Abstract
Major geologic and climatic changes during the Quaternary exerted a major role in shaping past and contemporary distribution of genetic diversity and structure of aquatic organisms in southern South America. In fact, the northern glacial limit along the Pacific coast, an area of major environmental changes in terms of topography, currents, and water salinity, represents a major biogeographic transition for marine and freshwater species. We used mitochondrial DNA sequences (D-loop) to investigate the consequences of Quaternary glacial cycles over the pattern of genetic diversity and structure of G. maculatus (Pisces: Galaxiidae) along two biogeographical provinces in the Chilean coast. Extreme levels of genetic diversity and strong phylogeographic structure characterize the species suggesting a low amount of influence of the last glacial cycle over its demography. However, we recognized contrasting patterns of genetic diversity and structure between main biogeographical areas here analyzed. Along the Intermediate Area (38°–41° S) each estuarine population constitutes a different unit. In contrast, Magellanic populations (43°–53° S) exhibited low levels of genetic differentiation. Contrasting patterns of genetic diversity and structure recorded in the species between the analyzed biogeographic areas are consistent with the marked differences in abiotic factors (i.e., different coastal configurations, Quaternary glacial histories, and oceanographic regimes) and to inherent characteristics of the species (i.e., salt-tolerance, physiology, and reproductive behavior).
Introduction
Quaternary glaciations, and especially the Last Glacial Maximum (LGM) 25–18 Ka, caused major shifts in the distribution of the biota, particularly at higher latitudes [1–3]. Until the last two decades, phylogeographic studies in higher latitude taxa were highly biased to Northern Hemisphere taxa and most of them were focused on terrestrial organisms [4,5]. However, during the last decade, more and more genetic data has been accumulated showing the role of the Quaternary glaciations in the southern tip of South America. Such studies include population-based and phylogeographic analyses in different Patagonian taxa from marine invertebrates [6–8] to mammals [9–11]. The glacial history of South America is generally well understood; ice sheet advances and retreats during the Quaternary generated major shifts in sea level, climate and landscape [12–15]. During the LGM the Patagonian Ice Sheet (∼ 480,000 km2) expanding from 35°S to 56°S covered most of the Pacific Magellanic fjords and channels with an ice-volume of more than 500,000 km3 [12,16–17]. Hence, the geomorphology of Patagonia during the last 2 million years has not always remained stable and major physical changes during Quaternary glacial periods markedly affected current patterns of genetic diversity and structure in aquatic organisms, particularly in freshwater and marine near-shore ecosystems [18,19]. In fact, radical glacial landscape changes resulted in periodic extinction of fauna associated to these ecosystems allowing the colonization of vacant niches, as well as creating opportunities for geographical isolation and speciation [7,20,21].
According to Zemlak [22], postglacial patterns of dispersal in southern South American fishes includes the presence of several independent Quaternary glacial refugia on the east side of the Andes, along the Patagonian Steppe [23–25]. Along the west slope of the Andes refugia could have been located both within [9,25–27] and outside of the glacier limits [24–26,28]. Absence of genetic structure and strong signal of recent demographic growth support the hypothesis of rapid postglacial expansion in shallow Magellanic marine benthic invertebrates [6,8]. Finally, there is evidence that some groups of marine organisms re-colonized the southern tip of South America from geographically distant ice-free regions [29,30].
The Family Galaxiidae includes ~ 50 species of Gondwanan-distributed fishes commonly found in cool-temperate regions in the Southern Hemisphere. All species inhabit freshwater ecosystems, although, some of them present a migratory life stage with a salt-tolerant larval phase known as whitebait. These diadromous species exhibit considerably greater dispersal capabilities than those that are non-migratory [19]. Among diadromous species, Galaxias maculatus is one of the most widely distributed freshwater fish in the planet and exhibits a Gondwanic distribution. Populations of G. maculatus are currently found in freshwater and marine ecosystems of southern Australia, Tasmania, New Zealand and surrounding islands, and in the temperate latitudes of South America and the Falkland/Malvinas Islands [31]. The transoceanic distribution of G. maculatus has been explained by vicariance [32] and dispersal [33] hypotheses, but molecular evidence suggests that populations dispersed from Australia to other locations following the West Wind Drift [34–36]. Along the Chilean coast, G. maculatus includes landlocked and migratory populations in freshwater, estuarine and marine ecosystems [37] in two of the three Chilean biogeographic areas, namely the Intermediate Area (30°S–42°S) and the Magellanic Province (42°S–56°S) [38]. Phylogeographic analyses in G. maculatus detected a small influence of the last glacial cycles over the genetic diversity of the species in South America. In fact, female populations size in G. maculatus seems to have remained relatively constant until ~ 500 Ka when populations sizes increased exponentially reaching contemporary levels [22]. More recently, Zemlak et al. [39] recognized that G. maculatus experienced a severe genetic bottleneck between 1.1 Ma and 0.6 Ma, a period when the Patagonian Ice Sheet reached its maximum northward expansion [13,15]. Following this, the species experienced a strong population recovery during the late Quaternary (400 Ka) probably associated to long and warm interglacial periods [39].
The Chilean coast represents an interesting area to evaluate the relative effect of glaciological history, oceanography, and habitat discontinuities for aquatic organisms. The presence of extensive ice sheets during the glacial periods of the Quaternary likely eradicated recurrently most of the suitable freshwater and brackish marine habitats along the Magellanic Province while the Intermediate Area was considerably less impacted. In this context, Galaxias maculatus constitutes a suitable model to infer how historical and contemporary climatic processes have shaped the distribution of the genetic diversity in the species. Here we evaluated the consequences of the Quaternary glacial cycles over the pattern of genetic diversity and structure in a diadromous species with special emphasis in coastal and estuarine populations along two biogeographic marine areas that were differentially affected by ice advances and retreats.
Materials and Methods
Ethics Statement
This work was conducted using puye (Galaxias maculatus) as model study, a common freshwater and brackish fish species along the Chilean coast. The species is not protected and is considered as vulnerable in Central Chile and as out of danger in the Magellanic Province. Permission to undertake field works and collect specimens was issued by the Chilean Fisheries Service Director (Pablo Galilea Carrillo), under the technical memorandum (427/2011). The Instituto de Ecología y Biodiversidad (IEB/12-2011) and Chilean Fisheries Service (SERNAPESCA 427/2011) ethic committees approved sampling protocols and experiments. For this, we compiled with local legislation and the Convention on Biological Diversity.
Sample collection, DNA extraction, PCR amplification and sequencing
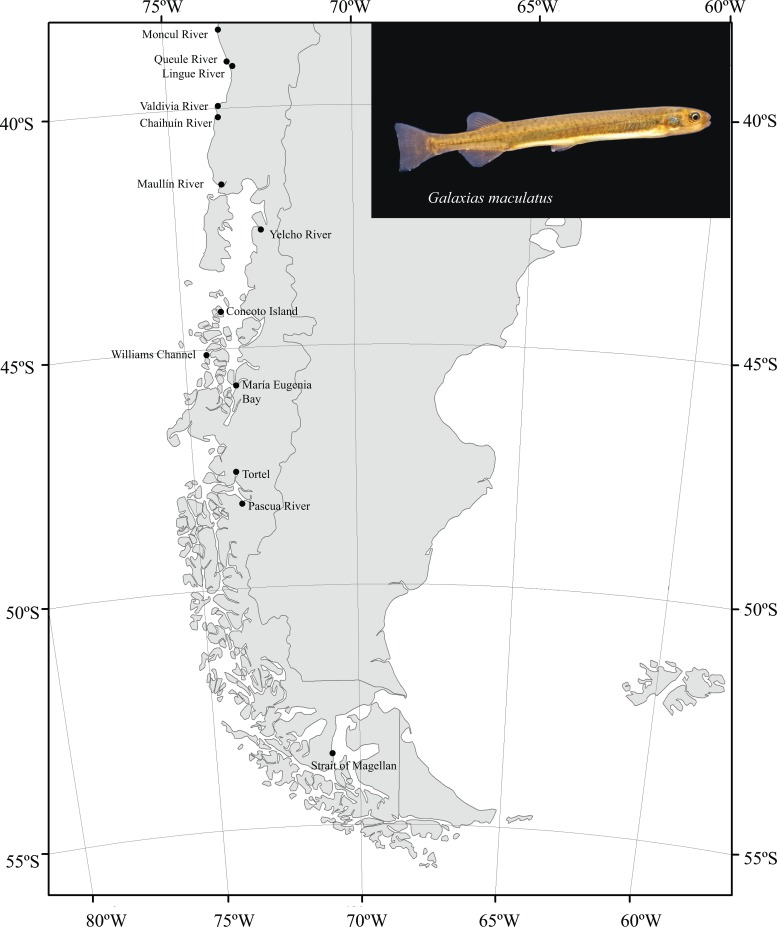
Individuals were collected between 2007–2014 in 13 estuarine localities along two main biogeographic areas in the Chilean Coast. Firstly, we included six localities from the Intermediate Area, between 38°S and 42°S, outside the direct influence of the LGM Patagonian Ice Sheet: Intermediate Area localities were: 1) Moncul River (38°42’S; 73°24’W); 2) Queule River (39°17’°S; 73°10’W); 3) Lingue River (39°26’S; 73°09’W); 4) Valdivia River (39°53’S; 73°24’W); 5) Chaihuín River (40°01’S; 73°27’W), and 6) Maullín River (41°35’S; 73°38’W). Secondly, we included seven localities from the Magellanic Province, between 43° S and 53° S, within the influence of the LGM Patagonian Ice Sheet. Magellanic localities were: 1) Yelcho River (43°03’S; 72°33’W), 2) Concoto Island (44°06’S; 73°43’W); 3) Williams Channel, Rivero Island (45°25’S; 74°21’W); 4) María Eugenia Bay (45°55’S; 73°31’W); 5) Tortel, Calén Fjord (47°45’S; 73°32’W); 6) Pascua River, Calén Fjord (48°15’S; 73°18’W); and 7) Strait of Magellan (53°14’S; 70°59’W; Fig 1).
Fig 1. Sampling localities of Galaxias maculatus along the Chilean coast.
Whole specimens were fixed in ethanol (95%) and DNA was prepared from muscle tissue using a salting-out method [40]. A partial fragment of the mitochondrial Control Region (D-loop) was amplified using specific primers GAL-F 5’–TAA CTC TCA TTA ACT AAA G– 3’ and GAL-R 5’–TGA TAG TAA AGT CAG CAA GCC– 3’) designed from the complete mitochondrial genome of the species (ACN: AP004104) [41]. PCR amplifications were performed in a 25 μl reaction containing 2.5 μl 10X Buffer (50 mM KCl, 10 mM Tris-HCl, pH 8.0), 1.0 μl of 50 mM MgCl2, 200 mM dNTPs, 0.5 μl of each primer (10 pg/μl), 1 U Taq (Invitrogen), 17.5 μl of double-distilled water and 5 ng of DNA. Thermal cycling parameters included an initial denaturation step at 94°C for 5 min, followed by 35 cycles at 94°C for 90 sec, 60.7°C for 90 sec, and 72°C for 90 sec, and a final 10 min extension at 72°C. PCR amplification products were purified using QIAquick Gel Extraction Kit (QIAGEN) and sequenced in both directions with an Automatic Sequencer ABI3730 x 1 at Macrogen Inc. (Seoul, Korea).
Genetic diversity and population structure in Galaxias maculatus
D-Loop sequences were edited using Proseq v. 3.5 [42] and aligned with ClustalW [43]. New D-loop haplotypes recorded in the species were deposited in GenBank under the Accession Numbers KP298433—KP298673 and KP336557—KP336665. We performed a DNA saturation analysis following Xia et al. [44] to evaluate how saturation of transitions is accumulated in relation to nucleotide divergence in the whole data set. Levels of genetic polymorphism were estimated using standard diversity indices including: the number of haplotypes (k), the number of segregating sites (S), haplotypic diversity (H), the average number of pairwise differences (П), and nucleotide diversity (π) for each locality, for each main area, and for the whole D-loop data set using DnaSP v.5.00.07 [45]. We performed neutrality statistical tests (Tajima’s D and Fu’s FS) for each locality, area, and for the whole data set to estimate whether D-loop sequences in G. maculatus deviate from expectations under a neutral model. Considering the high levels of genetic diversity previously estimated in the species [22,39,41] we determined the levels of genetic differentiation between the analyzed localities using mean pairwise differences (NST) following Pons and Petit [46] in Arlequin v. 3.5 [47]. The statistical significance of genetic differences between localities was estimated using permutation tests using 20,000 iterations of haplotype identities. Similarly, we estimated the levels of genetic differentiation of subpopulations using the nearest-neighbor statistic (Snn) that measure how often nearest neighbors (in sequence space) of sequences are from the same locality in geographic space [48]. The statistical significance of Snn analyses was determined through a permutation test using 10,000 iterations. We inferred the spatial genetic structure in the species by estimating the number and composition of groups that were most differentiated based on sequence data with an Analysis of Molecular Variance (AMOVA) following Excoffier et al. [47] in Arlequin. AMOVA is a popular method that uses multiple spatial scales in statistical methods to characterize spatial genetic structure by partitioning it into: within populations, among populations within groups and among groups. Finally, we performed a test for isolation by distance using a Mantel test with 1,000 permutations in Arlequin to determine the correlation between Slatkin’s linearized localities genetic differentiation [49] and the linear geographic distance (km) between populations measured using FOSSIL [50].
Demographic inference in G. maculatus
We constructed genealogical relationships in G. maculatus using Maximum Parsimony Networks in Hapview (http://www.cibiv.at). To estimate the pattern of demographic history in the species, we compared the distribution of pairwise differences between haplotypes (mismatch distribution) for each locality, main area, and for the whole data set, to the expected distribution under the sudden expansion growth model of Rogers and Harpending [51]. This analysis is a popular method since the amount of nucleotide differences between haplotypes depends on the length of time since they diverged. Alternatively, we reconstructed past population dynamics through time using a Bayesian Skyline Plot method implemented in BEAST v. 1.7 [52]. This method is fundamentally based on coalescent theory, which quantifies the relationship between a genealogy of the sequences and the demographic history of a population. In contrast to the previous method, it does not assume any demographic model. For comparison purposes, three substitution models (strict clock, uncorrelated lognormal and uncorrelated relaxed clock) were computed for the main areas here analyzed and compared statistically using a Bayes factor test [53] with TRACER v. 1.5 (http://beast.bio.ed.ac.uk/Tracer). The uncorrelaed lognormal model was the best fit for D-loop data in G. maculatus. We conducted three independent Bayesian MCMC runs using the GTR + I + G model, previously estimated using MrModeltest v. 2.3 (http://www.abc-se/~nylander), and a specific population level mutational rate estimated for G. maculatus by Salinas [54]. Two independent runs per area (Intermediate Area and Magellanic Province) were made for 50 x 106 generations (sampled every 1000 step), discarding a 10% of the trees as burn-in. The convergence of runs was confirmed with Tracer ensuring a minimum of 1000 effective sampling for each statistics (ESS). The results of the multiple runs were combined using LogCombiner [52]. The median and corresponding credibility intervals of the Bayesian skylines plots were depicted with Tracer.
Gene flow and connectivity
We compared different models of gene flow between the main areas of interest (Intermediate Area and Magellanic) with the software MIGRATE v.3.5 [55], to test for different scenarios. For this, we defined four candidate models constraining the presence of directionality of gene flow between the main areas. The first model allowed bidirectional gene flow between the Intermediate Area and the Magellanic Province (full island model). Models 2 and 3 were defined by asymmetric patterns of gene flow from the Intermediate Area to the Magellanic Province, and from the Magellanic Province to the Intermediate Area, respectively. Finally, model 4 assumed both main areas as the same panmictic population. All the analyses were run using a GTR + I + G substitution model and transition-transversion ratio of 2.9818 as previously estimated by jModeltest v2 [56]. The specific substitution rate for the selected marker in the species was set to constant, as suggested by the manual. Analysis consisted of one long chain with 500.000 recorded parameter steps, a sampling interval of 100 and a burn-in of 100.000 (10%), running multiple replicates (10 independent chains). A heated scheme (1.00, 1.50, 3.00, and 1000000.0) was used to calculate the marginal likelihoods for model comparisons. Comparisons among the different models were done using a Bayes Factors calculated by substracting the highest value of the log marginal likelihoods (lmL; Bezier curve approximation) from the lmL values of each models. Finally, the associated probability of each model in relation to others was estimated following Kaas and Rafery [57].
We estimated different demographic indices and evaluated two asymmetric isolation-with-migration models between main areas of interest (Intermediate Area and the Magellanic Province) using IMa2 software [58,59]: i) from the Intermediate Area to Patagonia (m N > S) and from Patagonia to the Intermediate Area (m S > N). We carried out separately several preliminary runs in the M mode (Markov Chain Monte Carlo; MCMC mode) of the software to set the best set of priors that ensure mixing and convergence. Uniform priors were used to estimate effective population size (Θ 1, Θ 2, and ancestral Θ a, Θ = 3000) and splitting time (t = 300), whereas an exponential prior (mean = 1) for gene flow (m) was adopted. We performed 80 x 106 MCMC steps sampling every 100 generations, with a burn-in of 8 x 106. D-loop sequences were assumed to mutate under the HKY mutation model following the author guidelines [59]. Once convergence was achieved under the M-mode, we used the same simulated genealogies under the L-Mode (Load Tree mode) to estimate the log maximum-likelihood and credibility intervals (95% under HPD) estimates for migration parameters using a Likelihood Ratio Test. Under the L-Mode we compared our data with a null model of no migration [58,59]. Finally, estimated parameters were re-scaled into years (t/μ) and effective rate of migration ((Θ x/ mx)/2) (rate at which genes come into population, per generation) using an overall substitution rate per year (m = 1.1 x 10−4) estimated on previous studies [54].
Results
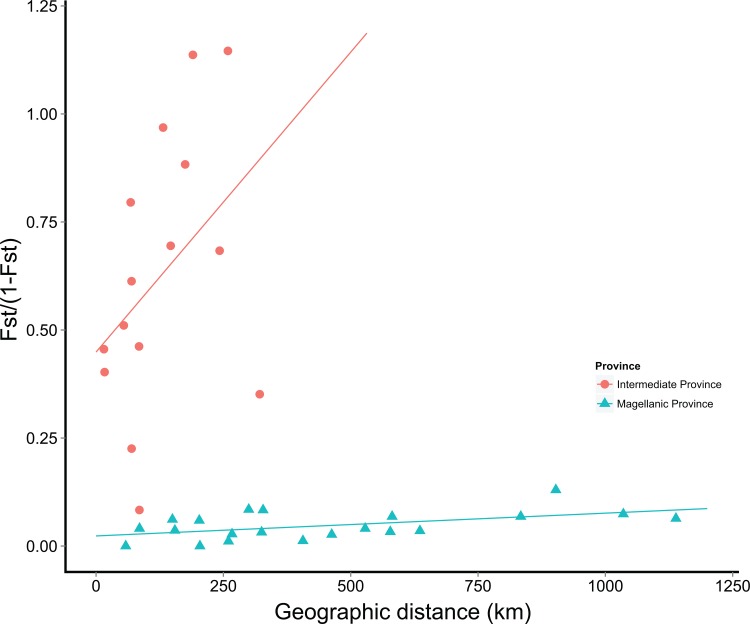
The whole D-loop data set in G. maculatus included 353 individuals and consisted in 926 nucleotide positions. As expected working with a non-coding region, several insertion and deletions were detected and were not considered in further analyses. Sequences were A-T rich (57.1%) compared to G-C content (42.9%). Levels of genetic diversity in the species were extremely high with 230 polymorphic characters (24.83%); and 156 of them (67.82%) were parsimoniously informative. In spite of the high level of genetic polymorphosm recorded in the species, D-loop sequences were not saturated. Haplotype diversity (H) ranged between 0.942 (Lingue River) and 1.000 (Moncul River, Queule River, Maullín River, Yelcho River, and María Eugenia Bay; Table 1). The average number of nucleotide differences (П) and nucleotide diversity (π) were very high in most of the localities and ranged between 30.55/0.033 (Chaihuín River) and 10.74/0.011 (Williams Channel). Levels of genetic diversity of G. maculatus were higher in Intermediate Area localities than in Magellanic ones, particular for П and π (Table 1). In fact, permutation test analyses (100,000 iterations) detected highly significant values for П (P = 0.00066) and π (P = 0.00063) in Intermediate Area populations compared to those from the Magellanic Province. Similarly, pairwise NST comparisons detected a high degree of genetic structure between Intermediate Area and Magellanic Province populations (Table 2). High levels of genetic structure, along a narrow geographical range (< 250 km), were detected among Intermediate Area localities. On contrast, low levels of genetic differentiation were recorded along the Magellanic Province (Table 2). Similarly, global Snn analyses in the species (Snn = 0.45) indicate moderate levels of phylogeographic signal in the whole data set. Nevertheless, Snn statistic recognized a higher degree of phylogeographic signal along the Intermediate Area localities (Snn = 0.63) than in Magellanic Province ones (Snn = 0.43). General pattern of genetic structure registered in G. maculatus was supported by AMOVA analyses detecting 7 groups with a maximal difference accounting for 37.98% of the total variance, and only 2.01% was due to within-group variation among localities (Table 3). AMOVA analysis recognized each one the Intermediate locality as a separate population while all the localities along the Magellanic Province were recognized as a single unit. A moderate and marginally correlation between genetic and geographic distances was detected among Intermediate Area populations (r = 0.398; P = 0.087) and the Magellanic Province ones (r = 0.45; P = 0.08) (Fig 2).
Table 1. Diversity índices and neutrality tests in Galaxias maculatus along it distribution in the Chilean coast.
| Locality | n | K | H | S | П | П | π | Tajima´s D | Fu´s FS | M. D. |
|---|---|---|---|---|---|---|---|---|---|---|
| Moncul River | 24 | 24 | 1.000 | 85 | 20.20 | 0.022 | -0.81 | -10.16*** | M | |
| Queule River | 23 | 23 | 1.000 | 85 | 22.93 | 0.025 | -0.64 | -8.55*** | M | |
| Lingue River | 24 | 18 | 0.942 | 78 | 23.23 | 0.025 | -0.010 | -3.53* | M | |
| Valdivia River | 28 | 24 | 0.989 | 68 | 24.79 | 0.027 | 1.03 | -3.76* | M | |
| Chaihuín River | 41 | 31 | 0.974 | 106 | 24.94 | 0.027 | -0.56 | -3.90* | M | |
| Maullín River | 27 | 27 | 1.000 | 112 | 30.55 | 0.033 | -0.28 | -9.12* | M | |
| Intermediate Area | 167 | 145 | 0.993 | 180 | 32.61 | 0.035 | -0.45 | -33.37 *** | M | |
| Yelcho River | 22 | 22 | 1.000 | 72 | 15.84 | 0.017 | -1.19 | -10.52*** | M | |
| Concoto Island | 26 | 24 | 0.994 | 54 | 10.88 | 0.011 | -1.09 | -12.27*** | M | |
| Williams Channel | 26 | 20 | 0.978 | 42 | 10.74 | 0.011 | -0.33 | -5.24*** | M | |
| María Eugenia Bay | 23 | 23 | 1.000 | 63 | 15.33 | 0.016 | -0.66 | -11.66*** | M | |
| Tortel | 27 | 26 | 0.997 | 71 | 15.88 | 0.017 | -1.01 | -11.95*** | M | |
| Pascua River | 45 | 43 | 0.998 | 75 | 13.24 | 0.014 | -1.28 | -32.40*** | M | |
| Strait of Magellan | 17 | 16 | 0.993 | 51 | 15.52 | 0.016 | -0.33 | -4.32** | M | |
| Magellanic Province | 186 | 170 | 0.998 | 142 | 14.11 | 0.015 | -1.73 | -261.20 *** | M | |
| Total | 353 | 310 | 0.998 | 218 | 25.03 | 0.027 | -1.27 | -33.94 *** | M |
n = number of analyzed individuals; k = number of haplotypes; S = polymorphic sites; H = haplotype diversity; П = average number of pairwise differences; π = nucleotide diversity. M.D. Mismatch Distribution, M = Multimodal.
*p<0.05
**p<0.01
*** p<0.001.
Table 2. Mean general pairwise values of differentiation (ФST) between Galaxias maculatus populations.
| Locality | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | *** | ||||||||||||
| 2 | 0.44 | *** | |||||||||||
| 3 | 0.31 | 0.28 | *** | ||||||||||
| 4 | 0.49 | 0.38 | 0.33 | *** | |||||||||
| 5 | 0.41 | 0.12 | 0.18 | 0.31 | *** | ||||||||
| 6 | 0.26 | 0.53 | 0.40 | 0.53 | 0.46 | *** | |||||||
| 7 | 0.60 | 0.11 | 0.42 | 0.46 | 0.12 | 0.66 | *** | ||||||
| 8 | 0.64 | 0.11 | 0.46 | 0.50 | 0.12 | 0.71 | 0.05 | *** | |||||
| 9 | 0.64 | 0.14 | 0.46 | 0.50 | 0.11 | 0.70 | 0.07 | 0.03 | *** | ||||
| 10 | 0.60 | 0.05 | 0.41 | 0.45 | 0.07 | 0.67 | 0.07 | 0.05 | 0.03 | *** | |||
| 11 | 0.60 | 0.06 | 0.42 | 0.45 | 0.08 | 0.66 | 0.03 | 0.01 | 0.02 | 0.00 | *** | ||
| 12 | 0.64 | 0.09 | 0.47 | 0.51 | 0.12 | 0.68 | 0.06 | 0.02 | 0.03 | 0.01 | 0.00 | *** | |
| 13 | 0.59 | 0.09 | 0.39 | 0.45 | 0.09 | 0.66 | 0.06 | 0.06 | 0.11 | 0.06 | 0.03 | 0.03 | *** |
Where: 1) Moncul River; 2) Queule River; 3) Lingue River; 4) Valdivia River; 5) Chaihuín River; 6) Maullín River; 7) Yelcho River; 8) Concoto Island; 9) Williams Channel; 10) María Eugenia Bay; 11) Tortel; 12) Pascua River; 13) Strait of Magellan. Statistically significant comparisons (after Bonferroni correction) and 100,000 iterations are marked in bold.
Table 3. Analysis of Molecular Variance (AMOVA) depicting the percentage of variation explained among groups (Moncul River, Queule River, Lingue River, Valdivia River, Chaihuín River, Maullín River, and Magellanic Province populations), among populations within groups, and within populations.
Where FSC represents differntiation within populations among groups while FCT represents differentiation among groups (*** p<0.001, ** p<0.01).
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation | |
|---|---|---|---|---|---|
| Among groups | 6 | 1571.864 | 6.14210 Va | 36.47 | |
| Among populations within groups | 6 | 88.683 | 0.18003 Vb | 1.10 | |
| Within populations | 340 | 3424.433 | 10.07186 Vc | 61.44 | |
| Total | 352 | 5084.980 | 16.39400 | ||
| Fixation Index | |||||
| FSC | 0.01756*** | ||||
| FCT | 0.37466*** | ||||
Fig 2. Relationship between linearized genetic differentiation (NST) and geographic distances (Km) along the Intermediate Area and the Magellanic Province.
Red circles and blue triangles represent pairwise values in the Intermediate Area and the Magellanic Province, respectively.
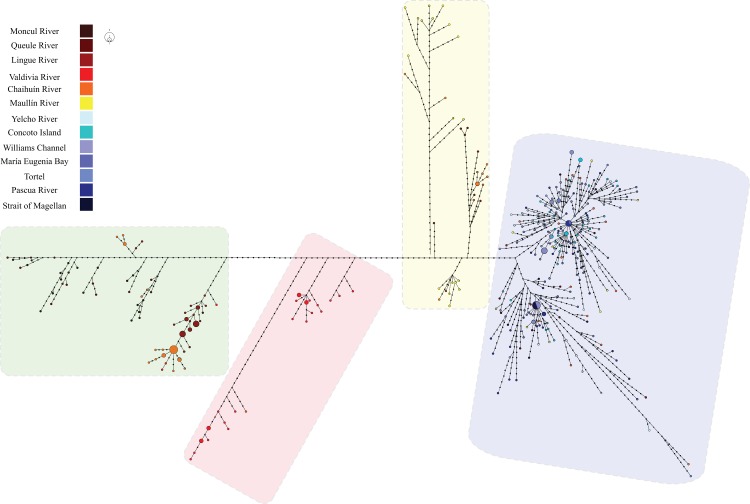
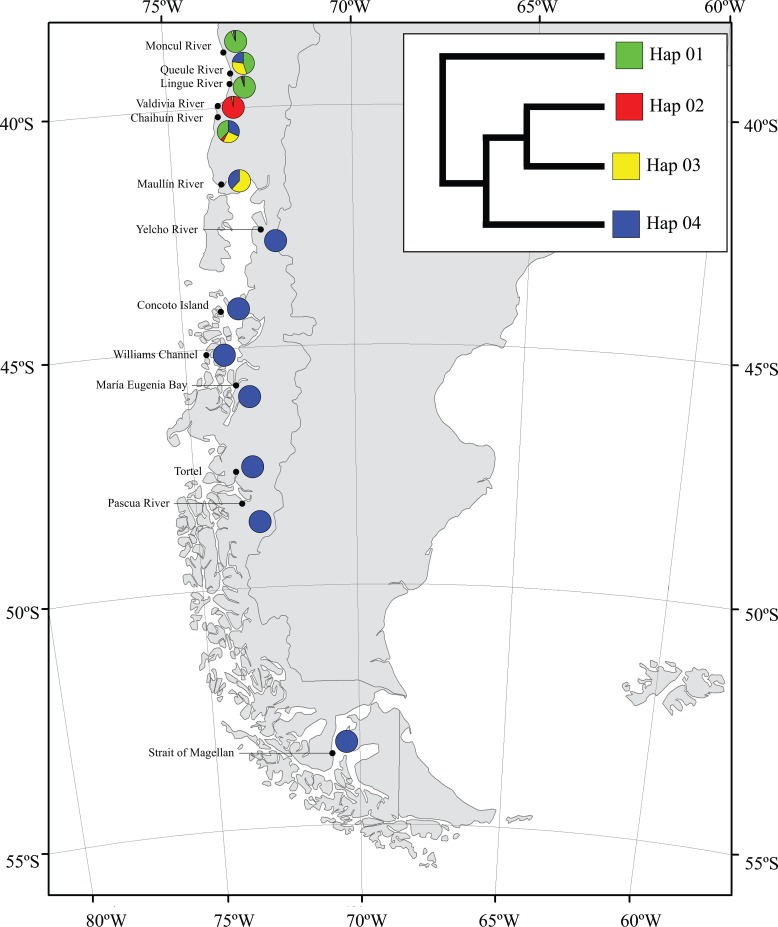
Maximum Parsimony haplotype network in G. maculatus recovered a total of 321 haplotypes and a very expanded genealogy (Fig 3). A total of 298 haplotypes (92.83%) were unique and only 23 haplotypes were present in more than two individuals. As previously recognized through mean standard diversity indices, Intermediate Area localities showed a more expanded genealogies than Magellanic Province ones. Four main groups of haplotypes (haplogroups) were detected in G. maculatus. Two of them (Hap 01 and Hap 03; Fig 4) include individuals collected at Intermediate Area localities from Moncul to Maullín. Haplogroup 02 was endemic at Valdivia River and was also present in a single specimen from Lingue River. The fourth haplogroup (Hap 04) includes individuals from Magellanic Province localities, from Yelcho River to the Strait of Magellan. Interestingly, all the diversity recorded in the Magellanic province fell within the Patagonian haplogroup 04 recorded by Zemlak et al [22]. Some individuals from Intermediate Area localities, particularly from Maullín, Chaihuín and Queule rivers, were also found within the southern haplogroup (Fig 4).
Fig 3. Maximum parsimony haplotype network including 353 individuals of Galaxias maculatus mtDNA D-loop sequences.
Each haplotype is represented by a colored circle the locality where was collected. The size of the circles are proportional to its frequency in the whole sampling effort. Colored areas mark the different recognized haplogroups where Green (Hap 01), Red (Hap 02), Yellow (Hap 03), and blue (Hap 04).
Fig 4. Distribution of the recognized haplogroups in G. maculatus along the sampling localities.
Tajima’s D and Fu’s FS neutrality tests showed similar trends at both sectors and for the whole D-loop data set in G. maculatus. Tajima’s D test was negative but not statistically significant for each locality, area, and for the whole D-loop data set. In contrast, Fu’s FS test was negative and statistically significant for all localities, area, and for the whole data set (Table 1). Furthermore, analyses of pairwise differences in G. maculatus recovered multimodal distribution of values with multiple peaks in each of the analyzed localities, area, and for the whole data set (Table 1).
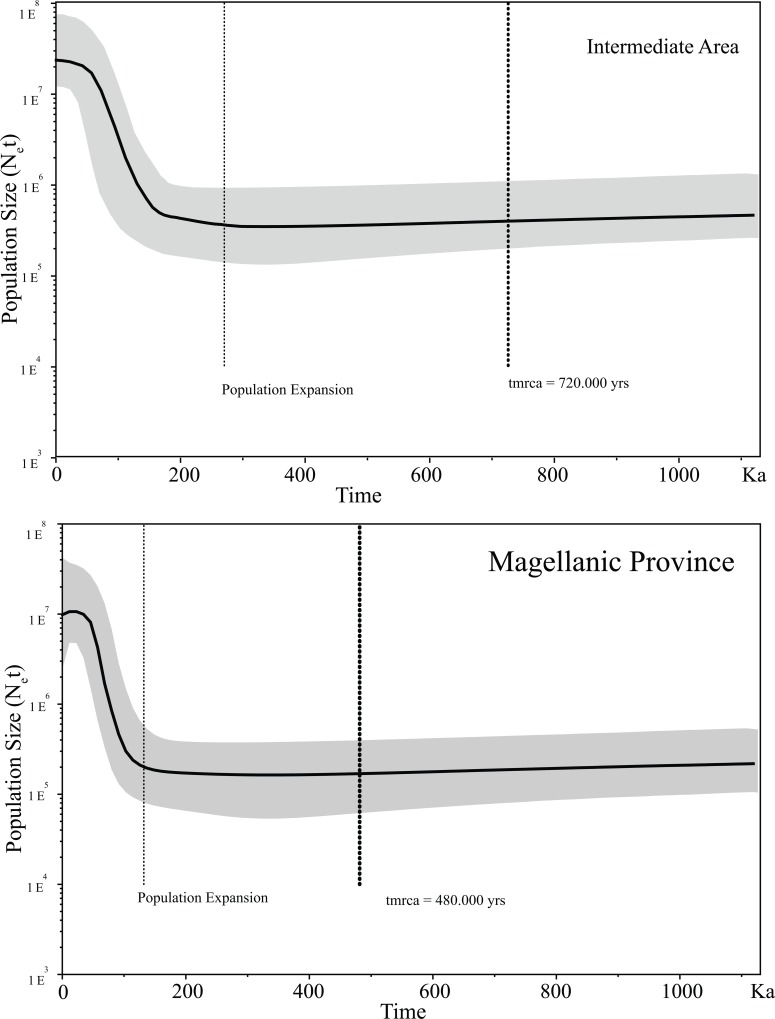
Bayesian Skyline plot analyses recognized differences in the times of the most recent common ancestor (tmrca) and population expansions between the Intermediate Area and the Magellanic Province. Based on these analyses, the trmca of the Intermediate Area occurred ∼ 72 Ka while the tmcra for the Magellanic Province occurred ∼ 48 Ka (Fig 5). Similarly, the onset of the population expansion in the Intermediate Area occurred ∼ 32 Ka against the ∼ 18 Ka estimated for the Magellanic Province.
Fig 5. Historical demographic trends of the effective population sizes (Ne) constructed using a Bayesian skyline plot approach based on D-loop haplotypes of Galaxias maculatus along its distribution in the Chilean coast (Intermediate Area and Magellanic Province).
The y-axis is the product of effective population size (Ne) and generation length in a log scale while the x-axis is the time 103 before present. The median estimate (solid line) and 95% highest probabilities density (HPD; grey areas) are shown. The thick dashed line represent the time of the most common ancestor (trmca) and the thin dashed line represents time for population expansion.
Gene flow analyses using MIGRATE and different migration models detected evidence of asymmetrical gene flow from the Magellanic Province to the Intermediate Area. Among the tested models the one from the Magellanic Province to the Intermediate Area received the highest probability (Table 4). Similarly, coalescent approach of isolation-with-migration implemented in IMA2 detected overall low levels of gene flow between the Intermediante Area and Magellanic Areas localities. However, the model predicted a most likely pattern of asymmetric migration with greater migration from the Magellanic Province to the Intermediate Area (Table 5).
Table 4. Log marginal likelihoods (lmL) and log Bayes factor (LBF) comparisons for different migration models between the Intermediate Area and the Magellanic Province in G. maculatus.
Where IA = Intermediate Area; MP = Magellanic Province.
| Model | Bezier lmL | LBF | Model prob | Model rank |
|---|---|---|---|---|
| 1. full migration | -8199.540 | -55.779 | <0.001 | 3 |
| 2. IA to MP | -8180.607 | -17.920 | <0.001 | 2 |
| 3. MP to IA | -8171.654 | 0 | 1 | 1 |
| 4. panmixia | -8397.543 | -451.779 | <0.001 | 4 |
Table 5. Estimation of isolation-with-migration model implemented in IMa2 including; migration rates (m) in each direction (mN->S, mS->N), estimated splitting time (τ) between provinces (in years ago), and effective population size of both provinces (ΘN, ΘS) and ancestral population size (ΘA) converted in demographic units (individuals).
| m N->S | m S->N | τ (YA) | ΘN | ΘS | ΘA | |
|---|---|---|---|---|---|---|
| Hight Point | 0.0005ns | 0.034ns | 28,347 | 1,636,744 | 637,824 | 2,992,261 |
| 95% HPD | 0.0005–0.1285 | 0.0025–0.1285 | 22,948–36,447 | 995,545–2,987,760 | 43,534–907,802 | 2,048,461–4,513,139 |
For each parameter, the most probable value (high point) and the 95% highest posterior density (95% HPD) of the marginal posterior probabilities are shown. Ns = non significant for the LTR for migration rates estimation. The convertion of scale in the τ and Θ parameter was carried on using a specific substitution rate of 12% per million year estimated by Salinas [52].
Discussion
Molecular-based genetic studies have become pivotal to further understand and unravel how Quaternary glacial cycles affected the distribution and demography of populations, species, and communities [3,4,60–65]. Evidence of postglacial recolonization of the Magellanic Province has been recorded in freshwater [22,25,66] and marine fishes [67,68], lizards [28], amphibians [27], mammals [9,10,69], marine invertebrates [6–8], and plants [70,71]. Traditional genetic models of glacial refugia and recolonization routes have been proposed to describe the response of populations, species, and communities to climatic changes [62–65]. The Expansion-Contraction Model propose that species would have become restricted to glacial refugia at lower latitude outside the influence of glacial ice advances during cooling periods. After this, they expanded their distribution towards previously glaciated areas at higher latitudes following the deglaciation process [62–64]. Therefore, unglaciated and refugial areas are expected to harbor higher levels of genetic diversity than peripheral, geologically altered, or newly founded regions. On contrast, formerly glaciated areas should exhibit evidence of recent postglacial demographic expansions. According to Maggs et al. [63], formerly glaciated areas should also exhibit small number of haplotypes dominating disproportionately large areas as a consequence of postglacial recolonization processes. Simultaneously, recolonized areas should exhibit low divergence among haplotypes and lower levels of genetic differentiation than refugial ones.
Main patterns of genetic diversity and structure in G. maculatus
Even if coastal and Andean South American samples of G. maculatus have been previously included in phylogeographic and demographic studies [22,39], here we present the first population-level study in the species along two biogeographical provinces that were differentially affected during the glacial cycles of the Quaternary. In contrast to previous studies that incorporated mainly river and lake specimens, our study focuses on estuarine and coastal populations. As previously determined in the species [22,39,41], G. maculatus harbors extremely high levels of genetic diversity along its distribution in the Chilean Pacific coast. In fact, our results further corroborate previous molecular studies in the species [22,41,72], pointing towards an extreme within-species genetic diversity among coastal and estuarine populations. In this study, out of 353 individuals we recognized 321 different haplotypes. Similarly, Zemlak [22] recorded a total of 273 different haplotypes, out of 299 individuals, while Waters et al. [72] recognized 139 different haplotypes out of 144 individuals from New Zealand. In spite of the high levels of genetic polymorphism recorded in this study, we also detected marked and significant differences in the average number of nucleotide differences and nucleotide diversity between main biogeographical areas here included. Therefore, higher levels of genetic diversity along formerly unglaciated areas (Intermediate Area) than in those located within the limits of the Patagonian Ice Sheet (Magellanic Province) detected in the species, supports the first prediction of the Expansion-Contraction model. Main differences recorded in terms of genetic diversity between the main biogeographical areas here included were also supported by population dynamics reconstructions showing a more recent population expansion in the Magellanic Province than in the Intermediate Area. Therefore, our results support the hypothesis that the Intermediate Area represents an older region and less affected by glacial advances and retreats than the Magellanic Province. Together with main patterns of genetic diversity recorded in the species we also found contrasting patterns of genetic structure between the main biogeographic areas here analyzed. Each one of the analyzed rivers along the Intermediate Area, expanding less than 250 km, represented a different genetic unit that generally coincided with the defined haplogroups of Zemlak et al. [22] at the same localities. Magellanic populations of G. maculatus, expanding for more than 1000 km, showed low levels and even absence of genetic differentiation. Again, major differences in terms of the genetic structure between formerly glaciated and unglaciated areas are adjusted to the predictions of the Expansion-Contraction model. Nevertheless, high levels of genetic diversity, measured as П and π, among Magellanic localities and the absence of dominant broadly distributed haplotypes in this area do not fit with the expectations of recent postglacial expansion from refugial areas at lower latitudes. Moreover, the presence of a markedly differentiated and highly diverse haplogroup (Hap04) in this Province does not support the idea of a recent postglacial recolonization. Such results contrast to those observed in other South American freshwater [24,25,73–76] and marine fishes [67,68]. In fact, Alò et al. [75] recorded low levels of genetic diversity and a clear genetic continuity between Intermediate Area and Magellanic province populations of Aplochiton (A. zebra, A. taenius and A. marinus). Most of these studies have demonstrated the important role of the last glacial cycle over the demography of Magellanic fishes with the presence of typical star-like haplotype genealogies as a consequence of recent colonization processes associated to strong founder effects. In this context, population genetic patterns in Patagonian terrestrial plants and vertebrates indicate that different processes and directional range shifts have generated a mosaic of phylogeographical patterns that are far more complex than a simple north to south described one [71]. In the particular case of G. maculatus, Zemlak et al. [22,39] proposed that the main patterns of genetic diversity and structure in the species suggest a low amount of influence of the last glacial cycle. In fact, levels of genetic diversity along the Magellanic Province likely reflect the existence of marine in situ or periglacial refugia during cooling periods of the Quaternary.
Patterns of genetic structure in G. maculatus along the analyzed areas
Low genetic differentiation along the Magellanic Province, a complex geographic and oceanographic area, may be a response of particular characteristics of the species. The southern fjords and channels ecosystems along the Pacific coast of Patagonia are considered as one of the largest estuarine ecosystem in the planet [77,78]. Subantarctic waters masses are mixed with freshwater ones from abundant precipitation, river flow, and glacial meltwater [79]. A low salinity surface layer generates a highly stable water column within the fjords whereas a well-mixed water column occurs in the gulfs and open channels. In this context, epipelagic G. maculatus larvae have been reported in ichthyological samples along the Magellanic Province [78]. Considering these findings and the pattern of genetic structure recorded in the species it seems like the Magellanic Province constitutes a continuous habitat for G. maculatus. In contrast, genetic structure along the Intermediate Area suggests low levels of connectivity between river basins. In this context, open sea areas between the coastal rivers here analyzed seem to be preventing the genetic continuity of the species along the Intermediate Area. Such results are quite surprising considering the high dispersal potential described for New Zealand coastal populations of G. maculatus [80]. In this context, mechanisms associated to larval retention or physiological limitation associated to salt tolerance may explain the complex pattern of genetic structure along the Intermediate Area. Otherwise, philopatric behavior could also generate small-scale genetic structure within the Intermediate Area localities, despite the high dispersal potential of the species [34].
Major differences in terms of genetic diversity and structure recorded in G. maculatus agree with the 42°S biogeographic break described along the Chilean coast resulting from different oceanographic, geologic, and climatic processes [38,77]. Along the Chilean coast, this break represents a boundary for major environmental changes in terms of topography, currents, and water salinity that represent a major transition for marine [38] and freshwater species [81]. Different biogeographic scenarios altered the species assemblages across this limit and likely generated distinct patterns of genetic diversity [82]. First, this break coincides with the separation of the West Wind Drift into two main oceanic currents along the Pacific coast [39]. Intermediate Area localities are washed by the northward flow of the Humboldt Current System [83] while the Magellanic Province is located within the influence of the Cape Horn Current, the southern branch of the West Wind Drift. In this context, the presence along Intermediate Area localities of haplotypes belonging to the Magellanic diversity (Hap 04) points towards asymmetric northward gene flow that could be associated to a latitudinal displacement of the West Wind Drift during glacial periods [84,85]. A concordance between phylogeographic and biogeographic patterns has been recorded in many areas of the globe indicating that the forces determining species distributions are also related to the spatial patterns of population genetic structure [86–90]. Therefore, local adaptation processes and distinctive characteristics of main biogeographic areas along the Chilean coast in terms of Quaternary glacial histories and oceanographic regimes are responsible for the general pattern of genetic structure recorded in G. maculatus.
Acknowledgments
This study was supported by a post-doctorate fellowship (Fondecyt 3120075) to CAG-W. All the specimens were captured under the Chilean legislation (Technical Memorandum P.INV N° 427/2011 SUBPESCA). At the same time, this research was supported by the Projects P05-002 ICM and PFB 023 (Insituto de Ecología y Biodiversidad IEB, Universidad de Chile), to E.P and CAG-W and projects Fondecyt 1110798 and 1151375 to LV-C.
Data Availability
D-loop haplotype sequences in G. maculatus are available at GenBank under the accession numbers KP298433–KP298673 and KP336557–KP336665.
Funding Statement
This study was supported by a post-doctorate fellowship (Fondecyt 3120075) to CAG-W. All the specimens were captured under the Chilean legislation (Technical Memorandum P.INV N° 427/2011 SUBPESCA). At the same time, this research was supported by the Projects P05-002 ICM and PFB 023 (Insituto de Ecología y Biodiversidad IEB, Universidad de Chile), to EP and CG-W and project Fondecyt 1110798 to LV-C.
References
- 1. Bennett KD, Tzedakis PC, Willis KJ (1991) Quaternary refugia of North European trees. J Biogeogr 18: 103–115. [Google Scholar]
- 2. Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7: 453–464. [DOI] [PubMed] [Google Scholar]
- 3. Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- 4. Hewitt GM (1996) Some genetic consequences of ice ages, and their role, in divergence and speciation. Biol J Linn Soc 58: 247–276. [Google Scholar]
- 5. Lessa EP, Cook JA, Patton JL (2003) Genetic footprints of demographic expansion in North America, but not Amazonia, during the late Quaternary. Proc Natl Acad Sci 100: 10331–10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Aranzamendi MC, Bastida R, Gardenal CN (2011) Different evolutionary histories in two sympatric limpets of the genus Nacella (Patellogastropoda) in the South-western Atlantic coast. Mar Biol 158: 2405–2418. [Google Scholar]
- 7. González-Wevar CA, Nakano T, Cañete JI, Poulin E (2011) Concerted genetic, morphological and ecological diversification in Nacella limpets in the Magellanic Province. Mol Ecol 20: 1936–1951. 10.1111/j.1365-294X.2011.05065.x [DOI] [PubMed] [Google Scholar]
- 8. González-Wevar CA, Hüne M, Cañete JI, Mansilla A, Nakano T, et al. (2012) Towards a model of postglacial biogeography in shallow marine species along the Patagonian Province: lessons from the limpet Nacella magellanica (Gmelin, 1791). BMC Evol Biol 12: 139 10.1186/1471-2148-12-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Himes CMT, Gallardo MH, Kenagy GJ (2008) Historical biogeography and post-glacial recolonization of South American temperate rain forest by the relictual marsupial Dromiciops gliroides . J Biogeogr 35: 1415–1424. [Google Scholar]
- 10. Lessa EP, D'Elía G, Pardiñas UFJ (2010) Genetic footprints of late Quaternary climate change in the diversity of Patagonian-Fueguian rodents. Mol Ecol 19: 3031–3037. 10.1111/j.1365-294X.2010.04734.x [DOI] [PubMed] [Google Scholar]
- 11. Palma RE, Boric-Bargetto D, Torres-Pérez F, Hernández CE, Yates TL (2012) Glaciation effects on the phylogeographic structure of Oligoryzomys longicaudatus (Rodentia: Sigmodontinae) in the Southern Andes. PLOS ONE 7: e32206 10.1371/journal.pone.0032206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hulton NRJ, Purves RS, McCulloch RD, Sugden DE, Bentley MJ (2002) The last glacial maximum and deglaciation in southern South America. Quat Sci Rev 21: 233–241. [Google Scholar]
- 13. Rabassa J, Coronato A, Salemme M (2005) Chronology of the Late Cenozoic Patagonian glaciations and their correlation with biostratigraphic units of the Pampean region (Argentina). J S Am Earth Sci 20: 81–103. [Google Scholar]
- 14. Hein AS, Hulton NRJ, Dunai TJ, Sugden DE, Kaplan MR, et al. (2010) The chronology of the Last Glacial Maximum and deglacial events in central Argentine Patagonia. Quat Sci Rev 29: 1212–1227. [Google Scholar]
- 15. Rabassa J, Coronato A, Martínez O (2011) Late Cenozoic glaciations in Patagonia and Tierra del Fuego: an updated review. Biol J Linn Soc 103: 316–335. [Google Scholar]
- 16. Clapperton CM (1993) Quaternary Geology and Geomorphology of South America Amsterdam: Elsevier. [Google Scholar]
- 17. McCulloch RD, Bentley MJ, Purves RS, Hulton NRJ, Sugden DE, et al. (2000) Climatic inferences from glacial and paleoecological evidence at the last glacial termination, southern South America. J Quat Sci 15: 409–417. [Google Scholar]
- 18. Valdovinos C, Navarrete SA, Marquet PA (2003) Mollusk species diversity in the Southern Pacific: why are there more species towards the pole? Ecography 26: 139–144. [Google Scholar]
- 19. Zattara EE, Premoli AC (2005) Genetic structuring in Andean landlocked populations of Galaxias maculatus: effects of biogeographic history. J Biogeogr 32: 5–14. [Google Scholar]
- 20. Marko PB, Hoffman JM, Emme SA, McGovern TM, Keever CC, et al. (2010) The “Expansion-Contraction” model of Pleistocene biogeography: rocky shores suffer a sea change? Mol Ecol 19: 146–169. [DOI] [PubMed] [Google Scholar]
- 21. Near TJ, Dornburg A, Kuhn KL, Eastman JT, Pennington JN, et al. (2012) Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc Natl Acad Sci 109: 3434–3439. 10.1073/pnas.1115169109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zemlak TS, Habit EM, Walde SJ, Carrea C, Ruzzante DE (2010) Surviving historical Patagonian landscapes and climate: molecular insights from Galaxias maculatus . BMC Evol Biol 10: 67 10.1186/1471-2148-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pastorino MJ, Gallo LA (2002) Quaternary evolutionary history of Austrocedrus chilensis, a cypress native to the Andean-Patagonian forest. J Biogeogr 29: 1167–1178. [Google Scholar]
- 24. Ruzzante DE, Walde SJ, Cussac VE, Dalebout ML, Seibert J, et al. (2006) Phylogeography of the Percichthyidae (Pisces) in Patagonia: roles of orogeny, glaciation, and volcanism. Mol Ecol 15: 2949–2968. [DOI] [PubMed] [Google Scholar]
- 25. Zemlak TS, Habit EM, Walde SJ, Battini MA, Adams EDM, et al. (2008) Across the southern Andes on fin: glacial refugia, drainage reversals and a secondary contact zone revealed by the phylogeographical signal of Galaxias platei in Patagonia. Mol Ecol 17: 5049–5061. 10.1111/j.1365-294X.2008.03987.x [DOI] [PubMed] [Google Scholar]
- 26. Xu J, Pérez-Lozada M, Jara CG, Crandall KA (2009) Pleistocene glaciation leaves deep signature on the freshwater crab Aegla alacalufiin Chilean Patagonia. Mol Ecol 18: 904–918. 10.1111/j.1365-294X.2008.04070.x [DOI] [PubMed] [Google Scholar]
- 27. Nuñez JJ, Wood NK, Rabanal FE, Fontanella FM, Sites JW Jr (2011) Amphibian phylogeography in the Antipodes: Refugia and postglacial colonization explain mitochondrial haplotype distribution in the Patagonian frog Eupsophus calcaratus (Cycloramphidae). Mol Phylogenet Evol 58: 343–352. 10.1016/j.ympev.2010.11.026 [DOI] [PubMed] [Google Scholar]
- 28. Victoriano PF, Ortiz JC, Benavides E, Adams BJ, Sites JW Jr (2008) Comparative phylogeography of codistributed species of Chilean Liolaemus (Squamata: Tropiduridae) from the central-southern Andean range. Mol Ecol 17: 2397–2416. 10.1111/j.1365-294X.2008.03741.x [DOI] [PubMed] [Google Scholar]
- 29. Fraser CI, Thiel M, Spencer HG, Waters JM (2010) Contemporary habitat discontinuity and historic glacial ice drive genetic divergence in Chilean kelp. BMC Evol Biol 10: 203 10.1186/1471-2148-10-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macaya EC, Zuccarello GC (2010) Genetic structure of the giant kelp Macrocystis pyrifera along the southeastern Pacific. Mar Ecol Prog Ser 420: 103–112. [Google Scholar]
- 31. McDowall RM (1970) The galaxiid fishes of New Zealand. Bull Mus Comp Zool. 139: 341–432. [Google Scholar]
- 32. Rosen DE (1978) Vicariant patterns and historical explanation in Biogeography. Syst Zool 27: 159–188. [Google Scholar]
- 33. McDowall RM (1978) Generalized tracks and dispersal in biogeography. Syst Zool 27: 88–104. [Google Scholar]
- 34. Waters JM, Burridge CP (1999) Extreme intraspecific mitochondrial DNA sequence divergence in Galaxias maculatus (Osteichthys: Galaxiidae), one of the world's most widespread freshwater fish. Mol Phylogenet Evol 11: 1–12. [DOI] [PubMed] [Google Scholar]
- 35. Waters JM, López JA, Wallis GP (2000) Molecular phylogenetics and biogeography of Galaxiid Fishes (Osteichthyes: Galaxiidae): dispersal, vicariance, and the position of Lepidogalaxias salamandroides . Syst Biol 49: 777–795. [DOI] [PubMed] [Google Scholar]
- 36. Burridge CP, McDowall RM, Craw D, Wilson MVH, Waters JM (2011) Marine dispersal as a pre-requisite for Gondwanan vicariance among elements of the galaxiid fish fauna. J Biogeogr 39: 306–321. [Google Scholar]
- 37. Barriga JP, Battini MA, Macchi PJ, Milano D, Cussac VE (2002) Spatial and temporal distribution of landlocked Galaxias maculatus and Galaxias platei (Pisces: Galaxiidae) in a lake in the South American Andes. NZ J Mar Fresh Res 36: 345–359. [Google Scholar]
- 38. Camus PA (2001). Biogeografía marina de Chile continental. Rev Chil Hist Nat 74: 587–617. [Google Scholar]
- 39. Zemlak TS, Walde SJ, Habit EM, Ruzzante DE (2011) Climate-induced changes to the ancestral population size of two Patagonian galaxiids: the influence of glacial cycling. Mol Ecol 20: 5280–5294. 10.1111/j.1365-294X.2011.05352.x [DOI] [PubMed] [Google Scholar]
- 40. Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nuc Acids Res 25: 4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González-Wevar CA, Salinas P, Hüne M, Segovia NI, Vargas-Chacoff L, et al. (2015) Contrasting genetic structure and diversity of Galaxias maculatus (Jenyns, 1848 along the Chilean coast: stock identification for fishery management. J Hered. 10.1093/jhered/esv005 [DOI] [PubMed]
- 42. Filatov DA (2009) Processing and population genetic analysis of multigenic datasets with ProSeq3 software. Bioinformatics 25: 3189–3190. 10.1093/bioinformatics/btp572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 44. Xia X, Xie Z (2001) DAMBE: Software package for data analysis in Molecular Biology and Evolution. J Hered 92: 371–373. [DOI] [PubMed] [Google Scholar]
- 45. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 46. Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered Versus unordered alleles. Genetics 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinf Online, 1: 37–50. [PMC free article] [PubMed] [Google Scholar]
- 48. Hudson RR (1990) A new statistic for detecting genetic differentiation. Genetics 155: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vavrek MJ (2011) fossil: palaeoecological and palaeogeographical analysis tools. Palaeontologia Electronica, 14:1T. [Google Scholar]
- 51. Rogers AR, Harpending H (1992) Population growth makes waves. Mol Biol Evol 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 52. Drummond AJ, Rambaut A (2007) BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol Biol 7: 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suchard MA, Weiss RE, Sinsheimer JS (2001) Bayesian Selection of Continous-Time Markov Chain Evolutionary Models. Mol Biol Evol 18: 1001–1013. [DOI] [PubMed] [Google Scholar]
- 54. Salinas MP (2011) Diversidad genetica de Galaxias maculatus en el Río Pascua y en el Fiordo Calén: colonización postglacial en una cuenca Patagónica y calibración del Reloj Molecular de la especie Tesis de Magíster en Ciencias con mención en Ecología y Biología Evolutiva, Facultad de Ciencias, Universidad de Chile, Santiago, Chile: p.56. [Google Scholar]
- 55. Beerli P, Felsenstein J (2001) “Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci 98: 4563–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaas RE, Raftery AE (1995) Bayes Factor. J Am Stat Assoc 90: 773–795. [Google Scholar]
- 58. Nielsen R, Wakeley J (2001) Distinguishing migration from isolation. A Markov chain Monte Carlo approach. Genetics 158: 885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hey J, Nielsen R (2007) Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci 104: 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Parmesan C (2006) Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol S 37: 1–35. [Google Scholar]
- 61. McMahon SM, Harrison SP, Armbruster WS, Bartlein PJ, Beale CM, et al. (2011) Improving assessment and modelling of climate change impacts on global terrestrial biodiversity. Trend Ecol Evol 26: 249–259. [DOI] [PubMed] [Google Scholar]
- 62. Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos T R Soc B 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maggs CA, Castihlo R, Foltz D, Henzler C, Jolly MT, et al. (2008) Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology, 89: S108–S122. [DOI] [PubMed] [Google Scholar]
- 64. Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol 23: 564–571. 10.1016/j.tree.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 65. Marko PB (2004) 'What‘s larvae got to do with it?’ Disparate patterns of post-glacial population structure in two benthic marine gastropods with identical dispersal potential. Mol Ecol 13: 597–611. [DOI] [PubMed] [Google Scholar]
- 66. Cussac V, Ortubay S, Iglesias G, Milano D, Lattuca M, et al. (2004) The distribution of South American galaxiid fishes: the role of biological traits and post-glacial history. J Biogeogr 31: 103–121. [Google Scholar]
- 67. Ceballos SG, Lessa EP, Victorio MF, Fernández DA (2011) Phylogeography of the sub-Antarctic notothenioid fish Eleginops maclovinus: evidence of population expansion. Mar Biol 159: 499–505. [Google Scholar]
- 68.Hüne M, González-Wevar C, Poulin E, Mansilla A, Fernández DA, Barrera-Oro E (2015) Low level of genetic divergente between Harpagifer fish species (Perciformes: Notothenioidei) points to a Quaternary colonization of Patagonia from the Antarctic Peninsula. Polar Biol. 10.1007/s00300-014-1623-6 [DOI]
- 69. Pardiñas UFJ, Teta P, D’Elia G, Lessa EP (2011) The evolutionary history of sigmodontine rodents in Patagonia and Tierra del Fuego. Biol J Linn Soc 103: 495–513. [Google Scholar]
- 70. Jakob SS, Martinez-Meyer E, Blattner FR (2009) Phylogeographic analyses and paleodistribution modeling indicate Pleistocene In Situ survival of Hordeum species (Poaceae) in Southern Patagonia without genetic or spatial restriction. Mol Biol Evol 26: 907–923. 10.1093/molbev/msp012 [DOI] [PubMed] [Google Scholar]
- 71. Sérsic AN, Cosacov A, Cocucci AA, Johnson LA, Pozner R, et al. (2011) Emerging phylogeographical patterns of plants and terrestrial vertebrates from Patagonia. Biol J Linn Soc 103: 475–494. [Google Scholar]
- 72. Waters JM, Disjkstra LH, Wallis GP (2000) Biogeography of a southern hemisphere freshwater fish: how important is marine dispersal? Mol Ecol 9: 1815–1821. [DOI] [PubMed] [Google Scholar]
- 73. Ruzzante DE, Walde SJ, Gosse JC, Cussac VE, Habit EM, et al. (2008) Climate control on ancestral population dynamics: insight from Patagonian fish phylogeography. Mol Ecol 17: 2234–2244. 10.1111/j.1365-294X.2008.03738.x [DOI] [PubMed] [Google Scholar]
- 74. Unmack PJ, Barriga JP, Battini MA, Habit EM, Johnson JB (2012) Phylogeography of the catfish Hatcheria macraei reveals a negligible role of drainage divides in structuring populations. Mol Ecol 21: 942–959. 10.1111/j.1365-294X.2011.05408.x [DOI] [PubMed] [Google Scholar]
- 75. Alò D, Correa C, Arias C, Cárdenas L (2013) Diversity of Aplochiton Fishes (Galaxiidea) and the taxonomic resurrection of A. marinus . PLOS ONE 8: e71577 10.1371/journal.pone.0071577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Unmack PJ, Bennin A, Habit EM, Victoriano PF, Johnson JB (2009) Impact of ocean barriers, topography, and glaciation on phylogeography of the catfish Trichomycterus areolatus (Teleostei: Trichomycteridae) in Chile. Biol J Linn Soc 97: 876–892. [Google Scholar]
- 77. Escribano R, Fernández M, Aranís A (2003) Physical processes and patterns of diversity of the Chilean eastern boundary pelagic and benthic marine ecosystems: an overview. Gayana 67: 190–205. [Google Scholar]
- 78. Bustos CA, Landaeta MF, Balbontín F (2010) Ichthyoplanknton spatial distribution and its relation with water column stratification in fjords of southern Chile (46°48’-50°09’S) in austral spring 1996 and 2008. Cont Shelf Res 31: 293–303. [Google Scholar]
- 79. Valdenegro A, Silva N (2003) Caracterización oceanográfica física y química de la zona de canales y fiordos australes de Chile entre el Estrecho de Magallanes y Cabo de Hornos (Cimar 3 Fiordos). Ciencia y Tecnología del Mar 26: 19–60. [Google Scholar]
- 80. McDowall RM, Robertson D, Saito R (1975) Ocurrence of Galaxiid larvae and juveniles in the sea. NZ J Mar Freshw Res 9: 1–9. [Google Scholar]
- 81. Dyer BS (2000) Systematic review and biogeography of the freshwater fishes of Chile. Estudios Oceanológicos 19: 77–98. [Google Scholar]
- 82. Vianna JA, Medina-Vogel G, Chehébar C, Sielfried W, Olavarría C, et al. (2011) Phylogeography of the Patagonian otter Lontra provocax: adaptive divergente to marine habitat or signature of southern glacial refugia? BMC Evol Biol 11: 53 10.1186/1471-2148-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thiel M, Macaya EC, Acuña E, Arntz WE, Bastias H, et al. (2007) The Humboldt Current System of Northern and Central Chile: oceanographic processes, ecological interactions and socioeconomic feedback. Annu Rev Oceanogr Mar Biol 45: 195–344. [Google Scholar]
- 84. Gersonde R, Crosta X, Abelmann A, Armand L (2005) Sea-surface temperature and sea ice distribution of the Southern Ocean at the EPILOG Last Glacial Maximum—a circum-Antarctic view based on siliceous microfossil records. Quat Sci Rev 24: 869–896. [Google Scholar]
- 85. Kemp AES, Grigorov I, Pearce RB, Naveira Garabato AC (2010) Migration of the Antarctic Polar Front through the mid-Pleistocene transition: evidence and climatic implications. Quat Sci Rev 29: 1993–2009. [Google Scholar]
- 86. Waters JM, Roy MS (2004) Out of Africa: The slow train to Australasia. Syst Biol 53: 18–24. [DOI] [PubMed] [Google Scholar]
- 87. Jolly MT, Jollivet D, Gentil F, Thiébaut E, Viard F (2005) Sharp genetic break between Atlantic and English Channel populations of the polychaete Pectinaria koreni, along the north coast of France. Heredity 94: 23–32. [DOI] [PubMed] [Google Scholar]
- 88. Goldstein SJ, Schiel DR, Gemmell NJ (2006) Comparative phylogeography of coastal limpets across a marine disjunction in New Zealand. Mol Ecol 15: 3259–3268. [DOI] [PubMed] [Google Scholar]
- 89. Taylor MS, Hellberg ME (2006) Comparative phylogeography of a genus of coral reef fishes: biogeographical and genetical concordance in the Caribbean. Mol Ecol 15: 695–707. [DOI] [PubMed] [Google Scholar]
- 90. Haye PA, Segovia NI, Muñoz-Herrera NC, Gálvez FE, Martínez A (2014) Phylogeographic structure in benthic marine invertebrates of the Southeaster Pacific coast of Chile with differing dispersal potential. PLoS ONE 9: e88613 10.1371/journal.pone.0088613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
D-loop haplotype sequences in G. maculatus are available at GenBank under the accession numbers KP298433–KP298673 and KP336557–KP336665.