Abstract
There is a rising resistance against antimony drugs, the gold-standard for treatment until some years ago. That is a serious problem due to the paucity of drugs in current clinical use. In a research to reveal how these drugs affect the parasite during treatment and to unravel the underlying basis for their resistance, we have employed metabolomics to study treatment in Leishmania infantum promastigotes. This was accomplished first through the untargeted analysis of metabolic snapshots of treated and untreated parasites both resistant and responders, utilizing a multiplatform approach to give the widest as possible coverage of the metabolome, and additionally through novel monitoring of the origin of the detected alterations through a 13C traceability experiment. Our data stress a multi-target metabolic alteration with treatment, affecting in particular the cell redox system that is essential to cope with detoxification and biosynthetic processes. Additionally, relevant changes were noted in amino acid metabolism. Our results are in agreement with other authors studying other Leishmania species.
Introduction
The term leishmaniasis encompasses the different clinical infections by protozoan species belonging to the genus Leishmania, whose importance for human health is only superseded by malaria. Worldwide, leishmaniasis accounts for 10–12 million infected people, with 2 million new cases per year and near 50,000 annual deaths [1]. Nowadays, treatment relies almost exclusively on chemotherapy; reduced to a small number of drugs with an effectiveness increasingly eroded by rising resistance, severe side-effects and unaffordable costs for massive implementation. Additionally, there is a shortfall of new candidates in the pipeline.
Since their introduction 60 years ago, organic pentavalent antimonials have constituted the gold standard for treatment until clinical resistance in Northwest India and Nepal reached an inadmissible rate of 60%, as reported even in patients without any prior treatment [2] leading to their progressive demotion by miltefosine or liposomal amphotericin B, especially for the visceral forms of the disease. Nevertheless, the current use of antimonials is still quite notorious in canine leishmaniasis in combination with allopurinol [3] and concerning human clinics, as monotherapy on some endemic areas of visceral leishmaniasis as well as on mucocutaneous and cutaneous forms of the disease [4], and finally as part of drug combination strategies [5], mostly based on clinical experience, rather than on knowledge of their mechanism of action and resistance and the targets involved. In this sense, a deeper insight into these items will ameliorate the clinical strategies to tackle the disease, in an attempt to avoid partial cross-resistance of antimonials with other drugs as observed in field studies [6–8]. This, together with vector transmission of the resistant phenotype aforementioned [9], and the improved fitness of resistant parasites inside the host [10], foresees a privileged expansion of the antimonial resistant parasites over the susceptible isolates [11].
Among anti Leishmania drugs, mechanisms of action of and resistance to antimonials are those with higher complexity. Their lethal mechanism is mostly based on deterioration of the thiol-dependent metabolism of the parasite [12–15], induced by the inhibition of trypanothione reductase and trypanothione depletion after its conjugation with Sb3+ [13], with a concomitant global dysfunction of the parasite metabolism and structural functionality [16–20].
The core of resistance to antimonials relies on a two-pronged strategy in Leishmania. First, the prevention of intracellular build-up of Sb3+, the toxic form of antimonials, by i) down expression of the parasite reductases responsible for the reduction of the administered prodrug, Sb5+, into Sb3+ [21,22], ii) impairment of the entry of external Sb3+ into the parasite through decreased levels of aquaglyceroporin 1 [23], and iii) decreasing the levels of intracellular Sb3+ by efflux pumps [8,17,24]. The second set of strategies were aimed to restore the redox power, crippled by the drug, by overexpression of enzymes of polyamine and trypanothione biosynthesis, with increment of intracellular levels of this metabolite and of glutathione, its precursor [25,26].
Furthermore, the final clinical outcome for antimonials encompassed additional factors such as the modulation of the physiology of the macrophage by these drugs [24,27,28], the strength of the immune status of the host [29] and, surprisingly, environmental factors, as the exposure of human population to low concentrations of As3+ in drinking water [30], that predisposed for antimonial resistance.
Unbiased “omics” studies of changes associated to the action of and resistance to antimonials have been carried out through genomics [17,31–33] and proteomics [17–20,34–36]. However, among the unbiased “omics” only metabolomics provides a direct snapshot of the final effect of the drug and how it challenges parasite metabolism [37–41]. The importance of this technique in Leishmania is highlighted by the utmost importance of the post-transcriptional control of gene expression operative in trypanosomatids compared with higher eukaryotes [42,43]. The potentiality of metabolomics to unravel the mechanism of action of antimonials is supported by previous works on L. donovani isolates from India [38,41] and on a reference strain of L. infantum from Spain [40], using for separation of metabolites ZIC-HILIC chromatography or capillary electrophoresis (CE), respectively.
The goal of this work is to test whether a metabolomic platform, gathering different complementary analytical techniques, will improve the metabolite coverage achieved by single separation techniques, applied to the mechanism of action and resistance to Sb3+ in Leishmania infantum. To achieve this purpose we have used exponential promastigotes from a reference strain of this species and its resulting Sb3+ resistant isolate, obtained by growing the parasites in vitro under a stepwise increasing concentration of Sb3+. Although a full extrapolation of the strategies adopted by the promastigote to cope with the metalloid stress into amastigotes will be unfeasible due to the extensive interstage metabolic rewiring, we surmise that most of the results obtained are shared by both stages; first, Sb3+,the oxidation stage of antimony used, is the toxic form of the metalloid for both stages; secondly, for a given strain the trait for antimonial-resistance is not only displayed by both stages of the parasite, but maintained through vector transmission [9,11]; finally, the results obtained point out to bioenergetic and redox metabolism, key processes with identical pathways, regardless of the stage of the life cycle of Leishmania. The use of promastigotes with a more active and, presumably, more flexible metabolism than the metacyclic form in a Leishmania species, will unveil the contribution of metabolic pathways unnoticed in metacyclic promastigotes. Additionally, the use of L. infantum strains may, in some degree, decrease the risk of previous antimonials exposure, as frequently happen with the anthoponotic transmission cycles for L. donovani. Moreover, the difficulty in growing anexic amastigotes of most Leishmania species means that the majority of the studies have to be done on cultured promastigotes cells [44].
Materials and Methods
Reagents and solvents
Methanol (LC-MS grade), heptane (GC-MS grade), chloroform (MS grade), acetonitrile (LC-MS grade), isopropanol (LC-MS grade), formic acid (MS grade), pyridine (silylation grade), C18:0 methyl ester and O-methoxyamine hydrochloride were purchased from Sigma-Aldrich (Taufkirchen, Germany); N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) plus 1% trimethylchlorosilane (TMCS) was purchased from Pierce Chemical Co (Rockford, IL, USA) and references masses purine and HP-0921 (hexakis-(1H,1H,3Htetrafluoropentoxy)-phosphazene) were from Agilent (atmospheric pressure inlet – time of flight (API-TOF) reference mass solution kit). Milli-Qplus 185 was provided by Millipore (Billerica, MA, USA).
Sample collection and preparation
L. infantum promastigotes strain M/CAN/ES/96/BCN150 were kindly provided by Dr. F. J. Carrion (Veterinary School, University of Madrid). Parasites were grown in RPMI-1640 medium supplemented with 10% heat inactivated fetal calf serum at 26°C. BCN150 L. infantum promastigotes resistant to Sb3+ were obtained from the parental strain by stepwise culture under increasing concentration of the drug until growth at 180 μmol × L-1 potassium antimony(III) tartrate was achieved. Large-scale culture was carried out in a roller apparatus (Gibco, Cell Culture) with an inoculum of 4 × 105 promastigotes × mL-1. Once they reached a mid- exponential phase of growth (8 × 106 promastigotes × mL-1) they were transferred into the same volume of fresh medium, and incubated or not with 120 μmol × L-1 potassium antimony(III) tartrate (a concentration causing 90% mortality in wild type parasites) for 12h. The BCN150 strain has an IC50 = 20.9 μmol × L-1 with respect to Sb3+. Once harvested, parasites were washed twice with Hanks medium at 4°C and immediately frozen in liquid N2 and kept at – 80°C until further processing.
The preparation for 13C analysis was done by growing the parasites in RPMI where L-arginine was substituted by 13C L-arginine (Sigma-Aldrich—Steinheim, Germany, 13C in all carbons) for 14h.
Investigated Groups
ST: susceptible strain treated, SNT: susceptible strain non–treated (controls) and R: resistant strain to Sb3+. Four replicates from each group were used in metabolic fingerprinting analysis.
LC-MS sample treatment
Samples corresponding to 4 × 107 L. infantum promastigotes were re-suspended in 200 μL MeOH/H2O (4/1, v/v) and disrupted in a TissueLyser LT (Qiagen, Germany) (2 glass balls of 2 mm, 5 min, 50 Hz). Solid parts were removed by centrifugation (centrifuge 5415 R Eppendorf) (15700 × g, 4°C, 20 min). The supernatant was collected and filtered through a 0.22 μm nylon filter (National Scientific – 197 Cardiff Valley Road, Rockwood, TN 37854, India) and then directly injected into the equipment.
CE-MS sample treatment
After disruption of the cellular pellets, as described for LC-MS (liquid chromatography-mass spectrometry), 150 μL of the supernatant were transferred into a new tube and evaporated to dryness in a SpeedVac SPD121P (Thermo Fisher Scientific, Waltham, MA) at 35°C. The metabolite extracts were re-suspended in 150 μL MilliQ water, centrifuged (15700 × g, 15 min, 4°C) and injected into the instrument.
GC-MS sample treatment and derivatization
Pellets corresponding to 4 x 107 L. infantum promastigotes were re-suspended and lysed by addition of 350 μL of cold (4°C) solution of methanol/chloroform/water (3/1/1, v/v/v), and processed for disruption as described for LC-MS samples. Two hundred μL of the resulting supernatant were transferred into a vial and evaporated to dryness in a SpeedVac SPD121P (Thermo Fisher Scientific, Waltham, MA.) at 35°C. For methoximation, 10 μL of O-methoxyamine hydrochloride (15 mg × mL-1) in pyridine were added to each GC (gas chromatography) vial and vortexed vigorously (FB 15024, Fisher Scientific, Spain). The vials were incubated in darkness at room temperature for 16 hours. Then, 10 μL of BSTFA with 1% TMCS (v/v) were added and samples were vortexed for 5 min, silylation was carried out for 1 h at 70°C and finally 100 μL of C18:0 methyl ester (10 mg × L-1 in heptane) were added as an internal standard and samples were mixed again by vortexing gently. Two blank samples were prepared by the same procedure of extraction and derivatization and analyzed at the beginning and at the end of the sequence.
Quality Controls
QC samples were prepared by pooling equal volumes of each sample from all groups. The same procedure was followed for the three techniques (before the derivatization step in GC-MS). They were analysed throughout the run to provide a measurement of the stability and performance of the system [45] as well as the reproducibility of the procedure for sample treatment.
Metabolomics fingerprinting by LC-ESI-QTOF-MS
The LC system consisted of a degasser, two binary pumps and autosampler (1200 series, Agilent); 15 μL of filtered supernatant sample were injected onto a reversed-phase column (Discovery HS C18 150 × 2.1 mm, 3 μm; Supelco) with a guard column (Discovery HS C18 20 × 2.1 mm, 3 μm; Supelco) kept at 40°C. The system was operated at a flow rate of 0.6 mL × min-1 with solvent A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The gradient started from 25% B to 95% B in 35 min, followed by 5 min of 95% of B and then returned to starting conditions in 1 min, with a re-equilibration step at 25% B for 9 min (total analysis time 50 minutes). Data were collected in positive ESI (electrospray) mode in separate runs on a QTOF (quadrupole time-of-flight) (Agilent 6520) operated in full scan mode from m/z 50 to 1000. The capillary voltage was 3000 V with a scan rate of 1.95 scans per second. The gas temperature was 330°C, the drying gas flow 10.5 L × min-1 and the nebulizer set up at 52 psi. The MS-TOF (time-of-flight) parameters were: fragmentor 175 V, skimmer 65 V and octopole radio frequency voltage (OCT RF Vpp) 750 V. During the analysis, two reference masses were used: 121.0509 (C5H4N4) and 922.0098 (C18H18O6N3P3F24). They were continuously infused into the system to allow constant mass correction. Samples were analyzed in a randomized run, during which time they were kept in the LC autosampler at 4°C.
Metabolomics fingerprinting by CE-ESI-TOF-MS
The equipment consisted of a Capillary Electrophoresis (7100 Agilent) coupled to a TOF Mass Spectrometry (6224 Agilent). The CE mode was controlled by ChemStation software (B.04.03, Agilent) and MS mode by Mass- Hunter Workstation Data Analysis (B.02.01, Agilent). The separation occurred in a fused-silica capillary (Agilent) (total length, 100 cm; internal diameter, 50 μm). It was carried out in normal polarity with a background electrolyte containing 0.8 mol × L-1 of formic acid solution in 10% methanol (v/v) at 20°C. New capillaries were pre-conditioned by flushing successively by NaOH 1.0 mol ×L-1 MilliQ water and background electrolyte, 30 min each. Before each analysis the capillary was conditioned by flushing of background electrolyte for 5 min. The sheath liquid (6 μL × min-1) was methanol/water (1/1, v/v) containing 1.0 mmol × L-1 formic acid with two reference masses: 121.0509 (C5H4N4) and 922.0098 (C18H18O6N3P3F24), to allow correction and higher accurate mass in the MS. Samples were hydrodynamically injected at 50 mbar for 50 s and stacked by applying background electrolyte at 100 mbar for 10 s. The separation voltage was 30 kV with 25 mbar of internal pressure and the analyses were carried out in 30 min. The MS parameters were optimized [40] by the study of signal-to-noise ratio of the ion 175.1179 Da (corresponding to L-arginine, previously identified in Leishmania); the best analytical conditions were: fragmentor 100 V, skimmer 65 V, octopole 750 V, nebulizer pressure 20 psi, drying gas temperature at 200°C and flow rate 12.0 L x min-1. The capillary voltage was 3500 V. Data were acquired in positive Dual-ESI mode with a full scan from m/z 80 to 1000 at a rate 1.02 scans per second.
Metabolomics fingerprinting by GC-EI-IT-MS
GC system (Agilent Technologies 7890A) consisted in an auto sampler (Agilent Technologies 7693) and an inert MSD with Quadrupole (Agilent Technologies 5975). 2 μL of the derivatized sample were injected through a GC-Column DB5-MS (30 m length, 0.25 mm internal diameter, 0.25 μm film 95% dimethylpolysiloxane / 5% diphenylpolysiloxane) with a pre-column (10 m J&W integrated with Agilent 122-5532G). The flow rate of the helium carrier gas was set at 1 mL × min-1 and the injector temperature 250°C. The split ratio was 1:10 flow into a Restek 20782 deactivated glass-wool split liner. The temperature gradient was programmed at 60°C (held for 1 min), with a ramping increase rate of 10°C × min-1 up to 325°C. Finally it was cooled down for 10 min before the next injection. The total analysis time was 37.5 min. The detector transfer line, filament source and quadrupole temperature were respectively set at 290°C, 230°C and 150°C. The electron ionization (EI) source was placed at 70 eV. The mass spectrometer operated in scan mode over a mass range of m/z 50–600 at a rate of 2 spectra per second. Peaks detection and spectra processing were obtained using Agilent ChemStation Software (G1701EA E.02.00.493, Agilent).
Data treatment
The resulting data files (LC-MS and CE-MS) were cleaned of background noise and unrelated ions by the Molecular Feature Extraction (MFE) tool in the Mass Hunter Qualitative Analysis software (B.04.00, Agilent).
Primary data treatment (filtering and alignment) was performed with Mass Profiler Professional software (B.02.01, Agilent). Features were filtered by selecting the data present for LC-MS and CE-MS in at least 50% of the QCs and in 75% of the samples in 1 of the 2 groups: ST vs. SNT and R vs. SNT, in order to assess the effects of the treatment, and to investigate the metabolic variations in resistance, respectively. Differences for individual metabolites were evaluated by comparison of these two groups using Mann-Whitney U (where p values < 0.05 were considered significant). The accurate masses representing statistical significant differences were searched against the METLIN database and MassTrix (Mass Translation into Pathways), a tool for data annotation. Treatment of GC-MS data was carried out as aforementioned for identification, and further deconvolution was performed. AMDIS (automated mass spectral deconvolution and identification v. 2.69) software was used to identify co-eluted compounds according to their retention index and retention time. Then, alignment and filtering steps (presence in at least 75% of the samples in at least one of the groups were done with Mass Profiler Professional software (B.02.01, Agilent), and statistical analysis for treatment (ST vs. SNT) and resistance (R vs. SNT) carried out as above. Masses with a p value < 0.05 were selected.
Compound identification
LC-MS analysis: the identity of the compounds selected according to their significance in class separation was further confirmed by LC-MS/MS using a QTOF (model 6520, Agilent). Experiments were repeated with identical chromatographic conditions of the primary analysis (described above). Ions were targeted for collision-induced dissociation (CID) fragmentation on the fly based on the previously determined accurate mass and retention time. Identity was confirmed by comparison of the structure of the proposed compound with the fragments obtained. If available, the resulting accurate mass data and isotopic distributions for the precursor and product ions were studied and compared with spectral data of reference compounds obtained under identical conditions, using MS/MS spectra in public database (METLIN- http://metlin.scripps.edu/) or against commercially available standards. A third way of identity confirmation was to check the accurate mass data and isotopic distributions for the precursor and product ions in MS Fragmentor software (12.01, ACD/Labs).
CE-MS analysis: the significant accurate masses were searched against the public databases METLIN: Metabolite and Tandem MS Database (http://metlin.scripps.edu/) and MassTrix: Mass Translation into Pathways (http://metabolomics.helmholtz-muenchen.de/masstrix2/).
GC-MS analysis: the compound identification was performed by using Fiehn RTL Library and the NIST mass spectra library version 2.0g, with the ChemStation software PBM algorithm (G1701EA E.02.00.493, Agilent).
Finally, the presence of the compounds labelled with 13C was confirmed by the analysis of its isotopic pattern distribution in LC-MS and CE-MS. The corresponding comparison between the extracted ions chromatograms/electropherograms were done in order to evaluate the discriminating presence of 13C for each compound belonging to the metabolic pathways of L-arginine in Leishmania.
Results and Discussion
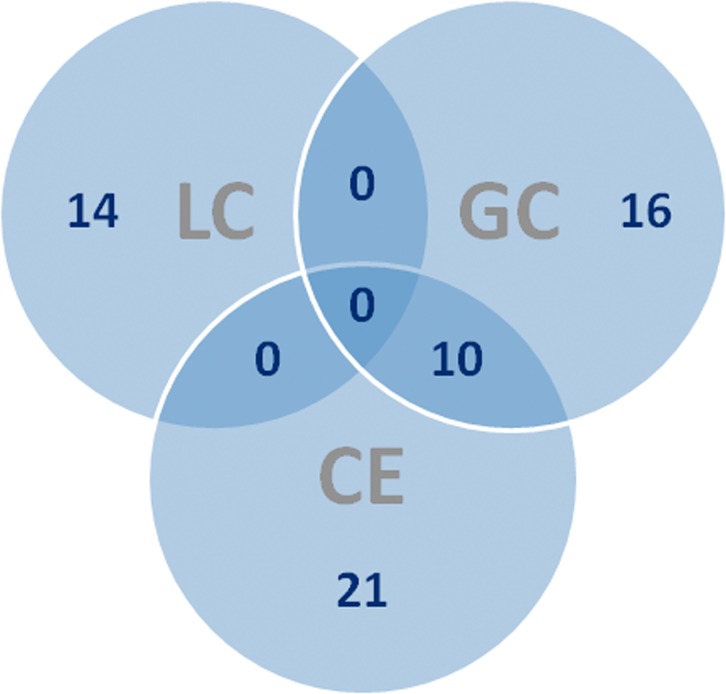
In this work, we have used three different analytical platforms to cover the widest range of metabolites from L. infantum promastigotes. Table 1 summarizes the number of features obtained for each step from the corresponding technique, while Fig 1 compiles the number of identified compounds for each technique. It is quite obvious how useful the multiplatform approach is; only 10 out of 61 compounds (16%) were identified by two techniques and none for the three platforms. This means that 84% of metabolites are exclusively identified by one technique; 35% for CE-MS, 26% for GC-MS and 23% from LC-MS/MS.
Table 1. Comparison of the different techniques and their respective number of features at the different steps of selection.
| LC-MS | CE-MS | GC-MS | |||||
|---|---|---|---|---|---|---|---|
| ST vs.SNT | R vs.SNT | ST vs.SNT | R vs.SNT | ST vs.SNT | R vs.SNT | ||
| Number of features | By alignment | 7299 | 7299 | 1072 | 1072 | 89 | 89 |
| After statistical treatment | 283 | 278 | 32 | 97 | 23 | 24 | |
| Identified | 14 | 31 | 26 | ||||
Fig 1. Venn diagram.
Venn diagram for the number of compounds identified in each technique.
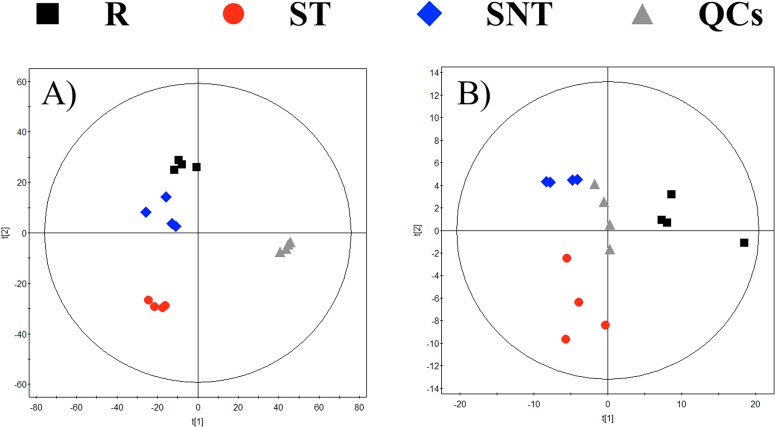
To assess the quality of the analytical techniques, we applied PCA (principal component analysis) models (built by SIMCAP+ software), as represented for LC-MS, CE-MS and GC-MS in Fig 2. The tight clustering of the QCs, located at the center of the plot, evidenced the robustness of the analytical procedure and that separations among the groups obeyed to a real biological differentiation.
Fig 2. PCA models.
PCA models for the whole data set filtered according to their presence in at leasts 50% of the QCs. Panels: A.- LC-MS. 2 components. R2 = 0.62. Q2 = -0.029. B.- CE-MS. 2 components. R2 = 0.872. Q2 = 0.694. C.- GC-MS. 2 components. R2 = 0.515. Q2 = 0.235. Each group is obtained from four samples, except R in CE-MS that consisted of three due to some problems during the sample treatment.
The results of putative and confirmed identification by LC-MS/MS, CE-MS and GC-MS are summarized in Table 2, while Table 3 shows the compounds identified by LC-MS/MS and their characteristic fragments.
Table 2. Compounds with statistical significance identified and their variation tendency for each of the comparisons.
| Compound name | Mass (Da) | Molecular formula | Mass error (ppm) | ST vs SNT | p-value | R vs SNT | p-value | CV in QC | Analytical technique | Confirmation | Biochemical nature |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (4,8,10-d18:3)Sphingosine | 295.2511 | C18H33NO2 | 2 | down | < 0.05 | 6 | LC-MS | Id MS/MS | Sphingolipids and spingoid bases | ||
| 2-Oxo-5-methylthiopentanoic acid | 162.0351 | C6H10O3S | 7 | down | < 0.01 | down | < 0.01 | 6 | CE-MS | Putative | Organic acids |
| 4-Hydroxy-proline / 5-amino-2-oxopentanoate | 131.0582 | C5H9NO3 | 0 | up | < 0.05 | 3 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| 131.0582 | C5H9NO3 | down | < 0.01 | 7 | GC-MS | Identified | Amino acids, peptides and conjugates | ||||
| 4-Oxoproline / L-1-Pyrroline-3-hydroxy-5-carboxylate / 1-Pyrroline-4-hydroxy-2-carboxylate | 129.0426 | C5H7NO3 | 1 | up | < 0.05 | 4 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| 2-Oxoarginine | 173.0800 | C6H11N3O3 | 0 | up | < 0.01 | up | < 0.01 | 12 | CE-MS | Putative | Organic acids |
| Adenosine | 267.0968 | C10H13N5O4 | up | < 0.05 | down | < 0.05 | 6 | GC-MS | Identified | Purines/pyrimidines and conjugates | |
| ADMA / SDMA | 202.1430 | C8H18N4O2 | 0 | up | < 0.01 | up | < 0.05 | 8 | CE-MS | Putative | Amino acids, peptides and conjugates |
| Alanine / Sarcosine | 89.0477 | C3H7NO2 | 6 | down | < 0.01 | up | < 0.05 | 4 | CE-MS | Putative | Amino acids, peptides and conjugates |
| Allantoic acid | 176.0546 | C4H8N4O4 | 10 | down | < 0.01 | down | < 0.01 | 8 | LC-MS | Id MS/MS | Organic acids |
| Arginine | 174.1117 | C6H14N4O2 | 1 | down | < 0.05 | up | < 0.05 | 2 | CE-MS | Id C13 | Amino acids, peptides and conjugates |
| Asparagine /N-carbamoyl Sarcosine | 132.0535 | C4H8N2O3 | 5 | up | < 0.05 | 4 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| 132.0535 | C4H8N2O3 | down | < 0.01 | 4 | GC-MS | Identified | Amino acids, peptides and conjugates | ||||
| Aspartic acid | 133.0375 | C4H7NO4 | down | < 0.05 | 6 | GC-MS | Identified | Amino acids, peptides and conjugates | |||
| Aspartyl-leucine | 246.1216 | C10H18N2O5 | 15 | up | < 0.05 | 10 | LC-MS | Id MS/MS | Amino acids, peptides and conjugates | ||
| Bis(glutathionyl)spermine | 780.3622 | C30H56N10O10S2 | 13 | down | < 0.01 | 17 | LC-MS | Id MS/MS | Amino acids, peptides and conjugates | ||
| C17 Sphinganine | 287.2824 | C17H37NO2 | 4 | up | < 0.05 | 5 | LC-MS | Id MS/MS | Sphingolipids and spingoid bases | ||
| Cellobiose | 342.1162 | C12H22O11 | down | < 0.05 | up | < 0.05 | 3 | GC-MS | Identified | Carbohydrates | |
| Choline | 103.0997 | C5H13NO | 3 | up | < 0.01 | up | < 0.05 | 3 | CE-MS | Putative | Amines |
| Citrulline | 175.0957 | C6H13N3O3 | 0 | down | < 0.01 | down | < 0.05 | 5 | CE-MS | Id C13 | Amino acids, peptides and conjugates |
| Cystathionine | 222.0674 | C7H14N2O4S | 2 | up | < 0.05 | up | < 0.01 | 0 | CE-MS | Putative | Amino acids, peptides and conjugates |
| Ergosterol | 396.3392 | C28H44O | up | < 0.01 | down | < 0.05 | 1 | GC-MS | Identified | Sterol and prenol lipids | |
| Glucose-6-phosphate | 260.0297 | C6H13O9P | down | < 0.05 | down | < 0.05 | 6 | GC-MS | Identified | Carbohydrates | |
| Glutamate / Isoglutamate / 2- Oxo-4 hydroxy-5-aminovalerate / L4-Hidroxy Glutamate semialdehyde | 147.0532 | C5H9NO4 | 0 | down | < 0.01 | up | < 0.05 | 15 | CE-MS | Putative | Amino acids, peptides and conjugates |
| Glutamine | 146.0691 | C5H10N2O3 | down | < 0.05 | down | < 0.05 | 6 | GC-MS | Identified | Amino acids, peptides and conjugates | |
| Glycerophosphocholine | 257.1028 | C8H20NO6P | 1 | up | < 0.01 | 27 | LC-MS | Id MS/MS | Glycerophospholipids | ||
| Guanine | 151.0494 | C5H5N5O | 0 | up | < 0.01 | 12 | CE-MS | Putative | Purines/pyrimidines and conjugates | ||
| Histidine | 155.0695 | C6H9N3O2 | 0 | up | < 0.01 | 6 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| 155.0695 | C6H9N3O2 | up | < 0.05 | 6 | GC-MS | Identified | Amino acids, peptides and conjugates | ||||
| Hypoxanthine | 136.0385 | C5H4N4O | 0 | down | < 0.05 | 5 | CE-MS | Putative | Purines/pyrimidines and conjugates | ||
| 136.0385 | C5H4N4O | up | < 0.01 | up | < 0.01 | 4 | GC-MS | Identified | Purines/pyrimidines and conjugates | ||
| Imidazole lactate | 156.0535 | C6H8N2O3 | 0 | down | < 0.01 | up | < 0.01 | 5 | CE-MS | Putative | Organic acids |
| Lauric acid | 200.1776 | C12H24O2 | 1 | up | < 0.05 | 5 | LC-MS | Id MS/MS | Fatty acyls | ||
| Lauric Acid ethyl ester | 228.2089 | C14H28O2 | 1 | down | < 0.05 | 5 | LC-MS | Id MS/MS | Fatty acyls | ||
| Leucine | 131.0946 | C6H13NO2 | 0 | down | < 0.05 | down | < 0.05 | 6 | CE-MS | Putative | Amino acids, peptides and conjugates |
| 131.0946 | C6H13NO2 | down | < 0.05 | 2 | GC-MS | Identified | Amino acids, peptides and conjugates | ||||
| Leucyl-Alanine | 202.1317 | C9H18N2O3 | 0 | up | < 0.05 | 7 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| Linoleic acid | 280.2402 | C18H32O2 | up | < 0.01 | up | < 0.01 | 3 | GC-MS | Identified | Fatty acyls | |
| Linolenic Acid | 278.2246 | C18H30O2 | 0 | up | < 0.05 | down | < 0.01 | 3 | LC-MS | Id MS/MS | Fatty acyls |
| Lysine | 146.1055 | C6H14N2O3 | 0 | up | < 0.05 | 1 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| 146.1055 | C6H14N2O3 | up | < 0.01 | up | < 0.01 | 5 | GC-MS | Identified | Amino acids, peptides and conjugates | ||
| Malic acid | 134.0215 | C4H6O5 | down | < 0.05 | down | < 0.01 | 4 | GC-MS | Identified | Organic acids | |
| Methylhistidine | 169.0851 | C7H11N3O2 | 0 | up | < 0.05 | 13 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| Myo-inositol | 180.0632 | C6H12O6 | down | < 0.01 | 4 | GC-MS | Identified | Carbohydrates | |||
| Myristic acid | 228.2089 | C14H28O2 | down | < 0.05 | up | < 0.05 | 5 | GC-MS | Identified | Fatty acyls | |
| Myristic Acid ethyl ester | 256.2402 | C16H32O2 | 1 | up | < 0.05 | 10 | LC-MS | Id MS/MS | Fatty acyls | ||
| N-Acetylaminobutanal/ Pipecolic acid | 129.0790 | C6H11NO2 | 0 | up | < 0.05 | 20 | CE-MS | Putative | Ketones and aldehydes | ||
| N-Acetyl-lysine | 188.1161 | C8H16N2O3 | 1 | up | < 0.05 | 12 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| Norleucine | 131.0946 | C6H13NO2 | 19 | down | < 0.05 | 8 | LC-MS | Id MS/MS | Amino acids, peptides and conjugates | ||
| Ornithine | 132.0899 | C5H12N2O2 | 0 | down | <0.05 | up | < 0.05 | 3 | CE-MS | Id C13 | Amino acids, peptides and conjugates |
| 132.0899 | C5H12N2O2 | down | < 0.05 | 3 | GC-MS | Identified | Amino acids, peptides and conjugates | ||||
| Palmitic Acid ethyl ester | 284.2715 | C18H36O2 | 3 | up | < 0.01 | 11 | LC-MS | Id MS/MS | Fatty acyls | ||
| Phosphocholine | 183.0660 | C5H14NO4P | 2 | up | 0.011 | down | < 0.01 | 28 | LC-MS | Id MS/MS | Amines |
| Phytosphingosine | 317.2930 | C18H39NO3 | 3 | up | 0.012 | 8 | LC-MS | Id MS/MS | Sphingolipids and spingoid bases | ||
| Proline | 115.0633 | C5H9NO2 | 1 | up | < 0.05 | 11 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| Putrescine | 88.1000 | C4H12N2 | 6 | down | < 0.01 | 4 | CE-MS | Putative | Amines | ||
| 88.1000 | C4H12N2 | up | < 0.01 | 5 | GC-MS | Identified | Amines | ||||
| Ribitol / Arabitol / Xylitol | 152.0685 | C5H12O5 | up | < 0.01 | up | < 0.05 | 4 | GC-MS | Identified | Carbohydrates | |
| Ribose / Lyxose | 150.0528 | C5H10O5 | down | < 0.05 | down | < 0.05 | 4 | GC-MS | Identified | Carbohydrates | |
| Ribose-5-phosphate | 228.0046 | C5H9O8P | up | < 0.01 | up | < 0.01 | 7 | GC-MS | Identified | Carbohydrates | |
| S-Adenosylhomocysteine | 384.1216 | C14H20N6O5S | 0 | up | < 0.05 | 24 | CE-MS | Putative | Purines/pyrimidines and conjugates | ||
| S-Adenosylmethionine | 398.1372 | C15H22N6O5S | 2 | down | < 0.05 | up | < 0.05 | 5 | CE-MS | Putative | Purines/pyrimidines and conjugates |
| Serine | 105.0426 | C3H7NO3 | down | < 0.01 | down | < 0.01 | 3 | GC-MS | Identified | Amino acids, peptides and conjugates | |
| Sorbitol / Mannitol | 182.0790 | C6H14O6 | down | < 0.01 | 3 | GC-MS | Identified | Carbohydrates | |||
| Threonine | 119.0582 | C4H9NO3 | up | < 0.01 | 6 | GC-MS | Identified | Amino acids, peptides and conjugates | |||
| Threonine / Homoserine | 119.0582 | C4H9NO3 | 0 | up | < 0.05 | 14 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| Trypanothione disulfide | 721.2887 | C27H47N9O10S2 | 0 | down | < 0.01 | up | < 0.05 | 1 | CE-MS | Putative | Amino acids, peptides and conjugates |
| Tyrosine | 181.0739 | C9H11NO3 | 0 | up | < 0.05 | 4 | CE-MS | Putative | Amino acids, peptides and conjugates | ||
| 181.0739 | C9H11NO3 | up | < 0.05 | NF | GC-MS | Identified | Amino acids, peptides and conjugates | ||||
| Valine / Betaine / 5-Aminopentanoate | 117.0790 | C5H11NO2 | 1 | down | < 0.01 | up | < 0.05 | 7 | CE-MS | Putative | Amino acids, peptides and conjugates |
| Xanthine | 152.0334 | C5H4N4O2 | up | < 0.05 | 2 | GC-MS | Identified | Purines/pyrimidines and conjugates |
Table 3. Compounds identified by LC-MS/MS and their characteristic fragments.
| Compound name | Mass (Da) | Molecular formula | Mass error (ppm) | Fragments |
|---|---|---|---|---|
| (4,8,10-d18:3) Sphingosine | 295.2511 | C18H33NO2 | 2 | 57.07, 81.07, 95.08, 121.10, 162.10, 204.11, 222.12, 279.23 |
| Allantoic acid | 176.0546 | C4H8N4O4 | 10 | 43.06, 60.06, 70.06, 71.05, 116.07, 130.09, 134.08, 175.19, 176.10, 177.10 |
| Aspartyl-leucine | 246.1216 | C10H18N2O5 | 15 | 70.03, 86.09, 132.10, 141.10, 155.11, 201.12, 247.13 |
| Bis(glutathionyl)spermine | 780.3622 | C30H56N10O10S2 | 13 | 70.06, 72.08, 86.09, 104.10, 143.03, 158.96, 184.07, 522.75 |
| C17 Sphinganine | 287.2824 | C17H37NO2 | 4 | 69.07, 90.05, 121.10, 164.87, 194.98, 196.97, 227.19, 254.25, 272.25 |
| Glycerophosphocholine | 257.1028 | C8H20NO6P | 1 | 60.08, 86.09, 104.11, 104.18, 105.11, 124.99, 184.07, 258.11 |
| Lauric acid | 200.1776 | C12H24O2 | 1 | 29.04, 41.04, 43.05, 55.05, 57.07, 69.07, 71.08, 89.06, 103.07, 117.09, 131.10, 151.86, 172.86, 183.85, 201.12 |
| Lauric Acid ethyl ester | 228.2089 | C14H28O2 | 1 | 43.05, 57.07, 71.09, 89.06, 103.07, 117.09, 229.22 |
| Linolenic Acid | 278.2246 | C18H30O2 | 0 | 55.06, 67.06, 81.07, 95.09, 109.10, 123.12, 137.13, 173.13, 279.23 |
| Myristic Acid ethyl ester | 256.2402 | C16H32O2 | 1 | 43.05, 57.07, 71.09, 89.06, 103.07, 117.09, 257.25 |
| Norleucine | 131.0946 | C6H13NO2 | 19 | 30.03, 41.04, 42.04, 43.06, 44.05, 57.07, 69.07, 86.09, 130.16, 131.16, 132.09 |
| Palmitic Acid ethyl ester | 284.2715 | C18H36O2 | 3 | 41.04, 43.06, 55.06, 57.07, 69.07, 71.09, 85.10, 89.06, 95.09, 103.08, 117.09, 135.12, 149.13, 173.15, 201.19, 229.21 |
| Phosphocholine | 183.0660 | C5H14NO4P | 2 | 45.04, 60.08, 86.1, 98.98, 124.99, 184.07 |
| Phytosphingosine | 317.2930 | C18H39NO3 | 3 | 41.04, 55.02, 71.05, 95.05, 113.06, 135.11, 171.10, 219.17, 252.26, 282.27, 300.28, 318.30 |
According to biochemical criteria, amino acids, peptides and conjugates were the largest category of identified compounds (Fig 3) accounting for at least 22% of the total obtained for the different techniques, followed by organic acids and amines. Nevertheless, the most important differences came from the comparison among techniques. In CE-MS the amino acids, peptides and conjugates were the largest group (67%) followed by purines, pyrimidines and their conjugates (13%) and by organic acids (10%). In GC-MS, amino acids (42%), carbohydrates (27%) and purines and pyrimidines (12%) were identified as the main groups; most importantly, the carbohydrates and the sterol and prenylated lipids were exclusively identified in this analytical platform. Finally, in LC-MS/MS, the most populated category corresponded to fatty acids (36%), followed by amino acids (22%), and sphingolipids and spingoid bases (21%). Fatty acids, sphingolipids and glycerophospholipids were uniquely identified by this technique.
Fig 3. Biochemical classification.
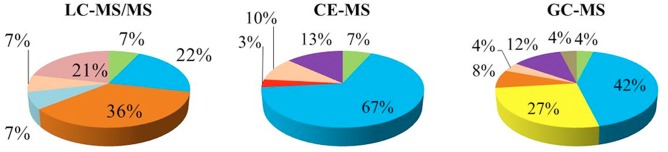
Biochemical classification of the identified compounds per each technique expressed as percentage. Green: amines. Cyan: amino acids, peptides and conjugates. Yellow: carbohydrates. Orange: fatty acids. Light blue: glycerophospholipids. Red: ketones and aldehydes. Ochre: organic acids. Purple: purines, pyrimidines and conjugates. Pink: sphingolipids and spingoid bases. Brown: sterol and prenol lipids.
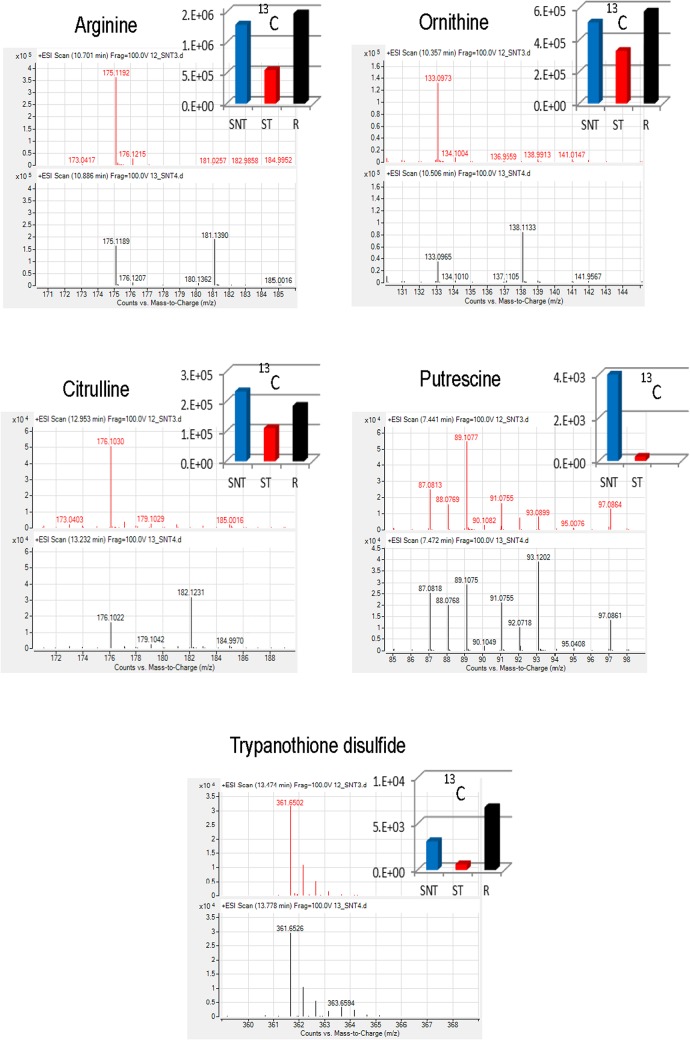
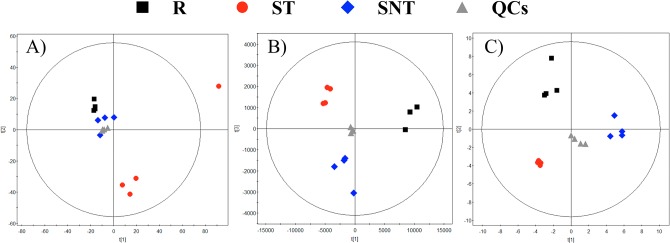
In order to pinpoint the metabolic origin of those compounds with statistically significant differences, the same experiment was repeated in the presence of the 13C isotopic form of L-arginine uniformly labelled in the culture medium. Once metabolomic analysis by LC-MS and CE-MS was carried out, multivariate analysis of the results showed a significant separation among the groups (Fig 4); this can be considered an additional biological validation of the first experiment. The list of confirmed compounds with 13C and their average abundance per group is shown in Fig 5.
Fig 4. PCA models for 13C.
PCA models for the whole data set filtered according to their presence in at leasts 50% of the QCs for the 13C arginine experiment. Panels: A LC-MS. 3 components. R2 = 0.428. Q2 = 0.107. B.- CE-MS. 2 components. R2 = 0.577. Q2 = 0.424.
Fig 5. 13C labeled compound.
13C labeled compound and their average abundance per group for the 13C-L-arginine experiment. Spectra from 12C and 13C samples appeared in red and black, respectively. Average abundance for each isotope appeared in the histogram.
Their most important effect consists in the deterioration of the thiol-dependent redox system of the parasite, leading to an increasing vulnerability against oxidative stress from its own metabolism, aggravated by the mitochondrial dysfunction caused by Sb3+ [46], and from the leishmanicidal mechanisms of the macrophage as host cell for Leishmania. This shortage of glutathione-derived metabolites may also affect glyoxal detoxification or ascorbate reduction pathways, as well as the levels of glutathionylated proteins [47,48].
The metabolic retooling of Leishmania to cope the metalloid stress can be gathered under major metabolic groups.
On one hand, when compared with untreated parasites (SNT), Sb3+ treated promastigotes (ST) underwent a depletion of metabolites (ornithine, putrescine, and trypanothione) across the arginine → polyamine → trypanothione axis, likely impairing its ability to control the homeostasis of its thiol-dependent intracellular redox environment [49]. It was previously proved that antimonial treatment increased the need of biosynthesis of trypanothione lost by its conjugation to Sb3+ to detoxify the parasite [13]. The maintenance of arginine levels, as initial biosynthetic precursor for polyamines, ruled out defects in its uptake, a highly regulated system, [50] as the origin for the levels of downstream metabolites of this pathway. Preliminary experiments of fluxomics, using 13C-arginine led to a significant incorporation of 13C label into ornithine, citrulline, putrescine and trypanothione disulfide (Fig 5). Furthermore, the level of thiols appeared consistently increased in antimonial resistant parasites. In this regard, the higher level of histidine, the biosynthetic precursor of ovothiol (N(1)-methyl-4-mercaptohistidine), may work as a compensation mechanism to recover an adequate redox power of the parasite. Unfortunately none significant variation for ovothiol was experimentally detected. In fact, glutathione-dependent thiols, but not ovothiol, increased in L. donovani antimonial resistant isolates [51]. The variation of metabolites inside this pathway with their corresponding trend for comparison of SNT with ST or R parasites is annotated in Figs 6 and 7.
Fig 6. Leishmania infantum altered metabolic pathways in ST vs SNT.
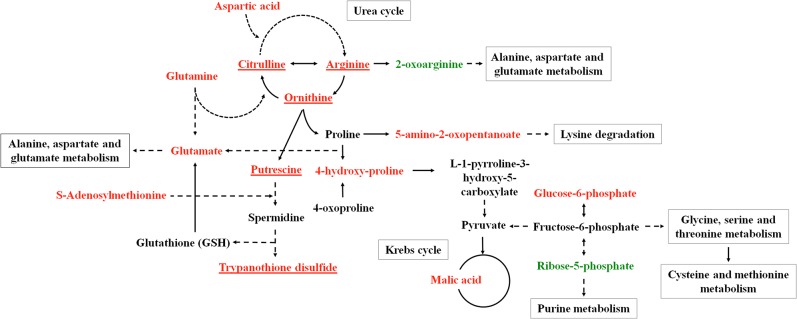
Leishmania infantum metabolic pathways with the higher divergence among the groups of parasites studied (ST vs SNT strains). Variation of the metabolites inside the metabolic pathways are represented in green (increase), red (decrease) or black (lack of statistical significance). Those that incorporates 13C appeared underlined.
Fig 7. Leishmania infantum altered metabolic pathways in R vs SNT.

Leishmania infantum metabolic pathways with the higher divergence among the groups of parasites studied (R vs SNT strains). Variation of the metabolites inside the metabolic pathways are represented in green (increase), red (decrease) or black (lack of statistical significance). Those that incorporates 13C appeared underlined.
A second observation in Sb3+ treated parasites concerns the decrease of metabolites used as bioenergetic substrates by the parasite trough glycolysis and oxidative phosphorylation, the latter being the main contributor [52]. Treated parasites (ST) showed decreased levels of i) glutamate and glutamine, two amino acids entering the TCA (tricarboxylic acid cycle) via α-ketoglutarate; ii) asparagine and aspartate that do via oxaloacetate, and iii) malate, a direct intermediate of the TCA. Furthermore, aspartate and glutamate, together with malate and α-ketoglutarate are involved in the electron transport from the cytoplasm into the mitochondria. In all, this discloses a dysfunctional Krebs cycle. Proline, another important amino acid as bioenergetic source in Leishmania, did not undergo a noticeable decrease.
A recent work on the metabolic basis of the stringent response of the amastigotes inside macrophages, disclosed a major role of mitochondrial metabolism in this stage [53], hence of a higher susceptibility of this form to antimonials [54], as supported by the decrease of the mitochondrial electrochemical potential in Sb3+ treated L. donovani amastigotes [55]. Furthermore, in ST parasites, the levels of two glycolytic intermediates, glucose-6-phosphate and dihydroxyacetone phosphate decreased, making unlikely a higher contribution of glycolysis to compensate for the deficiency in oxidative phosphorylation, as observed for other drugs [56].
If the increase of ribose-5-phosphate is indicative of a higher levels of other intermediates of the pentose phosphate pathway, a higher generation of NADPH, involved in the defense against the oxidative stress created by Sb3+, would be inferred [57].
None of the aforementioned metabolic shortages extend into nucleic acid bases, in fact in ST parasites the level of adenosine is even increased. It must to be kept in mind that purinic bases were acquired by savage mechanism; and that Leishmania is endowed both with a redundant and interwoven purine and pyrimidine metabolism with an easy interconversion among the molecular species, as well as with transporters with partial overlapping specificities for these bases [58,59].
The analysis of the metabolome for ST parasites suggests potential modification of their membrane composition and architecture. Concerning the lipid bilayer, antimonial treatment lead to a decrease in the ergosterol level, the main sterol in the plasma membrane; whether this is related to the concomitant decrease of leucine, ultimately responsible for ergosterol biosynthesis as its carbon skeleton is incorporated into the mevalonate pathway [60] will require further experiments. Secondly, myo-inositol also appeared decreased; 5–10% of the total Leishmania lipid is inositol phosphorylceramide [61], and myo-inositol the glycosidic moiety of the glycosylphosphatidylinositol anchor that tethers the main components of the plasma membrane to the phospholipid bilayer, such as lipophosphoglycan, Gp63, Gp46 or glycosylinositol phospholipids, with important roles in virulence and survival of the parasite [62]. The decrease in myristate may also impair the N-myristoylation of proteins, a modification required for the functionality of some proteins involved in membrane traffic and signaling [63]. Finally, the increase in choline and specially in glycerophosphocholine points towards an enhanced degradation of phospholipids by phospholipases. If so, an increased fatty acid availability is presumed, which may compensate as energetic fuel of the TCA activity, impaired by the general amino acid shortage. Likewise, concurrent to this membrane remodeling, alanine deficiency, one of the main osmolytes for Leishmania, will decrease the capacity of the parasite to cope with osmotic stress [64], which presumably will have an especial relevance for the survival inside the gut of the invertebrate host.
The changes in the metabolome of the resistant parasites showed an opposite trend to ST parasites: R parasites either maintained or increased the pools for metabolites that underwent a decrease in ST, in agreement with the increase in fitness in L. donovani antimonial resistant parasites [65]. Since macromolecular synthesis is tightly regulated, we infer that most of the changes observed can be interpreted under a metabolic perspective, in agreement with the high percentage of metabolic proteins with significant variation in their expression in antimonials resistance in L. infantum [17].
Metabolites allotted to arginine-polyamine-trypanothione pathway increased in R parasites over the untreated parasites (SNT) (Fig 7). This encompasses arginine, as the precursor at the top of the pathway, as well as ornithine, a metabolic intermediate, and trypanothione, the final product. In this trend, even the levels of citrulline and 2-oxoarginine, as beacons for minor diverting metabolism of arginine, also appeared decreased in R parasites.
In the same way we may interpret the rise of S-adenosyl methionine, required for spermidine biosynthesis and for the transulfuration in the biosynthesis of cysteine. Interestingly, the S-adenosylmethionine synthase was identified in proteomics of L. panamensis resistant to Sb3+ [36]. In other works overexpression of this enzyme leads to an increase in cysteine [66]. As this was not observed in our results, it may be likely due to a major allocation of this amino acid into the trypanothione biosynthesis. Moreover the levels of glutamate are increased; aside from its role inside the TCA cycle, is one of the amino acids involved in glutathione biosynthesis. Glycine and serine, linked through the triad glycine-serine-cysteine to glutathione biosynthesis, appeared unchanged or decreased. Due to the inconversion for these three amino acids [67], the demand of cysteine will be covered by a higher participation of trans-sulfuration pathway over de novo biosynthesis [68].
Similar to ST group, a decrease in glucose-6-phosphate and increase in ribose 5-phosphate were observed in R parasites However, R parasites showed a recovery of dihydroxyacetone phosphate level, a glycolytic metabolite located downstream glycolysis, so despite the decrease in glucose-6-phosphate, levels for glycolytic metabolites appeared to be enhanced in these parasites. In fact, for clinical- and in vitro-raised antimonial resistant Leishmania isolates from clinical or laboratory origins, higher levels of glycolytic and TCA cycle were disclose by proteomic studies [17,19]. In tune with this, R parasites showed an increment of proline and glutamate, both precursors for TCA intermediates, but the decrease in malate clashes against this scheme; its distribution among glycosome, mitochondria and cytoplasm may blur local variations, we lack a satisfactory explanation to account for this decrease.
Opposite to ST parasites, the signs for architectural remodeling of the membrane are subtler than for R parasites, as for these parasites glycerophosphocholine and myoinositol levels were similar to SNT and increased for myristic acid. Whereas in ST overall PUFA decreased, in R parasites linoleic showed the opposite trends. Nevertheless, the most remarkable changes in membrane components for resistant parasites are sphingolipid and sphingoid bases, C17-sphinganine increased whereas 4,8,12 d:18:3 sphingosine decreased. As in Leishmania myristoyl-CoA is preferred over palmitoyl-CoA for conjugation with serine [69], from a biosynthesis perspective a predominance of C16 over C17, thus the change in sphingoid bases will likely be acquired from scavenging from the serum of the medium.
Conclusions
We have demonstrated that Sb3+ mechanism of action and resistance is multifactorial, in agreement to other authors. The combination of the different systems involved gives rise to a heterogeneous pattern, where gene amplification, protein levels, and eventually the metabolome, will change according to the isolate, the species and the pathway to acquire resistance. In any case the metabolome fingerprinting strongly supports redox and bioenergetic metabolism as the axis for Sb3+ response. How this can be connected to the large percentage of proteins that underwent variation, is jeopardized as many of these were described as hypothetical proteins [17]. From our point of view, we have provided some valuable clues to solve part of this important puzzle in order to curtail parasite strategies aimed both to mitigate drug effects and to develop resistance. For these two purposes metabolomics may provide a strong support.
Acknowledgments
This work was supported by grants from Spanish Ministry of Economy and Competitiveness (CTQ2014-55279-R) (CB) and European Aeronautic Defence and Space Company – Construcciones Aeronáuticas SA and Brazilian Air Force (FAB) (GC and EC) and from FIS PS12-02706 and VI PN de I+D+I 2008–2011, Instituto de Salud Carlos III Subdirección General de Redes y Proyectos Cooperativos RD 12/0018/0007 (http://www.isciii.es/ISCIII/es/) and RD06/0021/0006-FEDER (LR). DR received a fellowship from the Spanish Ministry of Economy and Competitiveness (formerly MICINN). G. A. B. C. is thankful to FAPESP for a graduate fellowship (http://www.fapesp.br/en/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Data are publicly available in the link: http://dspace.ceu.es/handle/10637/6882.
Funding Statement
This work was supported by grants from Spanish Ministry of Economy and Competitiveness (CTQ2014-55279-R) (CB) and European Aeronautic Defence and Space Company – Construcciones Aeronáuticas SA and Brazilian Air Force (FAB) (GC and EC) and from FIS PS12-02706 and VI PN de I+D+I 2008-2011, Instituto de Salud Carlos III Subdirección General de Redes y Proyectos Cooperativos RD 12/0018/0007 (http://www.isciii.es/ISCIII/es/) and RD06/0021/0006-FEDER (LR). DR received a fellowship from the Spanish Ministry of Economy and Competitiveness (formerly MICINN). GABC is thankful to FAPESP for a graduate fellowship (http://www.fapesp.br/en/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. (2012) Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLoS One 7: e35671 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sundar S (2001) Drug resistance in Indian visceral leishmaniasis. Trop Med Int Health 6: 849–854. [DOI] [PubMed] [Google Scholar]
- 3. Solano-Gallego L, Miro G, Koutinas A, Cardoso L, Pennisi MG, Ferrer L, et al. (2011) LeishVet guidelines for the practical management of canine leishmaniosis. Parasit Vectors 4: 86 10.1186/1756-3305-4-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez U, Pinart M, Rengifo-Pardo M, Macaya A, Alvar J, Tweed JA (2009) Interventions for American cutaneous and mucocutaneous leishmaniasis. Cochrane Database Syst Rev: CD004834. [DOI] [PubMed]
- 5. van Griensven J, Balasegaram M, Meheus F, Alvar J, Lynen L, Boelaert M (2010) Combination therapy for visceral leishmaniasis. The Lancet infectious diseases 10: 184–194. 10.1016/S1473-3099(10)70011-6 [DOI] [PubMed] [Google Scholar]
- 6. Choudhury K, Zander D, Kube M, Reinhardt R, Clos J (2008) Identification of a Leishmania infantum gene mediating resistance to miltefosine and SbIII. Int J Parasitol 38: 1411–1423. 10.1016/j.ijpara.2008.03.005 [DOI] [PubMed] [Google Scholar]
- 7. Kumar D, Kulshrestha A, Singh R, Salotra P (2009) In vitro susceptibility of field isolates of Leishmania donovani to Miltefosine and amphotericin B: correlation with sodium antimony gluconate susceptibility and implications for treatment in areas of endemicity. Antimicrob Agents Chemother 53: 835–838. 10.1128/AAC.01233-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moreira DS, Monte Neto RL, Andrade JM, Santi AM, Reis PG, Frézard F, et al. (2013) Molecular characterization of the MRPA transporter and antimony uptake in four New World Leishmania spp. susceptible and resistant to antimony. Int J Parasitol Drugs Drug Resist 3: 143–153. 10.1016/j.ijpddr.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seblova V, Oury B, Eddaikra N, Ait-Oudhia K, Pratlong F, Gazanion E, et al. (2014) Transmission potential of antimony-resistant leishmania field isolates. Antimicrob Agents Chemother 58: 6273–6276. 10.1128/AAC.02406-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mukherjee B, Mukhopadhyay R, Bannerjee B, Chowdhury S, Mukherjee S, Naskar K, et al. (2013) Antimony-resistant but not antimony-sensitive Leishmania donovani up-regulates host IL-10 to overexpress multidrug-resistant protein 1. Proc Natl Acad Sci U S A 110: E575–582. 10.1073/pnas.1213839110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vanaerschot M, Huijben S, Van den Broeck F, Dujardin JC (2014) Drug resistance in vectorborne parasites: multiple actors and scenarios for an evolutionary arms race. FEMS Microbiol Rev 38: 41–55. 10.1111/1574-6976.12032 [DOI] [PubMed] [Google Scholar]
- 12. Ashutosh, Sundar S, Goyal N (2007) Molecular mechanisms of antimony resistance in Leishmania. J Med Microbiol 56: 143–153. [DOI] [PubMed] [Google Scholar]
- 13. Wyllie S, Cunningham ML, Fairlamb AH (2004) Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J Biol Chem 279: 39925–39932. [DOI] [PubMed] [Google Scholar]
- 14. Guimond C, Trudel N, Brochu C, Marquis N, El Fadili A, Peytavi R, et al. (2003) Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Res 31: 5886–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haimeur A, Guimond C, Pilote S, Mukhopadhyay R, Rosen BP, Poulin R, et al. (1999) Elevated levels of polyamines and trypanothione resulting from overexpression of the ornithine decarboxylase gene in arsenite-resistant Leishmania. Mol Microbiol 34: 726–735. [DOI] [PubMed] [Google Scholar]
- 16. Ashutosh, Garg M, Sundar S, Duncan R, Nakhasi HL, Goyal N (2012) Downregulation of mitogen-activated protein kinase 1 of Leishmania donovani field isolates is associated with antimony resistance. Antimicrob Agents Chemother 56: 518–525. 10.1128/AAC.00736-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brotherton MC, Bourassa S, Leprohon P, Legare D, Poirier GG, Droit A, et al. (2013) Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PLoS One 8: e81899 10.1371/journal.pone.0081899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Das S, Shah P, Baharia RK, Tandon R, Khare P, Sundar S, et al. (2013) Over-expression of 60s ribosomal L23a is associated with cellular proliferation in SAG resistant clinical isolates of Leishmania donovani. PLoS Negl Trop Dis 7: e2527 10.1371/journal.pntd.0002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biyani N, Singh AK, Mandal S, Chawla B, Madhubala R (2011) Differential expression of proteins in antimony-susceptible and-resistant isolates of Leishmania donovani. Mol Biochem Parasitol 179: 91–99. 10.1016/j.molbiopara.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 20. Kumar A, Sisodia B, Misra P, Sundar S, Shasany AK, Dube A (2010) Proteome mapping of overexpressed membrane-enriched and cytosolic proteins in sodium antimony gluconate (SAG) resistant clinical isolate of Leishmania donovani. Br J Clin Pharmacol 70: 609–617. 10.1111/j.1365-2125.2010.03716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou Y, Messier N, Ouellette M, Rosen BP, Mukhopadhyay R (2004) Leishmania major LmACR2 is a pentavalent antimony reductase that confers sensitivity to the drug pentostam. J Biol Chem 279: 37445–37451. [DOI] [PubMed] [Google Scholar]
- 22. Fyfe PK, Westrop GD, Silva AM, Coombs GH, Hunter WN (2012) Leishmania TDR1 structure, a unique trimeric glutathione transferase capable of deglutathionylation and antimonial prodrug activation. Proc Natl Acad Sci U S A 109: 11693–11698. 10.1073/pnas.1202593109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M (2005) Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol 57: 1690–1699. [DOI] [PubMed] [Google Scholar]
- 24. Gomez MA, Navas A, Marquez R, Rojas LJ, Vargas DA, Blanco VM, et al. (2014) Leishmania panamensis infection and antimonial drugs modulate expression of macrophage drug transporters and metabolizing enzymes: impact on intracellular parasite survival. J Antimicrob Chemother 69: 139–149. 10.1093/jac/dkt334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wyllie S, Mandal G, Singh N, Sundar S, Fairlamb AH, Chatterjee M (2010) Elevated levels of tryparedoxin peroxidase in antimony unresponsive Leishmania donovani field isolates. Mol Biochem Parasitol 173: 162–164. 10.1016/j.molbiopara.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wyllie S, Vickers TJ, Fairlamb AH (2008) Roles of trypanothione S-transferase and tryparedoxin peroxidase in resistance to antimonials. Antimicrob Agents Chemother 52: 1359–1365. 10.1128/AAC.01563-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muniz-Junqueira MI, de Paula-Coelho VN (2008) Meglumine antimonate directly increases phagocytosis, superoxide anion and TNF-alpha production, but only via TNF-alpha it indirectly increases nitric oxide production by phagocytes of healthy individuals, in vitro. Int Immunopharmacol 8: 1633–1638. 10.1016/j.intimp.2008.07.011 [DOI] [PubMed] [Google Scholar]
- 28. Pathak MK, Yi T (2001) Sodium stibogluconate is a potent inhibitor of protein tyrosine phosphatases and augments cytokine responses in hemopoietic cell lines. J Immunol 167: 3391–3397. [DOI] [PubMed] [Google Scholar]
- 29. Alvar J, Aparicio P, Aseffa A, Den Boer M, Canavate C, Dedet JP, et al. (2008) The relationship between leishmaniasis and AIDS: the second 10 years. Clin Microbiol Rev 21: 334–359, table of contents. 10.1128/CMR.00061-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perry MR, Wyllie S, Raab A, Feldmann J, Fairlamb AH (2013) Chronic exposure to arsenic in drinking water can lead to resistance to antimonial drugs in a mouse model of visceral leishmaniasis. Proc Natl Acad Sci U S A 110: 19932–19937. 10.1073/pnas.1311535110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Downing T, Stark O, Vanaerschot M, Imamura H, Sanders M, et al. (2012) Genome-wide SNP and microsatellite variation illuminate population-level epidemiology in the Leishmania donovani species complex. Infect Genet Evol 12: 149–159. 10.1016/j.meegid.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh N, Almeida R, Kothari H, Kumar P, Mandal G, Chatterjee M, et al. (2007) Differential gene expression analysis in antimony-unresponsive Indian kala azar (visceral leishmaniasis) clinical isolates by DNA microarray. Parasitology 134: 777–787. [DOI] [PubMed] [Google Scholar]
- 33. Vanaerschot M, Decuypere S, Downing T, Imamura H, Stark O, De Doncker S, et al. (2012) Genetic markers for SSG resistance in Leishmania donovani and SSG treatment failure in visceral leishmaniasis patients of the Indian subcontinent. J Infect Dis 206: 752–755. 10.1093/infdis/jis424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. El Fadili K, Drummelsmith J, Roy G, Jardim A, Ouellette M (2009) Down regulation of KMP-11 in Leishmania infantum axenic antimony resistant amastigotes as revealed by a proteomic screen. Exp Parasitol 123: 51–57. 10.1016/j.exppara.2009.05.013 [DOI] [PubMed] [Google Scholar]
- 35. Matrangolo FS, Liarte DB, Andrade LC, de Melo MF, Andrade JM, Ferreira RF, et al. (2013) Comparative proteomic analysis of antimony-resistant and-susceptible Leishmania braziliensis and Leishmania infantum chagasi lines. Mol Biochem Parasitol 190: 63–75. 10.1016/j.molbiopara.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 36. Walker J, Gongora R, Vasquez JJ, Drummelsmith J, Burchmore R, Roy G, et al. (2012) Discovery of factors linked to antimony resistance in Leishmania panamensis through differential proteome analysis. Mol Biochem Parasitol 183: 166–176. 10.1016/j.molbiopara.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 37. Vincent IM, Weidt S, Rivas L, Burgess K, Smith TK, Ouellette M (2014) Untargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolism. Int J Parasitol Drugs Drug Resist 4: 20–27. 10.1016/j.ijpddr.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. t'Kindt R, Scheltema RA, Jankevics A, Brunker K, Rijal S, Dujardin JC, et al. (2010) Metabolomics to unveil and understand phenotypic diversity between pathogen populations. PLoS Negl Trop Dis 4: e904 10.1371/journal.pntd.0000904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scheltema RA, Decuypere S, T'Kindt R, Dujardin JC, Coombs GH, Breitling R (2010) The potential of metabolomics for Leishmania research in the post-genomics era. Parasitology 137: 1291–1302. 10.1017/S0031182009992022 [DOI] [PubMed] [Google Scholar]
- 40. Canuto GA, Castilho-Martins EA, Tavares M, Lopez-Gonzalvez A, Rivas L, Barbas C (2012) CE-ESI-MS metabolic fingerprinting of Leishmania resistance to antimony treatment. Electrophoresis 33: 1901–1910. 10.1002/elps.201200007 [DOI] [PubMed] [Google Scholar]
- 41. Berg M, Mannaert A, Vanaerschot M, Van Der Auwera G, Dujardin JC (2013) (Post-) Genomic approaches to tackle drug resistance in Leishmania. Parasitology 140: 1492–1505. 10.1017/S0031182013000140 [DOI] [PubMed] [Google Scholar]
- 42. Requena JM (2012) Lights and shadows on gene organization and regulation of gene expression in Leishmania. Front Biosci 17: 2069–2085. [DOI] [PubMed] [Google Scholar]
- 43. De Gaudenzi JG, Noe G, Campo VA, Frasch AC, Cassola A (2011) Gene expression regulation in trypanosomatids. Essays Biochem 51: 31–46. 10.1042/bse0510031 [DOI] [PubMed] [Google Scholar]
- 44. Vincent IM, Barrett MP (2015) Metabolomic-Based Strategies for Anti-Parasite Drug Discovery. J Biomol Screen 20: 44–55. 10.1177/1087057114551519 [DOI] [PubMed] [Google Scholar]
- 45. Gika HG, Macpherson E, Theodoridis GA, Wilson ID (2008) Evaluation of the repeatability of ultra-performance liquid chromatography-TOF-MS for global metabolic profiling of human urine samples. J Chromatogr B Analyt Technol Biomed Life Sci 871: 299–305. 10.1016/j.jchromb.2008.05.048 [DOI] [PubMed] [Google Scholar]
- 46. Mehta A, Shaha C (2006) Mechanism of metalloid-induced death in Leishmania spp.: role of iron, reactive oxygen species, Ca2+, and glutathione. Free Radic Biol Med 40: 1857–1868. [DOI] [PubMed] [Google Scholar]
- 47. Flohe L (2012) The trypanothione system and its implications in the therapy of trypanosomatid diseases. Int J Med Microbiol 302: 216–220. 10.1016/j.ijmm.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 48. Manta B, Comini M, Medeiros A, Hugo M, Trujillo M, Radi R (2013) Trypanothione: a unique bis-glutathionyl derivative in trypanosomatids. Biochim Biophys Acta 1830: 3199–3216. 10.1016/j.bbagen.2013.01.013 [DOI] [PubMed] [Google Scholar]
- 49. Krauth-Siegel RL, Leroux AE (2012) Low-molecular-mass antioxidants in parasites. Antioxid Redox Signal 17: 583–607. 10.1089/ars.2011.4392 [DOI] [PubMed] [Google Scholar]
- 50. Castilho-Martins EA, Laranjeira da Silva MF, dos Santos MG, Muxel SM, Floeter-Winter LM (2011) Axenic Leishmania amazonensis promastigotes sense both the external and internal arginine pool distinctly regulating the two transporter-coding genes. PLoS One 6: e27818 10.1371/journal.pone.0027818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rai S, Bhaskar, Goel SK, Nath Dwivedi U, Sundar S, Goyal N (2013) Role of efflux pumps and intracellular thiols in natural antimony resistant isolates of Leishmania donovani. PLoS One 8: e74862 10.1371/journal.pone.0074862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tielens AG, Van Hellemond JJ (1998) Differences in energy metabolism between trypanosomatidae. Parasitol Today 14: 265–272. [DOI] [PubMed] [Google Scholar]
- 53. Saunders EC, Ng WW, Kloehn J, Chambers JM, Ng M, McConville MJ (2014) Induction of a stringent metabolic response in intracellular stages of Leishmania mexicana leads to increased dependence on mitochondrial metabolism. PLoS Pathog 10: e1003888 10.1371/journal.ppat.1003888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hart DT, Coombs GH (1982) Leishmania mexicana: energy metabolism of amastigotes and promastigotes. Exp Parasitol 54: 397–409. [DOI] [PubMed] [Google Scholar]
- 55. Sudhandiran G, Shaha C (2003) Antimonial-induced increase in intracellular Ca2+ through non-selective cation channels in the host and the parasite is responsible for apoptosis of intracellular Leishmania donovani amastigotes. J Biol Chem 278: 25120–25132. [DOI] [PubMed] [Google Scholar]
- 56. Manzano JI, Carvalho L, Perez-Victoria JM, Castanys S, Gamarro F (2011) Increased glycolytic ATP synthesis is associated with tafenoquine resistance in Leishmania major. Antimicrob Agents Chemother 55: 1045–1052. 10.1128/AAC.01545-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maugeri DA, Cazzulo JJ, Burchmore RJ, Barrett MP, Ogbunude PO (2003) Pentose phosphate metabolism in Leishmania mexicana. Mol Biochem Parasitol 130: 117–125. [DOI] [PubMed] [Google Scholar]
- 58. Boitz JM, Ullman B, Jardim A, Carter NS (2012) Purine salvage in Leishmania: complex or simple by design? Trends Parasitol 28: 345–352. 10.1016/j.pt.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carter NS, Yates P, Arendt CS, Boitz JM, Ullman B (2008) Purine and pyrimidine metabolism in Leishmania. Adv Exp Med Biol 625: 141–154. 10.1007/978-0-387-77570-8_12 [DOI] [PubMed] [Google Scholar]
- 60. Ginger ML, Chance ML, Sadler IH, Goad LJ (2001) The biosynthetic incorporation of the intact leucine skeleton into sterol by the trypanosomatid Leishmania mexicana. J Biol Chem 276: 11674–11682. [DOI] [PubMed] [Google Scholar]
- 61. Zhang K, Bangs JD, Beverley SM (2010) Sphingolipids in parasitic protozoa. Adv Exp Med Biol 688: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Novozhilova NM, Bovin NV (2010) Structure, functions, and biosynthesis of glycoconjugates of Leishmania spp. cell surface. Biochemistry (Mosc) 75: 686–694. [DOI] [PubMed] [Google Scholar]
- 63. Brannigan JA, Smith BA, Yu Z, Brzozowski AM, Hodgkinson MR, Maroof A, et al. (2010) N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J Mol Biol 396: 985–999. 10.1016/j.jmb.2009.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lefurgey A, Gannon M, Blum J, Ingram P (2005) Leishmania donovani amastigotes mobilize organic and inorganic osmolytes during regulatory volume decrease. J Eukaryot Microbiol 52: 277–289. [DOI] [PubMed] [Google Scholar]
- 65. Berg M, Vanaerschot M, Jankevics A, Cuypers B, Maes I, Mukherjee S, et al. (2013) Metabolic adaptations of Leishmania donovani in relation to differentiation, drug resistance, and drug pressure. Mol Microbiol 90: 428–442. 10.1111/mmi.12374 [DOI] [PubMed] [Google Scholar]
- 66. Perez-Pertejo Y, Reguera RM, Ordonez D, Balana-Fouce R (2006) Characterization of a methionine adenosyltransferase over-expressing strain in the trypanosomatid Leishmania donovani. Biochim Biophys Acta 1760: 10–19. [DOI] [PubMed] [Google Scholar]
- 67. Opperdoes FR, Michels PA (2008) The metabolic repertoire of Leishmania and implications for drug discovery In: Myler PJ, Fasel N, editors. Leishmania: After The Genome Norfolk, U.K Caister: Academic Press; pp. 123–158. [Google Scholar]
- 68. Williams RA, Westrop GD, Coombs GH (2009) Two pathways for cysteine biosynthesis in Leishmania major. Biochem J 420: 451–462. 10.1042/BJ20082441 [DOI] [PubMed] [Google Scholar]
- 69. Mina JG, Mosely JA, Ali HZ, Denny PW, Steel PG (2011) Exploring Leishmania major inositol phosphorylceramide synthase (LmjIPCS): insights into the ceramide binding domain. Org Biomol Chem 9: 1823–1830. 10.1039/c0ob00871k [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are publicly available in the link: http://dspace.ceu.es/handle/10637/6882.