Abstract
In bacteria, mechanisms that incorporate DNA into a genome without strand-transfer proteins such as RecA play a major role in generating novelty by horizontal gene transfer. We describe a new illegitimate recombination event in Escherichia coli K-12: RecA-independent homologous replacements, with very large (megabase-length) donor patches replacing recipient DNA. A previously uncharacterized gene (yjiP) increases the frequency of RecA-independent replacement recombination. To show this, we used conjugal DNA transfer, combining a classical conjugation donor, HfrH, with modern genome engineering methods and whole genome sequencing analysis to enable interrogation of genetic dependence of integration mechanisms and characterization of recombination products. As in classical experiments, genomic DNA transfer begins at a unique position in the donor, entering the recipient via conjugation; antibiotic resistance markers are then used to select recombinant progeny. Different configurations of this system were used to compare known mechanisms for stable DNA incorporation, including homologous recombination, F’-plasmid formation, and genome duplication. A genome island of interest known as the immigration control region was specifically replaced in a minority of recombinants, at a frequency of 3 X 10-12 CFU/recipient per hour.
Introduction
In prokaryotes, horizontal gene transfer (HGT; also called lateral gene transfer) is a massive force of evolutionary change and adaptation. It allows bacteria to adapt to ecological niches and survive in stressful conditions when traditional gene regulation is not sufficient [1]. This type of gene flow is so prevalent that it underlies many current topics in microbiology. For example, HGT is responsible for the global spread of antibiotic resistance [2,3], it shapes microbial communities like the human microbiome [4], and it is interconnected with biofilm formation [5], the CRISPR/cas immunity system [6], virulence [7,8], and other critical cellular functions. However, many aspects of HGT are still poorly understood and its overall effect on genomic evolution is the subject of active research [9].
The impact of HGT on genomic evolution is illustrated by variability of genome content within the E. coli species. In most E. coli strains, ~40% of the genome consists of variable gene islands [10,11]. These genetic elements typically encode niche-specific traits and account for the wide range of E. coli lifestyles [12]. The diversity among E. coli is so large that the pangenome, the full complement of genes discovered among E. coli strains, is estimated to be over 9 times larger than the core set of genes found in all E. coli [13]. The vast array of potential phenotypes encoded within the pangenome allows for the rapid development of new strains with niche-specific traits via genetic transfer [14,15].
For the purposes of this paper, we define HGT as the stable introduction of foreign DNA into a chromosome through a RecA-independent event [14]. This definition excludes RecA-mediated homologous recombination, which occurs at a much higher frequency than HGT. Although the literature does not always distinguish between homologous exchange and gene addition via HGT [16], we adopt a stricter definition to distinguish the distinct evolutionary consequences of the two processes. Homologous exchange tends to homogenize a population by selecting for the most favorable combination of traits within a community while HGT diversifies the population by introducing novel genetic material [17]. The two processes can complement each other. HGT can add a novel capability to one member of a population in a manner that homologous exchange cannot, and homologous exchange can then spread that capability more efficiently than the original HGT mechanisms that introduced it [14,18]. For example, a combination of HGT and homologous recombination is believed to have mediated the distribution of pathogenicity islands in E. coli that can turn a commensal strain into a virulent species [16].
A specific gene island of interest in Enterobacteriacea is known as the immigration control region (ICR). The ICR consists of a variable region of migratory genes surrounded by conserved framework genes (yjiPRS and yjiAXY) [19,20] (S1 Fig). This region shows signatures of high rates of homologous genetic exchange [14,21], but underlying that signature is a structure suggesting that it is also a likely target of site-specific replacement [22]. No mechanism for such replacement has been demonstrated.
Here we created a conjugal system to facilitate the study of RecA-independent horizontal transfer of the ICR in E. coli and relatives, and qualified its properties in the model strain K-12. This conjugal system proved to be an effective tool for discovering and analyzing novel mechanisms of horizontal gene transfer. Although it was designed to focus on a specific gene island (the ICR), it revealed a previously unreported capacity for large-chunk exchange in the absence of RecA. Most recombinants obtained arise from the replacement of large (0.1–2.4 Mb) segments of genomic DNA; about 1.4% have replaced only the ICR (~16 kb). We also identified the previously uncharacterized protein YjiP as an enhancer of recA-independent recombination.
Experimental Procedures
Strains, plasmids, and growth conditions
All strains, plasmids, and oligonucleotides used in this study are listed in S1 Table. Bacteria were grown in liquid Luria Broth (LB; 10 g/L Tryptone, 5 g/L yeast, 10 g/L NaCl) or rich broth (RB; 10 g/L Tryptone, 5 g/L yeast, 5 g/L NaCl, pH 7.2) medium at 37°C with vigorous shaking or on solid LB or RB medium containing 1.5% agar with appropriate selection. Ampicillin (Ap; 100 μg/mL) streptomycin (Sm; 100 μg/mL), kanamycin (Kn; 40 μg/mL), chloramphenicol (Cm; 30 μg/mL), tetracycline (Tc; 20 μg/mL), and nalidixic acid (Nl; 5 ug/mL in ΔrecA strains and 20 ug/mL in recA + strains) were used to select E. coli transformants. Plasmids were prepared from E. coli NEB Turbo cells. For temperature sensitive plasmids, 30°C incubation was used to permit plasmid replication and 42°C incubation was used to remove the plasmid.
Genetic and molecular techniques
Linear and plasmid DNA transformations were performed as described previously [23] as were transductions using the P1vir bacteriophage [24]. DNA constructs were created using the Gibson assembly kit (#E2611; New England Biolabs). Chromosomal gene deletions were generated using the λ Red recombinase system [25]. Unless otherwise stated, all PCR products were generated using E. coli MG1655 chromosomal DNA as a template and all strains were verified by PCR and/or sequence analysis (New England Biolabs DNA sequencing facility). PCR reactions used to generate sequencing templates or genetic constructions were performed with the Q5 High-Fidelity DNA Polymerase (#M0491; New England Biolabs) while diagnostic PCR reactions used the Hot Start Taq 2X Master Mix (#M0496; New England Biolabs). New England Biolabs produced all restriction enzymes and ligases used in these experiments.
Matings
Donor and recipient cultures were grown at 37°C to an OD600 of ~1.0 in RB with shaking. Mating was initiated by mixing the cultures in a 1:1 ratio. To ensure optimal stability of the F pilus, the mating mixtures were aerated with slow rolling in a tube roller. The duration of the mating was 0.25 h when the strains involved contained functional recA or repE genes because recombination efficiency was high. When the strains lacked both recA and repE, recombination efficiency was much lower, so mating duration was increased to 18 hours to yield sufficient recombinants. For rhamnose induction, rhamnose was added to recipients 3 hours before the start of the mating (when the mating duration was 15 minutes) or at the start of mating (when mating duration was 18 hours).
After mating, cells were diluted up to 10-6 and plated on antibiotic media selective for donors, for recipients, and for recombinants at dilutions that would yield at least 10–100 colonies per plate or as much as remained for recombinants. In matings with low recombination efficiency, up to 6 mL of the mating culture was centrifuged, re-suspended in residual broth, and spread out over multiple recombinant selective plates to obtain enough recombinants (1 mL culture per plate). The strain combinations, mating durations, and concentrations of antibiotics used for selection in all matings are described in S2 Table. Plates were incubated for 48h at 37°C and counted to calculate recombination efficiency and cell survival. Unmated donor and recipient cultures were subject to identical procedures to determine CFU/mL levels in unmated cells and to confirm that spontaneous mutants do not appear.
Characterization of recombinants
Recombinant colonies were purified once on the same selection media and tested for antibiotic resistance and UV sensitivity.
Antibiotic screens
Recombinants were streaked onto RB plates supplemented with kanamycin or chloramphenicol and incubated at 37°C. Strains that failed to grow or exhibited only sporadic colonies arising after 48 h were classified as sensitive to that antibiotic.
UV sensitivity screens
Recombinants were streaked horizontally across an RB plate alongside control rec + and rec - strains. Vertical portions of the plate were exposed to UV light (380 nm) for 0 to 60 seconds, then incubated in the dark for 24 h. Recombinants were classified as UV sensitive if they behaved similarly to the rec - control with only scattered colonies surviving at any exposure or as UV resistant if growth was confluent at UV exposure under 30 seconds with minor cell death at higher exposure times (as for the rec + control).
Testing the effect of rhamnose induction on cell viability
Cultures were grown to an OD600 of ~0.2 in RB and then split into multiple tubes. Cultures were treated with rhamnose concentrations ranging from 0–0.2% using a stock 20% L-rhamnose solution. Cells were grown at 37°C with constant shaking at high aeration levels for 3 hours. Every 30 minutes, a small aliquot of cells was diluted up to 10-6 and plated on streptomycin media. At 3 hours after rhamnose treatment, cells were split again and either mated with an equivalent volume of the donor strain ER3435 grown alongside the recipient or left untreated. Aliquots were plated after mating on streptomycin media at 30 minutes and 18 hours. Plates were incubated for 24h at 37°C and counted to determine cell survival. The CFU count for mated cells was doubled to account for dilution with the donor strain.
β-galactosidase assays
See supplemental methods (S1 File).
Genomic Sequencing
Illumina Sequencing
A detailed description of the Illumina Sequencing process can be found in the supplemental methods (S1 File).
Pacific Biosciences SMRT Sequencing
Genomic DNA samples were prepared for SMRT sequencing as previously described [26]. Briefly, genomic DNA from an overnight recombinant culture was purified using standard phenol-chloroform extraction methods and quantitated using a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, NY, USA). DNA samples were sheared to an average size of ~10 kb using Covaris G-tubes (Covaris, Woburn, MA, USA). Further DNA purification was achieved with MO BIO PowerClean DNA Clean-Up Kit (MO BIO Laboratories, Carlsbad, CA, USA) and AxyPrep Mag PCR Clean-up kit (Axygen, Union City, CA, USA). SMRTbell template libraries were prepared using the 10 kb preparation protocols provided by Pacific Biosciences (http://www.pacificbiosciences.com/samplenet/ProcedureChecklistLowInput10kbLibraryPreparationandSequencingMagBeadStation.pdf) and the reagents supplied in the SMRTbell Template Prep Kit 1.0 for DNA repair, ligation, and exonuclease digestion (Pacific Biosciences, Menlo Park, CA, USA). Products were verified for size using the Agilent 2100 Bioanalyzer and DNA 12000 Kits (Agilent Technologies, Santa Clara, CA, USA). Polymerase binding was accomplished with the DNA Polymerase Binding Kit 4.0, Magbeads, and the Magbead Buffer Kit (Pacific Biosciences, Menlo Park, CA, USA). Libraries were sequenced with the DNA Sequencing Kit 3.0 on a PacBio RS instrument and reads were analyzed with the Pacific Biosciences SMRT analysis software v2.2.0.
Variant visualization using Geneious
De novo assembled contigs were imported into Geneious 7.1.7 (Biomatters Ltd) and annotated. Edges were trimmed and the sequence was circularized to obtain a complete genome. Nucleotide #1 was set as the dnaA translation start site. Pairwise alignments of the two parents with each other and with each recombinant were made using Mauve and variants were visualized for each comparison. A more detailed explanation of this process can be found in the supplemental methods (S1 File). The annotated sequences are on deposit at NCBI: http://www.ncbi.nlm.nih.gov/bioproject/271807
Accession: PRJNA271807 ID: 271807
Statistical analysis
All experiments were performed with a minimum of three biological replicates. Unless otherwise noted, data is presented as mean ± standard error. Unpaired Student's t tests were used for statistical evaluation. A value of P ≤ 0.05 was considered statistically significant.
Results
The conjugal system
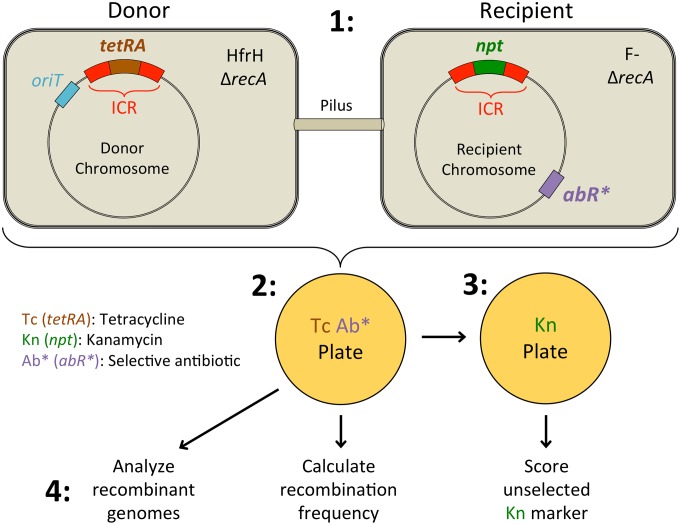
We designed a system that will efficiently introduce a donor copy of a variable gene island, the ICR, into a recipient cell with a different version of it (Fig 1, Table 1).
Fig 1. Model system for studying recombination events involving the ICR.
1: DNA transfer is mediated by a conjugal mating between an HfrH donor and a F- recipient. Transfer begins at oriT and proceeds in the direction of the nearby ICR. Both strains are ΔrecA to prevent homologous recombination. The donor contains a tetracycline resistance regulon (tetRA) within the ICR. The recipient carries unselected scoreable drug markers within the ICR, and counter-selection markers (resistance to streptomycin, nalidixic acid, and/or chloramphenicol) elsewhere (AbR*). During the mating, leading-strand DNA synthesis starting at oriT in donor displaces the complementary single strand, which is transferred to the recipient, where lagging-strand synthesis copies it. Transfer begins with oriT and moves clockwise through the tetRA-ICR construct. 2: Cells are plated on Tet + Ab* media, selecting for recombinants that have received a stable copy of tetRA in a recipient background. 3: Recombinants are tested for the unselected marker, kanamycin sensitivity, to see if the incorporated tetRA replaced npt). 4: In some experiments, parents and recombinants were analyzed by sequencing: Whole genomes were assembled and annotated, then aligned for identification of 450 segregating variant positions.
Table 1. Relevant features of the donor and recipient strains in the mating system.
| Donor Strain | Recipient Strain | ||
|---|---|---|---|
| Genotype | Function | Genotype | Function |
| oriT (HfrH) | Transfer of donor genomic DNA | F- | Cell can receive F DNA |
| traI-traM | F pilus and transfer replicon | recD1014 | Stabilize Invading DNA |
| ΔrecA | Prevent homologous recombination | ΔrecA | Prevent homologous recombination |
| ΔrepE* a | Prevent F plasmid formation | abR* b | Select for recipient cells |
| ICR(mrr)::tetAR | Select for donor DNA integration | ICR(yjiT-mrr)::npt or yjiP::cat yjiT::npt c | Unselected markers; Screen for ICR replacement |
a This mutation deletes the entire proximal F DNA segment from the donor strain, but if is referred to as ΔrepE* (rather than Δ(ygfA-ycaA)::(FRT)) for simplicity.
babR* refers to resistance markers for streptomycin (SmR; rpsL104), chloramphenicol (CmR; recA::cat(FRT)), and/or nalidixic acid (NlR; gyrA96)
cIn crosses 1–6 and 11–13, genotype is ICR(yjiT-mrr)::npt. In crosses 7–10 the genotype is yjiP::cat(FRT) yjiT::npt(FRT). See Fig 2 for a depiction of these genotypes.
The Donor
This system relies on Hfr conjugal transfer of the chromosome. A conjugal plasmid, the F (fertility) plasmid, is integrated near the locus of interest. DNA transfer begins at oriT within the F and proceeds in one direction (here, towards the adjacent ICR) at about 40 kb/min [27]. The entire donor genome is transferred in 100 min [28]. Transfer is rapid and efficient: most recipients in laboratory matings make contact with a donor within minutes [29] and subsequent DNA transfer occurs almost immediately [30]. In the HfrH strain we began with, oriT is ~250 kb from the ICR, ensuring that recipients will receive the donor ICR less than 15 minutes after mating is initiated. To prevent transfer of a wild-type copy of recA, the coding sequence was deleted from the donor as well. Selection for the donor ICR was enabled with an added tetracycline resistance marker (TcR; mrr::tetAtetR) (Fig 2). This disrupts a gene that is not a candidate for participation in recombination events.
Fig 2. The ICR region in the standard donors (ER3276 & ER3435), standard recipients (ER3436 & ER3460) and ICR proximal border recipients (ER3472, ER3473, ER3480, & ER3481) (not to scale).
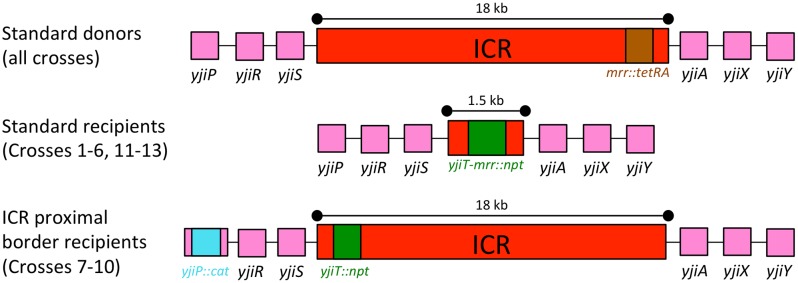
Recipients
The recipient ICR was treated as an unselected marker. In initial experiments, a kanamycin resistance cassette (KnR; Δ(yjiT-mrr)::npt(FRT)) replaces the entire ICR (Fig 2). Some of the later experiments involved recipients in which kanamycin and chloramphenicol resistance cassettes flank the proximal ICR border. In all recipients, distant drug resistance markers streptomycin (SmR; rpsL104), chloramphenicol (CmR; recA::cat(FRT)), and/or nalidixic acid (NlR; gyrA96) were used for counterselection. S2 Table lists antibiotic combinations used for selection in all matings. To stabilize the donor substrate in the recipient cell, the Exonuclease V activity of RecBCD was inactivated with a recD1014 mutation. This mutation increases recombination efficiency in RecA+ assays [30–33]. The (ΔrecA::cat(FRT)) marker removed recA from the recipient.
Validation experiments comparing the conjugal system
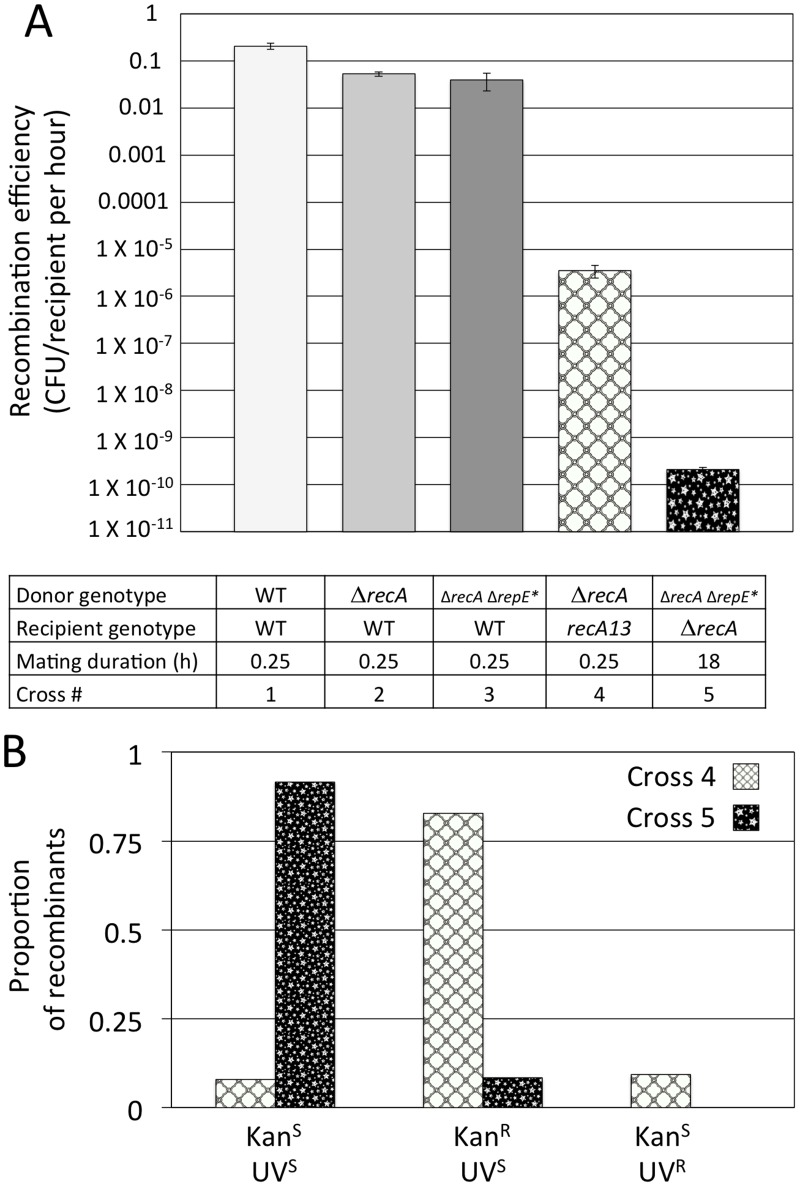
The plasticity of our conjugal system allows us to create multiple configurations that are each designed to study a different type of recombination event. This analysis provides us with a way to estimate the relative frequency of different mechanisms of genomic evolution. A complete analysis of the configurations tested can be found in S2 File, but a brief overview is provided here. The high efficiency of the system was confirmed with a RecA+ configuration in which ~20% of the recipients acquired tetracycline resistance from the donor per hour of mating (Fig 3A). Inactivating RecA reduced recombination efficiency ~10000 fold, but most recombinants from this configuration were created by the formation of novel F’-plasmids (S2 Fig). To suppress formation of these F' derivatives we deleted repE which is required for extrachromosomal vegetative replication [34] (S3 Fig). This further reduced mating efficiency to a low but measurable (2.1 ± 0.2) X 10-10 CFU/recipient per hour. Most recombinants were KnS, suggesting RecA-independent genomic replacement (Fig 3B).
Fig 3. The Effect of recA and repE on the mating system.
(A) The recombination efficiency per recipient per hour in mating crosses: 1) WT donor (ER3270) X WT recipient (ER1636), 2) ΔrecA donor (ER3276) X WT recipient (ER1636), 3) ΔrecA ΔrepE* donor (ER3435) X WT recipient (ER1636), 4) ΔrecA donor (ER3276) X recA recipient (ER3263), and 5) ΔrecA ΔrepE* donor (ER3435) X ΔrecA recipient (ER3436). See S2 Table for more details on individual crosses. The legend indicates status of recA and repE in the donor and recipient, mating duration, and which mating pair was used in each cross. Recombination efficiency is reduced by deleting recA from either the donor or recipient to eliminate homologous recombination, and with the ΔrepE* deletion in the donor to prevent F’-plasmid replication. All crosses were performed with a minimum of three biological replicates. Error bars represent standard error. (B) The proportion of recombinants that are kanamycin sensitive (KnS), kanamycin resistant (KnR), or UV resistant (UVR) from cross (4) ΔrecA donor X recA13 recipient and from cross (5) ΔrecA ΔrepE* donor X ΔrecA recipient. Most recombinants were KnR when the donor had repE in the leading F DNA because RepE allows for stabilization of donor DNA by plasmid replication (S3 Fig) [34]. In the absence of plasmid formation, crosses resulted primarily in KnS recombinants. In cross (4), UVR recombinants resulted from reversion of the ER3263 recipient’s recA13 point mutation allele [35,36]. To avoid this we deleted recA from recipient ER3436 for cross 5.
Regulated overproduction of candidate participant yjiP
After validating the system, we used it to test a candidate participant in RecA-independent genomic replacement. This was yjiP, an uncharacterized gene near the proximal border of the ICR (Fig 2). YjiP was hypothesized to contribute to site specific replacement at the ICR because it is highly conserved compared to the ICR contents [22], and it contains a PD-(D/E)XK phosphodiesterase domain suggesting nuclease activity [37]. Most E. coli strains encode a complete YjiP protein, but yjiP from E. coli K-12 (which our recipients are derived from) contains a point mutation that results in a premature stop codon. We synthesized a mutated K-12 yjiP lacking the premature stop codon (yjiPc, for yjiPcomplete), fused it to the rhamnose inducible promoter rhaBp, and integrated this rhaBp-yjiPc construct into the Tn7 attachment site of recipient strains [38]. The rhaBp promoter is essentially silent in the absence of rhamnose, and inducible 200-fold in its presence (S4A Fig).
A functional role for YjiP
YjiP expression increases the frequency of RecA-independent recombination
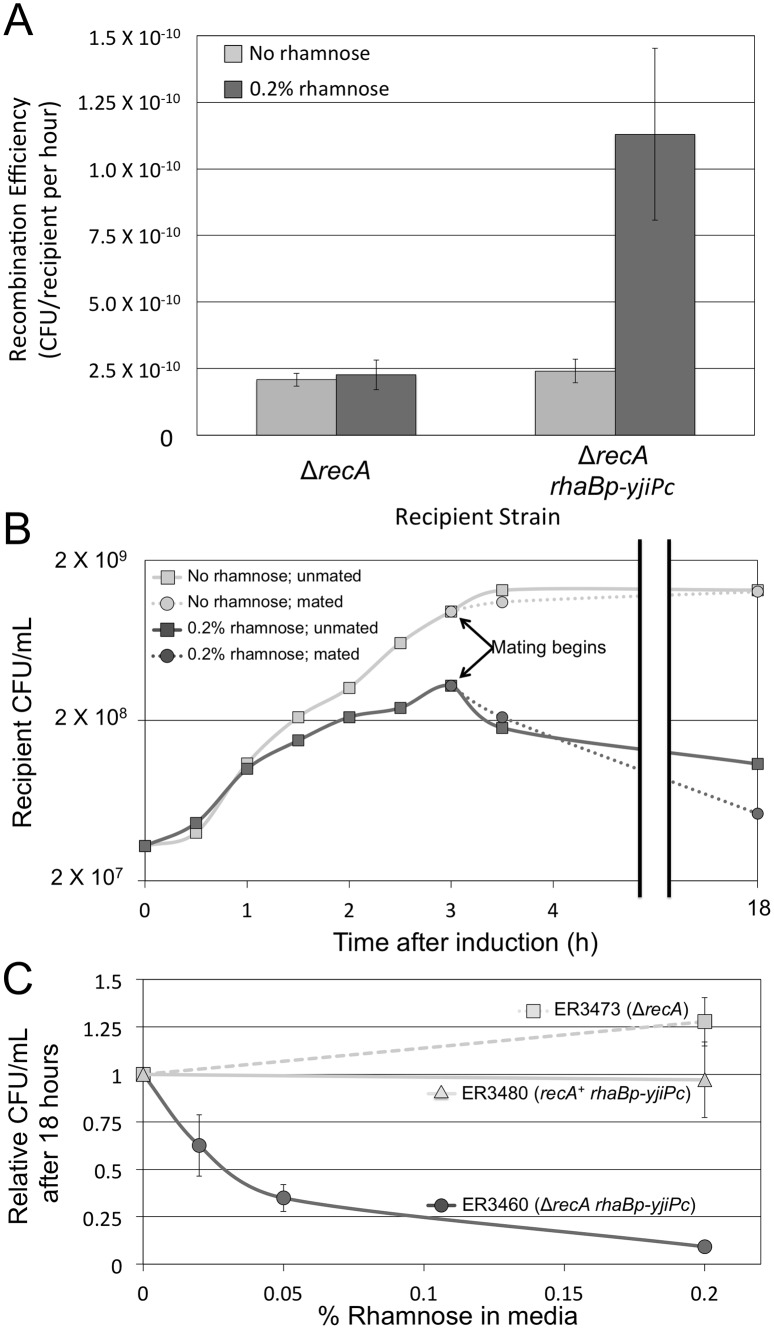
RecA-independent recombination efficiency increased significantly with increased yjiPc expression in the recipient. In a mating between the ΔrecA rhaBp-yjiPc recipient and the ΔrecA ΔrepE* donor (Cross 13), rhamnose addition increased recombination efficiency over 5 fold (Fig 4A). A similar increase in recombination efficiency upon yjiPc induction was observed when mating with a ΔrecA repE + donor, but yjiPc induction had no effect on recombination efficiency in a recA + recipient (Crosses 12 & 9 respectively, S4B and S4C Fig). A different ICR framework gene, yjiA, with a rhaBp-yjiA construct (ER3333), had no effect on recombination efficiency (Cross 11; S4C Fig). Thus, we conclude that yjiPc expression specifically increases the frequency of recA-independent recombination in our conjugal system.
Fig 4. YjiP promotes recA-independent recombination and reduces cell viability.
(A) The frequency of recombination during matings between the ΔrepE* ΔrecA donor and either the ΔrecA recipient (cross 6; ΔrecA) or a ΔrecA recipient with inducible overexpression of yjiP (cross 13; ΔrecA rhaBp-yjiPc), with and without 0.2% rhamnose. Recombination efficiency was calculated as the frequency of recombinant formation per viable recipient per hour in the mating mixture. Inducing yjiPc expression with rhamnose significantly increases recombination efficiency ~5 fold (P-value = 0.018), but rhamnose has no significant effect when the recipient lacks the rhaBp-yjiPc gene fusion. (B) Cell viability over time of the ΔrecA rhaBp-yjiPc recipient (ER3460) grown in 0.2% rhamnose. This experiment was performed three times with biological replicates, but only the results of a single representative trial are shown for clarity. Rhamnose was added at t = 0, and cells were mated after 3 hours of growth with ER3435. Untreated and unmated cells were also included as controls. Rhamnose-induced yjiPc expression reduces cell proliferation for the first three hours after treatment and begins to kill cells afterwards. Mating did not significantly affect cell viability. (C) Dose-response of cell killing: fraction starting titer for three strains at 18 hours as a function of inducer concentration. Strains were ΔrecA (ER3473), recA + rhaBp-yjiPc (ER3480), and ΔrecA rhaBp-yjiPc (ER3460) grown in various concentrations of rhamnose for 18 hours relative to an untreated control. Higher concentrations of rhamnose are increasingly lethal to ER3460; 0.2% rhamnose kills ~90% of the normally viable cells. All experiments in panels A and C were performed with a minimum of three biological replicates with error bars representing standard error.
YjiP expression reduces viability
In addition to increasing recombination efficiency, yjiPc expression reduced cell viability in ΔrecA recipients. The addition of 0.2% rhamnose, to the ΔrecA rhaBp-yjiPc recipient (ER3460) reduced the growth rate between 0–3 hours after treatment (as measured by CFU/mL on selective media) (Fig 4B). Cells began to die after 3 hours. At 3 hours, a surrogate reporter of expression potential (β-galactosidase) has reached a maximum (S4A Fig). By 18 hours CFU count was over 10 fold lower than an uninduced control. Cell death is dose-dependent when yjiPc is expressed; lower concentrations of rhamnose improved cell viability (Fig 4C) for the overexpresser, ER3460. Rhamnose did not significantly affect cell viability in a ΔrecA background without inducible yjiPc (ER3473) or when yjiPc is overexpressed in a recA + background (ER3480)
The mechanism by which yjiPc reduces cell viability is unclear, but clues suggest that DNA damage may occur. The non-viable cells do not appear to be lysing, as OD600 readings of a rhamnose treated culture are indistinguishable from a non-treated control (S4D Fig). The introduction of foreign DNA did not trigger cell death either: mating the yjiPc-expressing recipients with a donor strain did not immediately affect cell viability (Fig 4B). Mating itself may exacerbate this effect, as viability is reduced at the 18 hour timepoint.
We confirmed that yjiP overexpression results in DNA damage by testing its effect on the SOS response. A plasmid containing the rhamnose inducible yjiP construct was introduced into a rec + strain in which lacZ is fused to the DNA damage inducible dinD locus on the E. coli genome (dinDp-lacZ; [39]). Growing this strain (ER3544) on X-gal plates showed that the presence of rhamnose significantly increased lacZ expression (S4E Fig). Thus, we concluded that yjiP expression can cause enough DNA damage to result in an SOS response.
RecA-independent recombinants have replaced large segments of the recipient genome with the donor copy
Sequence characterization of recombinants
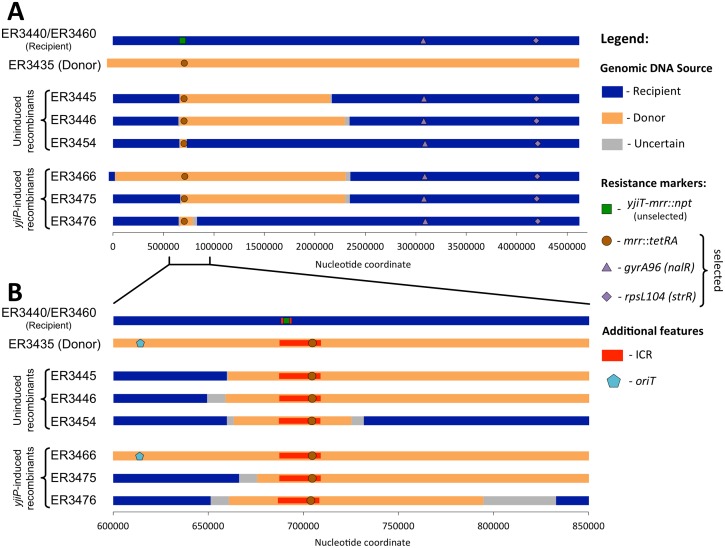
Variations between the donor and recipient genomes allowed us to characterize the genomic exchange events that created several ΔrecA recombinants. Although the donors and recipients are both K-12 descendants, the two lineages separated in the early 1950's and vary at about 450 positions, counting each insertion or deletion as a single position (about 1 variation per 10000 bp (S5 Fig Box 1)). We selected three uninduced KnS recombinants from Cross 5 and three yjiPc-induced KnS recombinants from Cross 13 for long-read Pac Bio SMRT sequencing and genome alignment (Fig 5). All recombinant genomes (uninduced: ER3445, ER3446, & ER3454; induced: ER3466, ER3475, & ER3476), along with the donor (ER3435) and recipient (ER3436) genomes were successfully sequenced and assembled de novo. Entire genomes were then aligned with Mauve [40] as implemented in the Geneious program package. The Genious alignment view enabled visualization of variant positions in the recombinant genomes and their assignment to recipient and donor (S5 Fig Boxes 2–4). The results are shown schematically in Fig 5.
Fig 5. Long Patch Replacements occur in RecA-independent crosses.
We compare (A) the entire genome and (B) the genomic region surrounding the ICR between the ΔrepE* ΔrecA donor (ER3435), the ΔrecA/ΔrecA rhaBp-yjiPc recipients (ER3440/ER3460), and six recombinants produced by mating these strains (Cross 6; uninduced recombinants, and Cross 13; yjiPc-induced recombinants). Each bar represents the genome for a single strain with the color indicating the origin of genomic DNA. See S5 Fig or an explanation of how the parent was identified for each position and the schematic created. Each recombinant exhibits a unique recombination patch that extends beyond the borders of the ICR. In four of the six cases, over 1.5 megabase of contiguous donor DNA replaced the corresponding recipient DNA. All markers are positioned to align with the common genomic backbone (i.e excluding indel variants such as F insertion). Coordinate 1 is placed at the translation start site of dnaA instead of the traditional coordinate #1 upstream of thrA.
This analysis revealed that all six recombinants were created by large replacements of recipient genomic DNA with the corresponding genomic DNA from the donor (Fig 5). The exchanged DNA always included the ICR, as demanded by selection for the antibiotic resistance marker. However, we were surprised to find that each exchange event replaced a unique segment of DNA that extended well beyond the borders of the ICR. In four of the recombinants, >1.5 Mb of donor DNA replaced the corresponding DNA on the recipient genome. The other two recombinants replaced less than 200 kb of DNA.
To understand how these large patch replacement events could occur in a RecA-deficient background, we focused our attention to the edges of DNA exchange in the six recombinants, which we refer to as crossover intervals. In this context, each recombinant results from 2 distinct crossover events: a proximal crossover in which the recipient genome connects to a segment of donor DNA proximal to the ICR, and a distal crossover in which that segment of donor DNA links to the recipient genome distal to the ICR. Because of the low density of informative variant positions distinguishing donor from recipient, these crossover intervals ranged from 0.5 to 50 kb.
The proximal crossover intervals are each different, but show some evidence of nonrandom distribution. The proximal crossover will be partially constrained to occur in the ~75 kb of chromosomal DNA between oriT and the ICR, since conjugal DNA transfer begins at oriT and proceeds in the direction of the ICR. Five of the six proximal crossovers occur in a 26 kb region between oriT and the ICR, which is only one third the size of the available segment (S6 Fig). The proximal crossover intervals for the three uninduced recombinants were particularly close together, spanning only 16.6 kb and overlapping each other. Although the number is small, this clustering suggests regional preference for the proximal crossover. In the sixth (yjiPc-induced) recombinant (ER3466) the proximal crossover occurred between the chromosomal replication origin and oriT. This can only occur if the donor transfers its entire chromosome, before transferring the ICR a second time via rolling-circle replication in the donor [41].
The distribution of distal crossovers also suggests a potential recombination hotspot. Three of the six crossovers (one uninduced and two yjiPc-induced) occurred in the same 44.5 kb interval located over 1.5 Mb away from the ICR. Since the distal crossover in this mating system could occur anywhere in the ~2.4 Mb between the ICR and the gyrA96 antibiotic resistance marker, the fact that 50% of the pooled set of sequenced recombinants show crossover in the same 44.5 kb (2% of available sequence) suggests a nonrandom event. We attribute this to the action of the XerCD recombinase at the dif site found in this interval (see Discussion).
We list the genes and features found in the crossover intervals observed in S3 Table. Some interesting ones associated with mobile elements, defining boundaries for all six recombinants, are briefly described here. The fimE/IS1 informative marker defines an edge of the proximal crossover interval in three recombinants, two uninduced (ER3445 & ER3454) and one yjiP-induced (ER3476). This marker consists of a copy of the IS1 mobile element that has inserted into the fimE gene in the recipient, but not in the donor (S6 Fig) [9]. An IS element variant, yjiC/IS5, also defines an edge of a fourth proximal crossover interval, for ER3475: an IS5 element has integrated in to yjiC in the recipient, but not in the donor [42]. A third marker associated with DNA mobility, and defining an edge of the proximal crossover for two recombinants (ER3445 and ER3454), is the invertible segment fimS [43,44]. This segment is oriented differently in the donor and the recipient. One of these recombinants (ER3454) exhibits a unique fimS sequence compared to both the donor and recipient. An interpretation of how these genetic elements may contribute to the recombination events can be found in the discussion.
The genome of one KnR recombinant from Cross 5 (ER3455) was also sequenced. Mapping reads from the sequencing against either the donor or recipient strains showed a large region of double coverage between the rrlE and rrsH ribosomal RNA genes (S7 Fig). Genome duplications are frequent in E. coli, especially between repetitive sequences like ribosomal operons [45], so we explored the possibility that such a duplication occurred. Using the Pacific Biosciences Bridgemapper program, we identified junction reads compatible with a tandem duplication mediated by these ribosomal operons. We infer that a duplication of this 820 kb region (including the ICR) occurred in the recipient strain prior to the recombination event with donor DNA. The mrr::tetRA transferred from the donor was then incorporated into one copy of the duplication, leaving the other copy of the duplication with the recipient ICR.
Genetic identification of ICR-only replacements
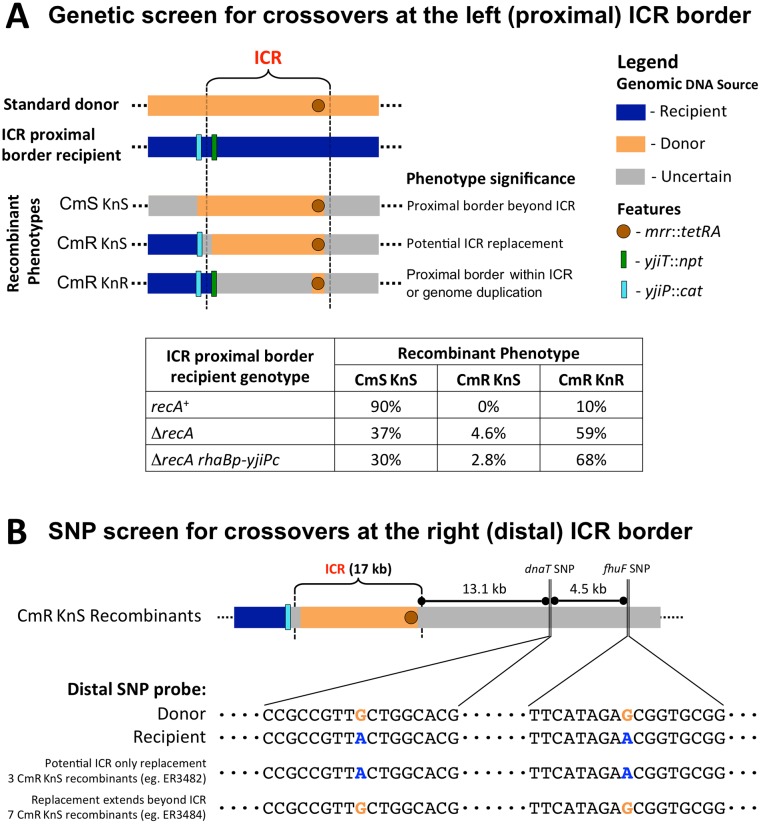
We were particularly interested in detecting recombinants in which only the ICR was exchanged. To estimate the frequency of such limited replacement events, we designed a new recipient marker configuration that could facilitate the search for them. These recipients (ER3472, Rec+ and ER3473, Rec-) carry two antibiotic resistance cassettes flanking the proximal edge of the ICR, with the wild-type sequence of the rest of the ICR. Specifically, a kanamycin resistance cassette replaced yjiT within the ICR and a chloramphenicol resistance cassette replaced yjiP outside of it (Fig 2). Treating these cassettes as unselected markers allows us to screen TcR SmR recombinants for those with the CmR KnS phenotype (Fig 6A), which will only arise from a recombination event in which one edge of the replacement event is located within the 2.5 kb stretch of DNA that contains the proximal edge of the ICR, between yjiP and yjiT. Since the purpose of these recipients is to focus on the proximal border of the ICR, we referred to them as ICR proximal border genetic probe recipients. We mated the recA + and ΔrecA recipients with this configuration with the ΔrecA ΔrepE* donor (Crosses 7 & 8, respectively). Over 100 recombinants from each mating were screened for kanamycin and chloramphenicol sensitivity. The possible recombinant configurations and frequencies of these among the recombinants are shown in Fig 6A.
Fig 6. Identification of recombinants that replace the ICR only.
(A) Schematic of the mating configuration designed to allow rapid genetic screening of the proximal ICR border status. In the recipient, A KnR cassette replaced a gene inside the proximal border of the ICR (yjiT::npt) and a CmR cassette replaced a nearby gene outside the proximal border (yjiP::cat). We call these ICR proximal border recipients. With this arrangement, a recombinant with donor DNA confined to the ICR will lose KnR but keep CmR and thus can be quickly identified by antibiotic screening. The embedded table displays the distribution of recombinant phenotypes in matings with different recipient backgrounds. The ΔrecA ΔrepE* donor was crossed with recA+, ΔrecA or ΔrecA yjiPc-rhaBp proximal border recipients (Crosses 7, 8, and 10 respectively); at least 100 recombinants from at least three independent matings of each cross were tested. The proximal crossover yielding complete donor replacement (top configuration) dominated Rec+, with some recombinants showing proximal crossover within the ICR (bottom configuration). In recA, a larger fraction showed proximal crossover within the ICR, and a minority showed proximal crossover within the ICR border region (middle configuration). When YjiPc was overexpressed this distribution did not change greatly. (B) Detecting distal crossover events with PCR and Sanger sequencing. The schematic shows SNP configurations at the distal ICR border. Sanger sequencing of the closest two SNPs distal to the ICR in the ten KnS, CmR recombinants from Cross 8 showed that seven shared the SNPs of the donor, while three (ER3482, ER3483, and ER3510) shared the SNPs of the recipient. For these three, the distal-side limit of the replacement event lies within a ~13 Kb region that includes the downstream edge of the ICR.
The control recA + cross (Cross 7) primarily produced recombinants that were sensitive to both drugs, suggesting that most recombination events replace a >20 kb segment of DNA that extends at least from yjiP to mrr. This agrees with the literature; recombinants from a Rec+ X Rec+ Hfr cross selecting for a marker within 500 kb of the oriT usually inherit donor DNA in a single large segment [31]. None of the recombinants from the rec + cross exhibited the KnS CmR phenotype resulting from a proximal crossover within the ~2.5 kb separating the two resistance cassettes. We infer that specific ICR replacement events are too rare to observe in a system that allows RecA-mediated recombination.
In the ΔrecA cross (Cross 8), the phenotypic ratios were quite different. Only 37% of the events replaced both recipient antibiotic markers (KnS CmS) (Fig 6A). These resemble the recA-independent large patch replacement events we previously characterized via genomic sequencing analysis (Fig 5). Most recombinants (59%) retained both recipient markers (KnR CmR). Three mechanisms could play a role in generating these. A randomly-placed proximal crossover of the same kind seen in Fig 5 might occur within the 16 kb between yjiT and mrr; a site-specific activity might replace a subset of the region; or a genome duplication event in the recipient population could be followed by replacement of the region in one copy, as was observed in recombinant ER3455 (S7 Fig). The fraction of presumptive duplication recombinants in the mating that produced ER3455 (Cross 5, see Fig 3B) is low (9%) compared with the present result (59%), arguing against a role for duplications in the recipient. We lack informative sites that would distinguish between the site-specific and random possibilities.
Ten recombinants out of the 217 screened (4.6%) display proximal crossovers yielding the CmR KnS phenotype compatible with specific ICR replacement. To determine the position of the distal crossover events in these recombinants, we used PCR interrogation of the two SNPs that distinguish the recipient from the donor at 13 and 18 kb distal to the ICR (Fig 6B). In seven of the recombinants, these two SNPs matched that of the donor suggesting that the integrated donor DNA extends beyond the distal side of the ICR in these recombinants. The other three recombinants (which were each obtained from independent matings) matched the SNPs of the recipient. Thus, three of 217 recombinants result from crossovers in both ICR border intervals (2.5 kb proximal and 13.1 kb distal).
These three recombinants (ER3482, ER3483, and ER3510) are compatible with occurrence of targeted recombination event at the ICR. They may represent an enriched class. If crossovers occurred randomly throughout the genome, a recombinant exhibiting both crossovers within the ICR border intervals is 1 in 6600 or 0.015% (See S3 File for an explanation). Our matings produced recombinants with this crossover pattern at a 100-fold higher frequency (3/217 or 1.4%). This is a frequency of 3 X 10-12 CFU/recipient per hour. At present, the mechanism of such enrichment is unclear.
We further investigated how YjiPc might influence the types of recA-independent recombination events by repeating our ICR proximal border genetic probe mating experiments with yjiPc induction. A ΔrecA yjiP::cat yjiT::kan rhaBp-yjiPc recipient was mated with the standard ΔrecA ΔrepE* donor in 0.2% rhamnose, and over 100 recombinants were screened for kanamycin and chloramphenicol sensitivity (Cross 10; Fig 6B). Of the recombinants tested, 30% were CmS KnS, 68% were CmR KnR, and 2.8% were CmR KnS. These phenotypic frequencies are quite similar to those of recombinants from the mating without yjiPc induction (37%, 58%, and 4.6% respectively in Cross 8). Thus, we conclude that although yjiPc is a promoter of recA-independent recombination, there is no evidence that YjiP mediates targeting of the replacement events by itself.
Discussion
In the present study, we developed a conjugal system to characterize mechanisms of horizontal gene transfer (HGT) starting with the model organism E. coli K-12. We discovered a novel low-frequency event class and implicated an uncharacterized protein family in promoting this class. We also identified genetic elements that may promote RecA-independent recombination.
Large RecA-independent genomic replacements are a novel recombinant class
Large RecA-independent genomic replacements (up to 2.4 Mb of the recipient chromosome) have not been reported previously. A recent paper explains how to use large genomic replacements to create chimeric genomes [46], but the study is in a recA + background. Other research groups study RecA-independent recombination, but focus on the replacement of a selective marker or at specific sites [47,48]. These are infrequent, and their detection here was dependent on whole-genome sequence assembly with the SMRT platform, a technology only recently available at reasonable cost.
To characterize the mechanism by which large replacements occur we combine our findings with the literature to draw some conclusions. Our first step in understanding this mechanism is to categorize this recombination event into one of the three classical recombination models: Break-join (homologous strand exchange), copy-choice (template exchange occurs during replication), or break-copy (a hybrid of the two) [49]. Crossovers in the six sequenced recombinants exhibiting large genomic replacements were all localized to regions of homology. Furthermore, sequencing did not reveal any “novel joints” in which donor DNA is connected to recipient DNA at a new point [50]. These observations support a copy choice model of homology-directed recombination, but do not refute the other models.
Several RecA-independent homology directed recombination mechanisms that follow the copy-choice model have been proposed by Lovett and coworkers [51]. These mechanisms involve a replication “slippage” event in which the polymerase temporarily dissociates from its original template allowing the nascent strand to anneal to a different homologous template before replication resumes. For our events, a DNA polymerase could switch homologs, for example on encountering a template nick. Resolution by for a distal crossover could be carried out in the same way, or another kind of resolution could reunite the replication complex with the recipient chromosome. In the context of this model, YjiP could promote recombination in several ways: by facilitating resolution of crossed strands (a structure specific nuclease activity), by nicking a template strand, or by exonuclease action at a broken fork to reveal a strand for annealing.
YjiP represents a novel class of DNA modifying enzymes
This study is the first experimental report to illuminate the role of YjiP. The small amount of bioinformatic evidence available is compatible with our demonstration that it can enhance RecA-independent recombination. It was classified as a putative transposase (Transposase_31) by the TigrFam database because of its distribution and conservation [52]. A family of related proteins is included in the Pfam database (PF04754) [53], where it is classified within a clan (PDDEXK domain) containing many phosphodiesterases [54].
YjiP homologs are prevalent in Enterobacteriaceae (E. coli, Salmonella, Yersinia, Shigella, ect.) and occur sporadically among other taxa [53,55]. Strains that have a copy of yjiP often contain multiple paralogs, which tend to be more similar to each other than to homologs in other species [52]. E. coli K-12 encodes 5 paralogs (YjiP, YadD, YfcI, YfaD, and YhgA); the yadD gene appears to have inserted into the panBCD operon relatively recently [56]. This distribution pattern is similar to that of mobile elements and their associated genes. Expression experiments in Escherichia coli also suggest a connection between yjiP and biofilms and quorum sensing [57]. A yjiP null strain exhibits ~50% reduction in biofilm formation [58] and yjiP is upregulated during the initial stages of biofilm construction. This connection is relevant to the biology of YjiP’s role in recombination because DNA transfer (a precondition for HGT) is enhanced in biofilms [5].
The putative phosphodiesterase activity of YjiPc provides a potential explanation for reduced viability seen in our experiments (Fig 4C & S4E Fig). An overexpressed nuclease could cause DNA damage, especially toxic in a cell unable to repair DNA via the efficient RecA-dependent pathways [59]. The observation that yjiPc overexpression does not reduce viability in a recA + background but does induce an SOS reporter supports this hypothesis.
We conclude that YjiP is part of an experimentally uncharacterized class of DNA modifying enzymes that contribute to HGT, consistent with the bioinformatic context described above. Its mechanism of action is unclear, but likely includes nuclease activity. YjiP’s function does not seem to be targeted to the ICR under our conditions. Notably, both F' formation and novel large replacements are stimulated by its expression. Other ICR framework genes (yjiRS and yjiAXY) might contribute targeting activity, resulting in SSR at this locus. Future studies will also focus on the YjiP paralogs in E. coli and how they might influence both recombination and cell viability.
Genetic elements that could affect recombination found in crossover intervals
A distinguishing aspect of this study is the inventory of 447 informative variant sequence positions. This allowed us to define proximal and distal crossover intervals where DNA exchange occurred in 6 recombinants (3 WT and 3 yjiPc induced). Several features found within these 12 crossover intervals deserve further discussion.
The sequence interval between the initial DNA entry (yhjP) and the selected marker (mrr::tetAR) defines the region in which the first (proximal) crossover is most likely to occur. There are six variant positions in this stretch, three containing sites of action for DNA transactions and coding sequences for the corresponding proteins. One of these elements is the invertible segment fimS together with the FimE and FimB recombinases that act on it, encoded in flanking genes [43,44]. The other two are the IS1 and IS5 elements encoding RecA-independent transposases (InsAB and InsH, respectively), and containing their sites of action. FimE, FimB, IS1, and IS5 all have the capacity to induce DNA breaks and ligate them at specific loci within the proximal crossover intervals. Normal action would result in inversion, transposition or adjacent deletion formation mediated by the mobile element. During conjugation, a large segment of homologous DNA is introduced; possibly such activity could contribute to the formation of an intermediate complex such that it connects recipient DNA to donor DNA, then to be acted on by YjiP or one of the four paralogs in K-12. In support of this theory is the fact that recombinant ER3454 has a novel fimS sequence compared to both the donor and the recipient, suggesting that the proximal crossover of this recombinant occurred within fimS. Such interactions have not been described previously, but since the events studied occur at very low frequencies, novel possibilities may be entertained.
For the second (distal) crossovers required to recover viable progeny, 50% of them occurred in the same 44.5 kb interval (S3 Table) suggesting the presence of a recombination hotspot. The most notable element found within this region was dif, located in the replication terminus [60]. This is the site of action of the site-specific XerCD recombinase. Xer-dif recombination decatenates chromosome copies during cell division [61][62]. This activity would therefore abridge any recombinants with distal crossovers past the dif site, by connecting the recipient homolog to the donor thus resolving a forked chromosome created by the proximal crossover that initiated the recombination event.
Relevance to evolutionary processes
This novel type of recombination could be significant factor in genomic evolution. The recent surge in bioinformatic studies has begun to show that natural chromosome transfer is much more prevalent than previously believed [63]. Novel Integrative conjugal elements that transfer chromosomal DNA are rapidly being discovered in a variety of bacteria ranging from Mycoplasma [64] to Streptococcus [65], to Yersinia [66]. Some of these systems are even less constrained and more efficient than Hfr conjugation from E. coli [66]. Although the RecA-independent events that we characterized are rare, the abundance of conjugation in nature gives these events ample opportunity to modify genomes. The events described here have also not shown a requirement for extensive homology and could occur between distantly related strains where homologous recombination would not be effective.
System qualification
Using different configurations of the conjugal system, we estimated relative event frequencies for known recombination events that we could identify (Table 2). These include homologous recombination, F’-plasmid formation, and large RecA-independent genomic replacement. Genome duplications mediated by ribosomal operons were also recovered as background events. We found that replacement of a gene island of particular interest (immigration control region (ICR)), likely occurs, but most of the recombination events we observed were not specific to the ICR.
Table 2. Frequencies of recombination events in the conjugal system.
| Recombination Event (frequency) | Recombination Subcategory | % of total Recombinants | yjiP induction increases frequency? |
|---|---|---|---|
| Homologous Recombination (0.21 CFU/recipient per hour) | Recombination beyond ICR borders | 90% | No |
| Recombination within ICR or into genomic duplication | 10% | No | |
| Site specific ICR replacement | 0.0% | NA | |
| F'-plasmid formation (3.5 X 10-6 CFU/recipient per hour) | Recipient ICR replaced | 9.6% | Yes |
| Recipient ICR maintained | 90% | Yes | |
| RecA-independent genomic exchange (2.1 X 10-10 CFU/recipient per hour) | Recombination beyond ICR borders | 40% | Yes |
| Recombination within ICR | 50% | Yes | |
| Genome duplication before recombination | 8.5% | Yes | |
| Site specific ICR replacement | 1.4% | Yes |
The conjugal system is an effective genetic tool for analyzing HGT
The conjugal system we developed has proven to be an extremely versatile tool for studying HGT. It is robust to the point where rare recombination events can be isolated and has the flexibility to focus on specific genetic elements, gene islands, or proteins of interest. It can also be expanded to analyze HGT between more distantly related strains where homology-based recombination is much less of a factor. We hope that other labs studying genomic evolution, DNA recombination, or other related fields will recognize the utility of our system and begin to use it for their own research purposes.
Supporting Information
The content of the ICR region tends to be highly variable, even among closely related strains. Boxes of the same color in different strains represent orthologous genes. Genes were judged orthologous if nucleotide similarity was >65%. Type IA and Type IB restriction systems are paralogous. This variability is in sharp contrast to the conserved framework genes (yjiPRS & yjiAXY) suggesting that a site-specific mechanism could be involved in exchange of all or part of the ICR. Image adapted from [22].
(TIF)
Four recombinant genomes from the ΔrecA donor X recA13 recipient mating (Cross 4) were sequenced with Illumina and aligned to an in silico model of the donor (ER3276) genomic sequence. The relative coverage of (A) the entire genome and (B) the genomic region surrounding the F factor and ICR loci is shown. Disagreement between the donor reads and the in silico model revealed the presence of a deletion ((Δori1 = Δ(pifA-yddA)) covering one of the two vegetative origins of the F plasmid. All recombinants exhibit an approximate 2-3X increase of coverage, from the start of the proximal F region to a variable point beyond the ICR. We infer the presence of an F’ plasmid carrying the overrepresented region. Each F’-plasmid is distinct in the extent of genomic DNA that it carries. See S2 File for discussion.
(TIF)
(A) DNA transferred from oriT as far as the selected ICR by the original ΔrecA donor (ER3276) and by the donor in which the leading F-DNA has been deleted (ER3435). The leading F DNA deletion is denoted ΔrepE*; noted that the deletion removes 49 genes including the toxin-antitoxin systems ccdAB and flmAB. (B) Depiction of the recombinants derived from each donor. Genetic and sequence analysis revealed an F’-plasmid with a stable copy of tetRA in all recombinants created by the original donor. The chromosomal ICR typically kept its npt cassette, but a small proportion of recombinants replaced it with a tetRA cassette. In recombinants created with the ΔrepE* donor, the F’-plasmid did not form and the yjiT-mrr::npt marker on the recipient genome was usually replaced with the mrr::tetRA marker from the donor.
(TIF)
(A) Expression dynamics of a surrogate reporter. The β-galactosidase activity of a rhaBp-lacZ construct in E. coli (ER3340) after induction with rhamnose. Cultures were grown at 37°C with shaking and treated with 0.2% rhamnose at an OD600 of 0.2. β-galactosidase assays of culture samples were then taken at regular intervals over the next 6.5 hours. The addition of rhamnose increased the accumulation of β-galactosidase ~200 fold compared to an untreated control and took about 225 min to reach full expression. (B) The frequency of recombination during matings between the ΔrecA ΔrepE* donor and a rec + rhaBp-yjiPc recipient (cross 9) with and without 0.2% rhamnose. Recombination efficiency was calculated as the frequency of recombinant formation per viable recipient per hour in the mating mixture. Inducing yjiP expression with rhamnose did not significantly affect recombination efficiency in a rec + background. (C) The change in recombination efficiency due to rhamnose treatment in matings between the ΔrecA repE + donor and a recA13 recipient (Cross 4) or between that donor and recA13 recipients with with rhamnose inducible copies of either yjiA (cross 11; rhaBp-yjiA) or yjiPc (cross 12; rhaBp-yjiPc). As with the other matings, inducing yjiPc expression increased recombination efficiency around 4 fold, but rhamnose had no effect on the control recipient or the yjiA inducible recipient. (D) Cell growth of a recA13 recipient containing the rhaBp-yjiPc construct (ER3336) treated with and without 0.2% rhamnose as measured by OD600 readings. Although yjiPc induction reduces the ability of recipients to form colonies on selective media (Fig 4B–4C), OD600 readings remain unaffected by rhamnose treatment. (E) yjiP overexpression induces an SOS response in E. coli. A rec + strain carrying both the rhaBp-yjiPc construct and a dinDp-lacZ reporter of the SOS response (ER3544; [39]) on X-gal media with and without 0.2% rhamnose. Colonies were substantially more blue in the presence of rhamnose than in its absence.
(TIF)
The donor and recipient genomes display about 1 variation per 10 kb (box 1). When a recombinant genome (ER3445) is aligned to the recipient genome using Mauve, variations are observed where donor genomic DNA has been incorporated (box 2). The inverse pattern is seen when the recombinant is compared with the donor (box 3). This display allows assignment of DNA stretches to each parent (box 4). Since variations are separated by a ~10000 bps this analysis still leaves a small region of DNA of uncertain origin (grey coloring). Presumably, the DNA crossover events occur in these uncertain intervals, designated "crossover intervals".
(TIF)
Variations that match the donor genome are orange and those matching the recipient genome are blue. The fimS invertible segment, labelled fimInv here, is present in opposite orientations in donor and recipient. The gray FimInv* element in recombinant ER3454 is a unique sequence compared to both the donor and the recipient. This analysis was performed with the Geneious R7 software and was used to determine the proximal crossover intervals described in S3 Table.
(TIF)
Sequences were assembled using the RS_Bridgemapper.1 algorithm with either the (A) ΔrecA ΔrepE* donor (ER3435) or (B) ΔrecA (ER3440) recipient genomes as the reference sequence. An area of 2X coverage between the rrlE and rrsH ribosomal subunit encoding genes indicates that a duplication of this region is present. When compared with the donor, the ICR drops to single copy levels. This recombinant contains no F DNA, and has lost the Rac prophage. We infer that the duplication occurred in the recipient strain prior to the recombination event with donor DNA. The duplicated region then integrated the mrr::tetRA transferred from the donor into one copy of the recipient’s ICR, leaving the other ICR with the yjiT-mrr::npt construct unaffected.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Alexey Fomenkov and Brian Anton for assistance with SMRT sequencing, and Laurence Ettwiller and Brad Langhorst for guidance on use of bioinformatic tools. Christine Ponkratz assisted with the dinD reporter assays. We greatly appreciated the help of Paul Morrison and Zachary Herbert of DFCI MBCF in acquiring Illumina Nextera sequences. We are grateful for discussions and advice from Mehmet Berkmen, Jim Samuelson, K. Brooks Low, Susan Lovett and John Roth. Marion Sibley carried out the initial exploratory strain constructions and crosses. We thank the anonymous reviewers whose suggestions greatly improved the manuscript.
We are especially grateful to Don Comb and New England Biolabs for underwriting this research and the many years of projects from which it stemmed.
Data Availability
All sequence files are available from the NCBI Bioproject database, accession number(s) PRJNA271807 ID: 271807.
Funding Statement
New England Biolabs provided financial support in the form of salaries for authors AWK, CD, CRR, EAR, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Ragan MA, Beiko RG (2009) Lateral genetic transfer: open issues. Philos Trans R Soc Lond B Biol Sci 364: 2241–2251. 10.1098/rstb.2009.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de la Cruz F, Davies J (2000) Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol 8: 128–133. [DOI] [PubMed] [Google Scholar]
- 3. Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. (2013) The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis 13: 155–165. 10.1016/S1473-3099(12)70317-1 [DOI] [PubMed] [Google Scholar]
- 4. Tamames J, Moya A (2008) Estimating the extent of horizontal gene transfer in metagenomic sequences. BMC Genomics 9: 136 10.1186/1471-2164-9-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Madsen JS, Burmolle M, Hansen LH, Sorensen SJ (2012) The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65: 183–195. 10.1111/j.1574-695X.2012.00960.x [DOI] [PubMed] [Google Scholar]
- 6. Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moreno Switt AI, den Bakker HC, Cummings CA, Rodriguez-Rivera LD, Govoni G, Raneiri ML, et al. (2012) Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One 7: e41247 10.1371/journal.pone.0041247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiles TJ, Norton JP, Smith SN, Lewis AJ, Mobley HL, Casjens SR, et al. (2013) A phyletically rare gene promotes the niche-specific fitness of an E. coli pathogen during bacteremia. PLoS Pathog 9: e1003175 10.1371/journal.ppat.1003175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darmon E, Leach DR (2014) Bacterial genome instability. Microbiol Mol Biol Rev 78: 1–39. 10.1128/MMBR.00035-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, et al. (2008) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190: 6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chaudhuri RR, Henderson IR (2012) The evolution of the Escherichia coli phylogeny. Infect Genet Evol 12: 214–226. 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 13. Vieira G, Sabarly V, Bourguignon P-Y, Durot M, Le Fèvre F, Mornico D, et al. (2011) Core and panmetabolism in Escherichia coli. Journal of Bacteriology 193: 1461–1472. 10.1128/JB.01192-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Didelot X, Meric G, Falush D, Darling AE (2012) Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. BMC Genomics 13: 256 10.1186/1471-2164-13-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT (2011) Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A 108: 7200–7205. 10.1073/pnas.1015622108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, et al. (2009) Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 5: e1000257 10.1371/journal.ppat.1000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence JG, Retchless AC (2009) The interplay of homologous recombination and horizontal gene transfer in bacterial speciation. Methods Mol Biol 532: 29–53. 10.1007/978-1-60327-853-9_3 [DOI] [PubMed] [Google Scholar]
- 18. Tenaillon O, Skurnik D, Picard B, Denamur E (2010) The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8: 207–217. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 19. Raleigh EA (1992) Organization and function of the mcrBC genes of Escherichia coli K-12. Mol Microbiol 6: 1079–1086. [DOI] [PubMed] [Google Scholar]
- 20. Murray NE (2002) 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology 148: 3–20. [DOI] [PubMed] [Google Scholar]
- 21. Milkman R, Jaeger E, McBride RD (2003) Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics 163: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sibley MH, Raleigh EA (2004) Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucleic Acids Res 32: 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanahan D, Jessee J, Bloom FR (1991) Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol 204: 63–113. [DOI] [PubMed] [Google Scholar]
- 24. Sternberg NL, Maurer R (1991) Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol 204: 18–43. [DOI] [PubMed] [Google Scholar]
- 25. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW (2010) A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res 38: e159 10.1093/nar/gkq543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bachmann BJ, Low KB, Taylor AL (1976) Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev 40: 116–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bachmann BJ (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36: 525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Achtman M, Morelli G, Schwuchow S (1978) Cell-cell interactions in conjugating Escherichia coli: role of F pili and fate of mating aggregates. J Bacteriol 135: 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M (2008) Direct visualization of horizontal gene transfer. Science 319: 1533–1536. 10.1126/science.1153498 [DOI] [PubMed] [Google Scholar]
- 31. Lloyd RG, Buckman C (1995) Conjugational recombination in Escherichia coli: genetic analysis of recombinant formation in Hfr x F- crosses. Genetics 139: 1123–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB (2004) Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432: 187–193. [DOI] [PubMed] [Google Scholar]
- 33. Amundsen SK, Taylor AF, Chaudhury AM, Smith GR (1986) recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci U S A 83: 5558–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Masson L, Ray DS (1986) Mechanism of autonomous control of the Escherichia coli F plasmid: different complexes of the initiator/repressor protein are bound to its operator and to an F plasmid replication origin. Nucleic Acids Res 14: 5693–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kowalczykowski SC (1991) Biochemical and biological function of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie 73: 289–304. [DOI] [PubMed] [Google Scholar]
- 36. Willetts NS, Clark AJ, Low B (1969) Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol 97: 244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K (2012) Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res 40: 7016–7045. 10.1093/nar/gks382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sibley MH, Raleigh EA (2012) A versatile element for gene addition in bacterial chromosomes. Nucleic Acids Res 40: e19 10.1093/nar/gkr1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piekarowicz A, Yuan R, Stein DC (1991) A new method for the rapid identification of genes encoding restriction and modification enzymes. Nucleic Acids Res 19: 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lanka E, Wilkins BM (1995) DNA processing reactions in bacterial conjugation. Annu Rev Biochem 64: 141–169. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Z, Saier MH Jr (2009) A novel mechanism of transposon-mediated gene activation. PLoS Genet 5: e1000689 10.1371/journal.pgen.1000689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McClain MS, Blomfield IC, Eisenstein BI (1991) Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol 173: 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCusker MP, Turner EC, Dorman CJ (2008) DNA sequence heterogeneity in Fim tyrosine-integrase recombinase-binding elements and functional motif asymmetries determine the directionality of the fim genetic switch in Escherichia coli K-12. Mol Microbiol 67: 171–187. [DOI] [PubMed] [Google Scholar]
- 45. Andersson DI, Hughes D (2009) Gene amplification and adaptive evolution in bacteria. Annu Rev Genet 43: 167–195. 10.1146/annurev-genet-102108-134805 [DOI] [PubMed] [Google Scholar]
- 46. Ma NJ, Moonan DW, Isaacs FJ (2014) Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering. Nat Protoc 9: 2285–2300. 10.1038/nprot.2014.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dutra BE, Sutera VA Jr, Lovett ST (2007) RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci U S A 104: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schlake T, Bode J (1994) Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33: 12746–12751. [DOI] [PubMed] [Google Scholar]
- 49. Kuzminov A, Stahl FW (1999) Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev 13: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chayot R, Montagne B, Mazel D, Ricchetti M (2010) An end-joining repair mechanism in Escherichia coli. Proc Natl Acad Sci U S A 107: 2141–2146. 10.1073/pnas.0906355107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bzymek M, Lovett ST (2001) Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A 98: 8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, et al. (2001) TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29: 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kosinski J, Feder M, Bujnicki JM (2005) The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics 6: 172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–568. 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Merkel WK, Nichols BP (1996) Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol Lett 143: 247–252. [DOI] [PubMed] [Google Scholar]
- 57. Zhou J, Rudd KE (2013) EcoGene 3.0. Nucleic Acids Res 41: D613–624. 10.1093/nar/gks1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herzberg M, Kaye IK, Peti W, Wood TK (2006) YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol 188: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cox MM (1999) Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol 63: 311–366. [DOI] [PubMed] [Google Scholar]
- 60. Blakely G, May G, McCulloch R, Arciszewska LK, Burke M, Lovett ST, et al. (1993) Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75: 351–361. [DOI] [PubMed] [Google Scholar]
- 61. Grainge I, Lesterlin C, Sherratt DJ (2011) Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res 39: 5140–5148. 10.1093/nar/gkr078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, Arciszewska LK (2006) Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol Microbiol 59: 1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP (2011) The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7: e1002222 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dordet-Frisoni E, Sagne E, Baranowski E, Breton M, Nouvel LX, Blanchard A, et al. (2014) Chromosomal transfers in mycoplasmas: when minimal genomes go mobile. MBio 5: e01958 10.1128/mBio.01958-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Puymege A, Bertin S, Guedon G, Payot S (2015) Analysis of Streptococcus agalactiae pan-genome for prevalence, diversity and functionality of integrative and conjugative or mobilizable elements integrated in the tRNA gene. Mol Genet Genomics. [DOI] [PubMed] [Google Scholar]
- 66. Lesic B, Zouine M, Ducos-Galand M, Huon C, Rosso ML, Prevost MC, et al. (2012) A natural system of chromosome transfer in Yersinia pseudotuberculosis. PLoS Genet 8: e1002529 10.1371/journal.pgen.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The content of the ICR region tends to be highly variable, even among closely related strains. Boxes of the same color in different strains represent orthologous genes. Genes were judged orthologous if nucleotide similarity was >65%. Type IA and Type IB restriction systems are paralogous. This variability is in sharp contrast to the conserved framework genes (yjiPRS & yjiAXY) suggesting that a site-specific mechanism could be involved in exchange of all or part of the ICR. Image adapted from [22].
(TIF)
Four recombinant genomes from the ΔrecA donor X recA13 recipient mating (Cross 4) were sequenced with Illumina and aligned to an in silico model of the donor (ER3276) genomic sequence. The relative coverage of (A) the entire genome and (B) the genomic region surrounding the F factor and ICR loci is shown. Disagreement between the donor reads and the in silico model revealed the presence of a deletion ((Δori1 = Δ(pifA-yddA)) covering one of the two vegetative origins of the F plasmid. All recombinants exhibit an approximate 2-3X increase of coverage, from the start of the proximal F region to a variable point beyond the ICR. We infer the presence of an F’ plasmid carrying the overrepresented region. Each F’-plasmid is distinct in the extent of genomic DNA that it carries. See S2 File for discussion.
(TIF)
(A) DNA transferred from oriT as far as the selected ICR by the original ΔrecA donor (ER3276) and by the donor in which the leading F-DNA has been deleted (ER3435). The leading F DNA deletion is denoted ΔrepE*; noted that the deletion removes 49 genes including the toxin-antitoxin systems ccdAB and flmAB. (B) Depiction of the recombinants derived from each donor. Genetic and sequence analysis revealed an F’-plasmid with a stable copy of tetRA in all recombinants created by the original donor. The chromosomal ICR typically kept its npt cassette, but a small proportion of recombinants replaced it with a tetRA cassette. In recombinants created with the ΔrepE* donor, the F’-plasmid did not form and the yjiT-mrr::npt marker on the recipient genome was usually replaced with the mrr::tetRA marker from the donor.
(TIF)
(A) Expression dynamics of a surrogate reporter. The β-galactosidase activity of a rhaBp-lacZ construct in E. coli (ER3340) after induction with rhamnose. Cultures were grown at 37°C with shaking and treated with 0.2% rhamnose at an OD600 of 0.2. β-galactosidase assays of culture samples were then taken at regular intervals over the next 6.5 hours. The addition of rhamnose increased the accumulation of β-galactosidase ~200 fold compared to an untreated control and took about 225 min to reach full expression. (B) The frequency of recombination during matings between the ΔrecA ΔrepE* donor and a rec + rhaBp-yjiPc recipient (cross 9) with and without 0.2% rhamnose. Recombination efficiency was calculated as the frequency of recombinant formation per viable recipient per hour in the mating mixture. Inducing yjiP expression with rhamnose did not significantly affect recombination efficiency in a rec + background. (C) The change in recombination efficiency due to rhamnose treatment in matings between the ΔrecA repE + donor and a recA13 recipient (Cross 4) or between that donor and recA13 recipients with with rhamnose inducible copies of either yjiA (cross 11; rhaBp-yjiA) or yjiPc (cross 12; rhaBp-yjiPc). As with the other matings, inducing yjiPc expression increased recombination efficiency around 4 fold, but rhamnose had no effect on the control recipient or the yjiA inducible recipient. (D) Cell growth of a recA13 recipient containing the rhaBp-yjiPc construct (ER3336) treated with and without 0.2% rhamnose as measured by OD600 readings. Although yjiPc induction reduces the ability of recipients to form colonies on selective media (Fig 4B–4C), OD600 readings remain unaffected by rhamnose treatment. (E) yjiP overexpression induces an SOS response in E. coli. A rec + strain carrying both the rhaBp-yjiPc construct and a dinDp-lacZ reporter of the SOS response (ER3544; [39]) on X-gal media with and without 0.2% rhamnose. Colonies were substantially more blue in the presence of rhamnose than in its absence.
(TIF)
The donor and recipient genomes display about 1 variation per 10 kb (box 1). When a recombinant genome (ER3445) is aligned to the recipient genome using Mauve, variations are observed where donor genomic DNA has been incorporated (box 2). The inverse pattern is seen when the recombinant is compared with the donor (box 3). This display allows assignment of DNA stretches to each parent (box 4). Since variations are separated by a ~10000 bps this analysis still leaves a small region of DNA of uncertain origin (grey coloring). Presumably, the DNA crossover events occur in these uncertain intervals, designated "crossover intervals".
(TIF)
Variations that match the donor genome are orange and those matching the recipient genome are blue. The fimS invertible segment, labelled fimInv here, is present in opposite orientations in donor and recipient. The gray FimInv* element in recombinant ER3454 is a unique sequence compared to both the donor and the recipient. This analysis was performed with the Geneious R7 software and was used to determine the proximal crossover intervals described in S3 Table.
(TIF)
Sequences were assembled using the RS_Bridgemapper.1 algorithm with either the (A) ΔrecA ΔrepE* donor (ER3435) or (B) ΔrecA (ER3440) recipient genomes as the reference sequence. An area of 2X coverage between the rrlE and rrsH ribosomal subunit encoding genes indicates that a duplication of this region is present. When compared with the donor, the ICR drops to single copy levels. This recombinant contains no F DNA, and has lost the Rac prophage. We infer that the duplication occurred in the recipient strain prior to the recombination event with donor DNA. The duplicated region then integrated the mrr::tetRA transferred from the donor into one copy of the recipient’s ICR, leaving the other ICR with the yjiT-mrr::npt construct unaffected.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All sequence files are available from the NCBI Bioproject database, accession number(s) PRJNA271807 ID: 271807.