 100 nM. At various time intervals post-transfection, cells were lysed and BCDIN3 expression levels were monitored by western blotting (see the Supplemental Experimental Procedures for a description of the antibodies used).
RNA blot analyses of total RNAs from HeLa cells, HEK293 cells treated with siRNA, or BCDIN3-TAP eluate were carried out as previously described (Li et al., 2007) (see the Supplemental Experimental Procedures for further details).
Sequence Analysis
The proteins belonging to the Bin3 family (NCBI’s conserved domain database; pfam06859) have the following accession numbers: H. sapiens, NP_062552; M. musculus, NP_659162; D. melanogaster, NP_724468; C. elegans, NP_496572; A. thaliana, NP_568752; and S. pombe, NP_596220.
Other Methods
See the Supplemental Experimental Procedures for methods relating to gel filtration chromatography, immunodepletion, RNA methyltransferase, dephosphorylation, and decapping assays.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory and Jacques Archambault for helpful discussions and comments. The SART3/Tip110 antibody was a kind gift from J.J. He. We thank Denis Faubert for mass spectrometry analysis and S. Clarke for helpful discussions. This work is supported by grants from the Canadian Institutes for Health Research (CIHR), Genome Canada, Génome Québec, Genome Prairie-Alberta, the Ontario Genomics Institute, the Fonds de la recherche en santé du Québec (FRSQ), and the Canadian Foundation for Innovation. G.G.P. holds a Canada Research Chair in Targeted Proteomics. B. Chabot holds a Canada Research Chair in Functional Genomics. C.J. and C.T. hold studentships from the CIHR and the FRSQ. G.C. holds a Charles H. Best fellowship.
Footnotes
Supplemental Data include Supplemental Discussion, Supplemental Experimental Procedures, Supplemental References, two figures, and one table and can be found with this article online at http://www.molecule.org/cgi/content/full/27/2/262/DC1/.
References
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005;33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- Cougot N, van Dijk E, Babajko S, Seraphin B. Captabolism. Trends Biochem Sci. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- Gupta S, Busch RK, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J Biol Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- Gurney T, Jr, Eliceiri GL. Intracellular distribution of low molecular weight RNA species in HeLa cells. J Cell Biol. 1980;87:398–403. doi: 10.1083/jcb.87.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyl-transferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Kurokawa Y, Matsuura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Lembo F, Pero R, Angrisano T, Vitiello C, Iuliano R, Bruni CB, Chiariotti L. MBDin, a novel MBD2-interacting protein, relieves MBD2 repression potential and reactivates transcription from methylated promoters. Mol Cell Biol. 2003;23:1656–1665. doi: 10.1128/MCB.23.5.1656-1665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–2512. doi: 10.1093/nar/gkm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MH, Busch RK, Buckley B, Reddy R. Characterization of antibodies against methyl-pppN cap structure: plant U3 small nucleolar RNA is recognized by these antibodies. Nucleic Acids Res. 1992;20:4299–4304. doi: 10.1093/nar/20.16.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li J, Kim BO, Pace BS, He JJ. HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J Biol Chem. 2002;277:23854–23863. doi: 10.1074/jbc.M200773200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kim BO, Kao C, Jung C, Dalton JT, He JJ. Tip110, the human immunodeficiency virus type 1 (HIV-1) Tat-interacting protein of 110 kDa as a negative regulator of androgen receptor (AR) transcriptional activation. J Biol Chem. 2004;279:21766–21773. doi: 10.1074/jbc.M314321200. [DOI] [PubMed] [Google Scholar]
- Lu SC. S-Adenosylmethionine. Int J Biochem Cell Biol. 2000;32:391–395. doi: 10.1016/s1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- Medenbach J, Schreiner S, Liu S, Luhrmann R, Bindereif A. Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol Cell Biol. 2004;24:7392–7401. doi: 10.1128/MCB.24.17.7392-7401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- Nitta M, Saijo M, Kodo N, Matsuda T, Nakatsu Y, Tamai H, Tanaka K. A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 2000;28:4212–4218. doi: 10.1093/nar/28.21.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Reddy R, Singh R, Shimba S. Methylated cap structures in eukaryotic RNAs: structure, synthesis and functions. Pharmacol Ther. 1992;54:249–267. doi: 10.1016/0163-7258(92)90002-h. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Shimba S, Buckley B, Reddy R, Kiss T, Filipowicz W. Cap structure of U3 small nucleolar RNA in animal and plant cells is different. gamma-Monomethyl phosphate cap structure in plant RNA. J Biol Chem. 1992;267:13772–13777. [PubMed] [Google Scholar]
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Shinshi H, Miwa M, Sugimura T. Enzyme cleaving the 5′-terminal methylated blocked structure of messenger RNA. FEBS Lett. 1976;65:254–257. doi: 10.1016/0014-5793(76)80492-9. [DOI] [PubMed] [Google Scholar]
- Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones. 2004;9:109–121. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Ueland PM. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacol Rev. 1982;34:223–253. [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Nakao M, Shichijo S, Sasatomi T, Takasu H, Matsumoto H, Mori K, Hayashi A, Yamana H, Shirouzu K, Itoh K. Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res. 1999;59:4056–4063. [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol Cell Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J Biol Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang Q, Zhang H, Liu Q, Du X, Richter M, Greene MI. UXT is a novel centrosomal protein essential for cell viability. Mol Biol Cell. 2005;16:5857–5865. doi: 10.1091/mbc.E05-08-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yik JH. The yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zhu W, Hanes SD. Identification of drosophila bicoid-interacting proteins using a custom two-hybrid selection. Gene. 2000;245:329–339. doi: 10.1016/s0378-1119(00)00048-2. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.