Abstract
Chondrosarcomas are highly resistant to conventional radiation and chemotherapy, and surgical removal is the only option for curative treatment. Consequently, there is nothing to offer patients with inoperable tumours and metastatic disease. The aim of this study is to investigate genes involved in cell cycle control: CDK4, CDKN2A/p16, cyclin D1, p21, p53, MDM2 and c-MYC, which may point towards new therapeutic strategies. The pRb pathway was targeted using CDKN2A/p16 overexpressing vectors and shRNA against CDK4 in chondrosarcoma cell lines OUMS27, SW1353, and CH2879. Cell survival and proliferation were assessed. CDK4, MDM2 and c-MYC expression levels were investigated by qPCR and immunohistochemistry (IHC) in 34 fresh frozen and 90 FFPE samples of enchondroma and chondrosarcoma patients. On a subset of 29 high-grade chondrosarcomas IHC for cyclin D1, p21 and p53 was performed. The overexpression of CDKN2A/p16 and knockdown of CDK4 by shRNA in OUMS27, SW1353 and CH2879 resulted in a significant decrease in cell viability and proliferation and a decreased ability to form colonies in vitro. Expression of CDK4 and MDM2 was associated with high-grade chondrosarcoma both at the mRNA and protein level. Combining these results with the expression of cyclin D1 and the previously shown loss of CDKN2A/p16 expression show that the majority (96%; 28/29) of high-grade chondrosarcomas contain alterations in the pRb pathway. This suggests a role for the use of CDK4 inhibitors as a treatment of metastatic or inoperable high-grade chondrosarcoma.
Keywords: chondrosarcoma, bone tumour, cell cycle, pRb-pathway, shRNA
Introduction
Chondrosarcoma of bone is a malignant cartilage-forming tumour that is notorious for its resistance to conventional chemotherapy and radiation therapy. The majority of tumours arise in the medullar cavity of bone and are designated primary central chondrosarcomas (80–85%) [1]. For < 1% of chondrosarcomas, there is clinical evidence that they arose secondary to a pre-existing (benign) enchondroma [1, 2]. Enchondromas occur mostly as solitary lesions, although they may occur as multiple lesions in the context of non-hereditary enchondromatosis (Ollier disease).
Chondrosarcomas are histologically divided into three grades, which is currently the only objective predictor of metastasis. While grade I tumours rarely metastasize and the 10-year survival rate is 83%, patients with grade III tumours develop metastatic disease in up to 71% of the cases and the 10-year survival rate decreases to 29%[3]. Marginal or intralesional excision of tumours can result in local recurrence. Thirteen percent of recurrent chondrosarcomas are of a higher grade than the primary tumour [4]. Currently, surgical removal of the tumour is the only option for curative treatment. There is no treatment to offer patients with metastatic disease or inoperable tumours in the extremities or pelvis. Elucidating the molecular background of high-grade chondrosarcomas and the involved pathways that lead to tumour progression may help identify targets for future therapeutic strategies to improve clinical outcome.
In contrast to other solid tumours, central chondrosarcomas harbour relatively few, consistent, numerical genomic alterations; however, the amplification of 12q13 [5, 6] and deletion of 9p21 [7–9] are two consistent genetic aberrations. Using array comparative genomic hybridization (CGH), we previously showed amplification of 12q13 in 6 of 21 (29%) central chondrosarcomas, which correlated with high histological grade [6], as was also suggested by others [9]. Several genes in this region are of importance for cell cycle control including CDK4 and MDM2, players in the pRb and p53 pathway, respectively. Defects in these pathways are found at high rates in almost all types of human cancer [10, 11]. Combining the array CGH results with those of our genome-wide expression profiling experiments showed overexpression of the CDK4 proto-oncogene in tumours with 12q13 amplification [6]. CDK4 controls progression through the cell cycle by regulating the transit of the cell through the G1 restriction point. This occurs by hyper-phosphorylation of pRb, leading to the release of E2F transcription factors. To accomplish this, CDK4 forms a complex with cyclin D1. This complex is tightly regulated by the inhibitory protein CDKN2A (CDKN2A/p16), which is encoded by the INK4A-ARF locus located on chromosome 9p21. Inhibition of the pRb-mediated cell cycle control through amplification of cyclin D1 or CDK4 and/or loss of expression of CDKN2A/p16/INK4A has been observed in many tumours [12]. Despite LOH of 13q14 has been found in a subset of chondrosarcoma [13, 14], i.e. in 10 of 28 tumours by Yamaguchi et al.[5], pRb mutations were not found [5]. Ropke et al. showed pRb expression in 16 of 17 chondrosarcomas by immunohistochemistry [14].
We and others previously demonstrated that loss of CDKN2A/p16 protein expression is correlated with increasing histological grade in central chondrosarcoma [7, 15, 16]. Cyclin D1 was previously shown to be expressed in 25 of 34 (73%) high-grade central chondrosarcomas [17].
In addition to CDK4, the 12q13 gene region harbours the MDM2 gene that is frequently found to be co-amplified with CDK4[18]. The MDM2 gene encodes an E3 ubiquitin ligase involved in the degradation of p53 protein. The tumour suppressor protein p53 is activated upon various forms of stress, including aberrant mitogenic signalling, resulting in cell cycle arrest and/or the induction of apoptosis [11]. p53 mutations have been found in a subset of chondrosarcomas, and are mostly associated with aggressive behaviour (reviewed in Rozeman et al., 2002 [19]). Amplification of MDM2 is frequently found in sarcomas (reviewed in Sandberg et al., 2004 [20]).
In addition to 12q13 and 9p21 alterations, Morrison et al. reported amplification of the oncogene c-MYC (8q24) in about 33% of high-grade chondrosarcomas [21]. However, these results could not be reproduced in other series [6]. c-MYC among others, drives cells into S phase [22]. Slight differences in c-MYC expression were reported between enchondromatosis-related and solitary chondrosarcomas [23].
The aim of our study was to investigate whether the pRb and p53 pathways harbour potential targets for therapy of inoperable or metastatic chondrosarcomas. Because 12q13 and 8q24 amplifications and 9p21 deletions suggest an important role for cell cycle regulators, especially those in the pRb and p53 pathways, we present the first in vitro evidence for an important role of C DKN2A/p16 and CDK4 in chondrosarcoma cell survival and proliferation. Subsequently, we validated the expression of CDK4, MDM2 and c-MYC at the mRNA and protein level in a large series of central chondrosarcomas.
Materials and methods
Cell culture
Chondrosarcoma cell lines derived from chondrosarcoma grade II (SW1353, American Type Culture Collection, Manassas, VA), and chondrosarcoma grade III (CH2879 [24] and OUMS27 [25]) were cultured in RPMI 1640 (Gibco, Invitrogen Life-Technologies, Scotland, UK). The breast carcinoma cell line MCF7 was grown in Dulbecco’s modified Eagle medium. Media for both cell lines were supplemented with 10% heat-inactivated foetal calf serum (Gibco). Cells were grown at 37°C in a humidified incubator with 95% air and 5% CO2. The cartilaginous phenotype was confirmed by RT-PCR, showing mRNA expression of collagens I, 2B, 3 and 10; Aggrecan; and SOX9[26].
Overexpressing and short hairpin (sh) RNA lentiviral vectors
The CDKN2A/p16-expressing lentiviral vector (kindly provided by Dr. R. Hoeben, department of Molecular Cell Biology, Leiden University Medical Center) has been described previously [27]. To generate vectors expressing shRNA against CDK4, oligonucleotides (for sequences see Supplementary Table S1) were cloned into the pTER vector [28]. Subsequently, fragments containing the H1-promoter and cloned oligonucleotides were recloned into the lentiviral pRRL-CMV-GFP vector [29]. Production of lentiviruses by transfection into 293T cells has been described previously [29]. For infection of the chondrosarcoma cell lines, 105 cells were seeded into 6-cm dishes and allowed to attach overnight. Virus was quantitated by antigen capture ELISA measuring HIV p24 levels (ZeptoMetrix Corporation, NY). This value has been converted to an infectious titre using the approximation that 1 ng of p24 equals 2500 infectious units (multiplicity of infection (MOI)). To obtain overexpression, cells were infected with the CDKN2A/p16-expressing lentivirus with an MOI of 1. An empty vector was used as a negative control for infections. To obtain specific knockdown, a mixture of three CDK4-shRNA-expressing lentiviral vectors was used (MOI 3); shRNA against murine MDM4 was used as a control. Cells were transduced in the presence of 8 μg/ml polybrene (Sigma Aldrich, Zwijndrecht, the Netherlands). Microscopic evaluation of green fluorescent protein (GFP) expression three days post-transduction showed 80–90% transduction efficiency for all conditions.
Immunoblotting
Proteins were extracted from cell cultures using Giordano lysis buffer (50 mm Tris-HCl pH 7.5, 250 mm NaCl, 0.1% Triton X-100, 5 mm EDTA, and 15% glycerol). Protein concentrations were measured using a Bradford assay (Bio-rad Laboratories, Hercules, CA, USA). Ten micrograms of total protein lysate from each sample was separated on SDS-PAGE. Lysates of normal human skin fibroblast cell line VH10, which was density-arrested and serum starved during two weeks, and subsequently reseeded in 20% serum, served as positive control for hyper-phosphorylated pRB (p-pRb). Two p16 negative melanoma cell lines were used as a control for loss of p16 staining in the chondrosarcoma cell lines. Proteins were transferred onto polyvinylidene difluoride membranes (Immobilon-P, Millipore, Billerica, MA, USA). Equal protein loading was verified by a tubulin staining. The membranes were pre-incubated with blocking solution (10% Non-fat dry milk in Tris buffered saline pH 8.0, 0.2% Tween-20). After incubation with primary (Supplementary Table S2) and secondary antibodies, the membranes were developed with Super Signal West Dura (Pierce Biotechnology, Rockford, IL, USA) and visualized by exposure to X-ray films or via the Chemigenius XE3 (Syngene, Cambridge, UK).
Proliferation assays
Cell counts were performed in duplicate using a Bürker chamber. A WST-1 colorimetric assay (Roche Diagnostics GmbH, Penzberg, Germany) was used to measure metabolic activity that represented the amount of viable cells. Briefly, cells were seeded into 96-well flat-bottom plates (1000 cells/well), each condition in quadruplicate. On days 3 and 6 post-transduction, the metabolic activity of the cells was measured on a Victor3 Multilabel Counter 1420-042 (Perkin Elmer, MA, USA) at 450 nm.
Clonogenic survival assay
Cells (1000, 5000 and 10000) were plated on 6-well plates. Cells were allowed to form colonies over a period of 14 days and subsequently fixed with methanol/acetic acid and stained using Giemsa.
Patient material
Conventional central chondrosarcomas were selected based on accepted clinicopathological and radiological criteria [1]. Peripheral-, juxtacortical-, mesenchymal-, dedifferentiated- and clear-cell chondrosarcomas were excluded. In total, specimens from 105 patients were studied including 45 high-grade chondrosarcomas. The clinical details are outlined in Table 1.
Table 1.
Clinicopathological data of the 105 enchondromas and chondrosarcomas
| Enchondromas | Chondrosarcomas | |||
|---|---|---|---|---|
| FFPE | Freshfrozen | FFPE | Fresh frozen | |
| Total number of tumours | 20 | 7* | 70 | 27 |
| Grade I | – | – | 25 | 11 |
| Grade II | – | – | 28** | 7 |
| Grade III | – | – | 17** | 9 |
| Male | 11 | 3 | 36 | 17 |
| Female | 9 | 4 | 34 | 10 |
| Enchondromatosis | 6 | 5 | 7 | 11 |
| Median age at diagnosis years (range) | 33.6 (11–66.4) | 18(12–37) | 51.4(17.8–84) | 40(17.8–84) |
| Median follow-up months (range) | 100 (6–221) | 84(5–247) | ||
Abbreviations: FFPE, Formalin fixed paraffin embedded.
All fresh frozen enchondromas were located in the phalanx.
A subset of 29 FFPE high-grade chondrosarcomas (grade II and III) was selected to study pRb and p53 pathway (Table 3).
Histological grading was performed according to Evans [3]. All specimens were handled according to the ethical guidelines described in ‘Code for Proper secondary Use of Human Tissue in The Netherlands’ of the Dutch Federation of Medical Scientific Societies.
Quantitative real-time reverse transcriptase PCR (qPCR)
Fresh frozen tumour tissue was available for RNA isolation, performed as described previously, from 34 cases [30]. Growth plate samples (n= 4) were used as controls. mRNA expression of CDK4, MDM2 and c-MYC (for primer sequences see Supplementary Table S3) were studied usingquantitative RT-PCR, as previously described [31]. Four control genes (CYPA, CPSF6, SRPR and HNRPH1) were selected because of their invariable expression in chondrosarcoma [31]. As a reference for normalization and statistical analysis, a mixture of 15 cell lines [23] was included. Normalization was performed using GENORM [32].
Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded material from 90 tumours was used, including 45 high-grade (grade II and III) chondrosarcomas to study CDK4, MDM2 and c-MYC by IHC. Twenty-nine of these 45 high-grade chondrosarcomas were previously investigated for CDKN2A/p16[15] protein expression (Table 1 and 3) and 14 of these were negative for p16. To obtain a full overview of the pRb pathway in high-grade chondrosarcomas, we further studied these 29 tumours for expression of other players in the pRb and p53 pathway by means of IHC for cyclin D1, p21 and p53. For 19 tumours, corresponding fresh frozen tissue was available (Table 1). Details of the primary antibodies used are described in Supplementary Table S2. As negative controls, slides were incubated in PBS/BSA 1% without primary specific antibodies. An IHC protocol optimized for cartilaginous tissue was applied to avoid detachment of sections [13]. Antigen retrieval was performed using citrate buffer at 98°C for 20 min in a water bath. Slides were independently semi-quantitatively scored for nuclear staining, as described previously [13] by two observers (YS, JVMG). Both were blinded to the clinicopathological data. Scores were given for intensity (1 = weak, 2 = moderate, 3 = strong) and for the percentage of positive cells (1 = 0–24%, 2 = 25–49%,3 = 50–74% and 4 = 75–100%). To avoid tumours with single positive cells being regarded as positive, cut-off levels for statistical analysis were applied (sum of score CDK4 and p53≥ 4, and of cyclin D1, MDM2 andc-MYC≥ 3. Tumours were regarded as negative for p21 with a sum of score ≤ 1, similar to the previous p16 staining [15]).
Table 3.
Twenty-nine high-grade chondrosarcomas and associated alterations in the key players of the pRb and p53 pathways
| L-number | pRb pathway | p53 pathway | |||||
|---|---|---|---|---|---|---|---|
| p16 | CDK4 | Cyclin D1 | p21 | MDM2 | p53 | ||
| 1 | 147 | + | − | − | − | n/a | − |
| 2 | 164 | + | + | + | + | − | − |
| 3 | 171 | − | + | − | + | − | − |
| 4 | 172 | − | + | + | + | n/a | − |
| 5 | 181 | − | + | − | − | − | − |
| 6 | 182 | + | + | − | + | − | − |
| 7 | 184 | − | + | + | + | + | + |
| 8 | 187 | − | + | + | − | − | − |
| 9 | 190 | − | + | − | − | n/a | − |
| 10 | 250 | + | − | + | + | − | + |
| 11 | 253 | − | − | n/a | − | + | n/a |
| 12 | 260 | + | − | + | + | − | + |
| 13 | 265 | + | − | + | + | + | + |
| 14 | 266 | − | − | + | − | − | + |
| 15 | 278 | + | + | − | + | + | − |
| 16 | 286 | − | − | − | + | n/a | − |
| 17 | 304 | + | − | + | + | + | − |
| 18 | 333 | − | − | + | + | − | + |
| 19 | 536 | + | − | + | − | − | − |
| 20 | 629 | + | + | + | + | − | + |
| 21 | 654 | − | − | + | + | − | + |
| 22 | 795 | − | + | + | + | + | + |
| 23 | 802 | + | + | − | + | − | − |
| 24 | 813 | + | + | n/a | n/a | − | n/a |
| 25 | 822 | + | − | + | + | − | + |
| 26 | 861 | − | + | − | − | n/a | − |
| 27 | 903 | + | − | + | + | − | − |
| 28 | 908 | + | + | + | + | + | + |
| 29 | 1066 | − | + | − | + | + | − |
| Total | Negative | Positive | Positive | Negative | Positive | Positive | |
| 14/29(48%) | 16/29 (55%) | 17/27(62%) | 8/28(28%) | 8/24(33%) | 11/27(41%) | ||
| Summary | 28/29(96%) | 21/29 (72%) | |||||
+ and – indicate positive and negative results, respectively. Immunohistochemical results for p16 were published previously [15]. n/a: data not available.
Statistical analysis
Normalized expression levels of different tumour groups were compared with growth plates using the Student’s t-test or one-way ANOVA with Bonferroni correction, after log10 transformation. Correlation between immunohistochemical staining and histological grade was analysed using Pearson chi-square. Immunohistochemical data were correlated with follow up by calculating the Kaplan–Meier curves and corresponding log rank tests. P-values < 0.05 were considered significant.
Results
Functional analysis of the pRb pathway in vitro
Immunoblotting showed an absence of CDKN2A/p16 in all three chondrosarcoma cell lines, while pRb was mainly present in its inactive, hyper-phosphorylated form (Fig. 1A). Overexpression of CDKN2A/p16 caused a shift of hyper-phosphorylated pRb to hypophosphorylated pRb (1B), and a decrease in total pRb levels in SW1353, OUMS27 and CH2879. The relative number of cells decreased in SW1353, OUMS27 and CH2879 upon overexpression of CDKN2A/p16 (P= 0.035, 0.002, and 0.014, respectively; Fig. 1C). In all cell lines, the WST-1 assay detected decreased metabolic activity, referred to as cell viability, to almost half of the metabolic activity of the controls for OUMS27 and CH2879 (P= 0.003 and P= 0.0455; Fig. 1D); this was less pronounced in SW1353 (P= 0.059).
Figure 1.
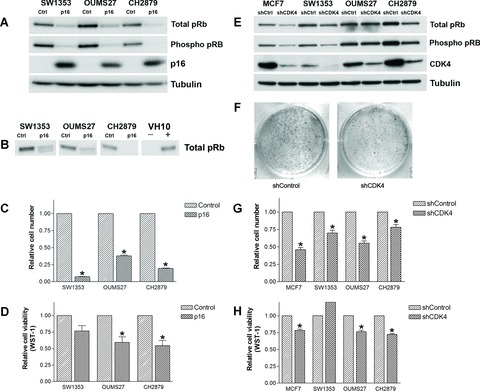
Immunoblot analysis of chondrosarcoma cell lines upon the overexpression of CDKN2A/p16 (A) or knock-down of CDK4 (E). (B) The characteristic band shift of pRb on 7.5% gel in the chondrosarcoma cell lines SW1353 and OUMS27. VH10 that was density-arrested and serum starved and subsequently reseeded in 20% serum (+) served as a control for hyper-phosporylated pRb. (F) The decreased ability of OUMS27 to form colonies after knockdown of CDK4. Relative cell numbers (C, G) and cell viability (D, H) compared with controls are shown (both measured 6 days after treatment). *significant differences.
All three cell lines showed high levels of CDK4 mRNA expression (see Supplementary Fig. S1). shRNA targeting CDK4 in the chondrosarcoma cell lines and MCF7 was effective (Fig. 1E). Decrease in hyper-phosphorylated pRb expression is indicative of cell cycle arrest and was found upon knockdown of CDK4 in all the cell lines, although to a lesser extent in OUMS27. Again, a reduction of total pRb with hypo-phosphorylation of existing pRb is observed. Enhanced degradation of pRb has been described previously after the inhibition of cyclin D1 in p21WAF1 expressing lung cancer cells [33]. Interestingly, the CH2879 cells, in which the effect on pRb levels is most prominent (Fig. 1E), show a relatively high level of p21WAF1, correlating with the wild-type p53 status in these cells (data not shown). The number of cells in the CDK4 shRNA transduced cells was significantly decreased (SW1353 P= 0.002; OUMS27 P= 0.003; CH2879 P= 0.015; Fig. 1G). Metabolic activity was decreased in CDK4 shRNA transduced OUMS27 and CH2879 cells (P= 0.0002 and P < 0.0001, respectively; Fig. 1H). In SW1353 an increase in metabolic activity was observed, analogous to the non-significant changes after overexpression of p16. SW1353 has a less cartilaginous appearance in vitro[34], despite its expression of typical cartilage mRNAs, and has a higher rate of proliferation than OUMS27 and CH2879, which may explain the divergent results in the WST-1 assay for this cell line. The capacity of all chondrosarcoma cell lines to form colonies in vitro was reduced upon knockdown of CDK4 (e.g. OUMS27, Fig. 1F).
Expression of pRb and p53 components in clinical samples
Quantitative PCR
The increase in CDK4 and MDM2 mRNA expression correlates with increasing histological grade (Fig. 2A and C) (Pearson R= 0.684, P < 0.0001 and R= 0.508, P= 0.007, respectively). Expression of MDM2 was significantly higher in tumours demonstrating 12q13 amplification at array-CGH [6] than in tumours without amplification (Student’s t-test P= 0.044, confidence interval [−0.83; −0.014], supplementary Fig. S2). c-MYC mRNA expression was not associated with histological grade (data not shown). In enchondromatosis-related tumours, c-MYC mRNA expression was significantly higher than in solitary tumours (Student’s t-test P= 0.011, confidence interval [−0.80; −0.116]; Supplementary Fig. S3).
Figure 2.
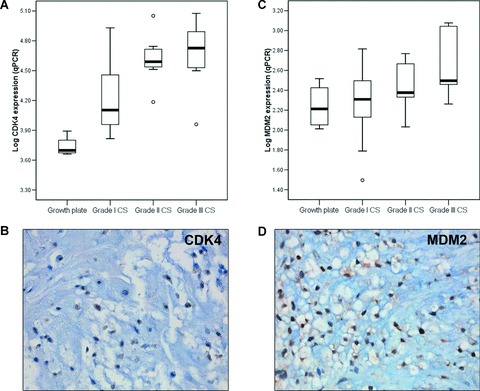
CDK4 (A) and MDM2 (C) mRNA expression levels relative to the growth plate are shown. Nuclear protein expression of CDK4 (B) and MDM2 (D) was determined (magnification 40×).
Immunohistochemistry
Results of CDK4, MDM2 and c-MYC staining on the series of 90 FFPE sections are shown in Table 2. Nuclear expression of CDK4 and MDM2 protein, as illustrated by Figure 2B and D, was correlated with increasing histological grade in chondrosarcomas (Pearson’s R= 0.368, P= 0.009 and Fig. 2E, R= 0.356, P= 0.007, respectively). Of the 29 high-grade chondrosarcomas that were previously studied for CDKN2A/p16 protein expression [15] (Table 3) and that were selected for further study, cyclin D1 was expressed in 62% (17/27). Moreover, 8 of 28 tumours were negative for p21, a CDK4/Cyclin D1 inhibitor activated by p53. Eight of 24 high-grade chondrosarcomas (33%) were positive for MDM2 and in 11 of 27 p53 was overexpressed. These results emphasize that aberrations in the pRb pathway occur in the majority (28/29, 96%) of high-grade chondrosarcomas. In fact, central cartilaginous tumours harbouring aberrations in the pRb pathway had shorter disease-free survival (log rank test P= 0.018), although this was not independent of histological grade.
Table 2.
Immunohistochemical staining of 90 FFPE samples of enchondroma and chondrosarcoma patients
| CDK4 | MDM2 | c-MYC | ||||
|---|---|---|---|---|---|---|
| Enchondroma | 6/12 | 50% | 0/20 | 0% | 0/20 | 0% |
| Chondrosarcomagrade I | 4/20 | 20% | 1/19 | 5% | 3/25 | 12% |
| Chondrosarcomagrade II | 11/21 | 52% | 7/25 | 28% | 2/28 | 7% |
| Chondrosarcomagrade III | 6/9 | 67% | 6/13 | 46% | 3/17 | 18% |
While a correlation between mRNA expression of CDK4 and MDM2 with 12q13 amplification was evident, only in two of four tumours with an amplification (L795 and L1066) CDK4 and MDM2 protein expression were found (data not shown). Nuclear c-MYC protein expression was found in 0 of 20 enchondromas and in only 8 of 70 (11%) chondrosarcomas (Table 2). The difference in c-MYC mRNA expression between enchondromatosis and solitary tumours was also not confirmed at the protein level(χ2P= 0.983).
Discussion
The aim of our study was to investigate whether the pRb and p53 pathways harbour potential targets for therapy of inoperable or metastatic chondrosarcomas. We present the first in vitro evidence for an important role of CDKN2A/p16 and CDK4 in chondrosarcoma cell survival and proliferation.
Unfortunately, there is nothing with curative intent to offer patients with inoperable or metastatic high-grade chondrosarcoma.12q13 and 8q24 amplifications and 9p21 deletions are reported and suggest an important role for cell cycle regulators. Therefore, we investigated whether the pRb and p53 pathways carry a specific target that could be used for future targeted therapy of high-grade central chondrosarcoma, similar to the attempts currently being made for other types of cancers. We demonstrate alterations of the pRb pathway in the vast majority of high-grade central chondrosarcomas.
Increased CDK4 expression, both at the mRNA and protein level, was found in 16 of 29 high-grade central chondrosarcomas and correlated with increasing histological grade and, consequently, poor prognosis. Increased CDK4 expression was also shown previously in a pRb-negative chondrosarcoma cell line by Asp et al. [16]. Reducing CDK4 expression in chondrosarcoma cell lines resulted in decreased survival and cell proliferation, confirming the important role of CDK4 in chondrosarcoma progression.
Loss of p16 was previously shown in chondrosarcoma specimens and cell lines by Asp et al. [7, 16] and by us [15]. We now show the functional implications of the p16 loss in central chondrosarcoma by overexpressing p16 in three p16-negative chondrosarcoma cell lines. We found decreased cell growth upon p16 overexpression, which is probably caused by senescence, since increased apoptosis was not observed (data not shown).
Other players in the pRb pathway are also affected in central chondrosarcoma, as was also reported previously[7, 14–17, 35]. We show that the pRb pathway is affected in 96% of the high-grade central chondrosarcomas, either by a decrease in the amount of CDKN2A/p16 (48%), an increase in the amount of CDK4 (55%) or expression of cyclin D1 (62%) (Table 3, Fig. 3). In addition, we show that MDM2 overexpression is correlated with increased histological grade. Thirteen of 38 (34%) high-grade tumours showed staining for MDM2, indicating that p53 degradation through MDM2 is also associated with tumour progression in a subset of central chondrosarcomas. Thus, alterations in p53 and pRb pathways are non-redundant in high-grade central chondrosarcoma. Crosstalk between the pRb and p53 pathway occurs via the p53 response gene, p21WAF1. The p21WAF1 protein can inhibit the CDK4-cyclin D1 complex upon overexpression (Fig. 3). Surprisingly, p21 expression was also shown to be associated with increasing grade in chondrosarcoma [36].
Figure 3.
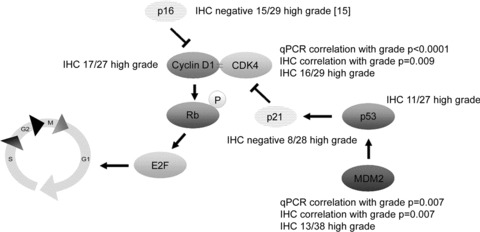
Changes in the pRb and p53 pathway promoting cell cycle passage in high-grade central chondrosarcoma, as found in the present and previous studies. Correlations are positive, unless stated otherwise.
The reported amplification of the c-MYC locus (8q24) [21] could not be verified in our previous arrayCGH experiments [6]. cDNA microarray demonstrated differences in mRNA expression of the oncogene c-MYC in enchondromatosis-related versus solitary tumours [23], which we confirmed in the present study using qPCR. c-MYC overexpression is observed in a large number of malignancies [22]. Based on the low c-MYC protein expression levels we detected in chondrosarcoma, the importance of oncogene c-MYC in central chondrosarcoma development or progression is questionable.
Chondrosarcomas are highly insensitive to classical chemotherapeutics that interfere with the cell cycle, like methotrexate and 5-fluorouracil, and to radiation therapy. Surgery is currently the only therapeutic option. We now show that central chondrosarcomas gain defects in the pRb pathway upon progression in grade, and that in the majority of high-grade chondrosarcomas either CDKN2A/p16 expression is inhibited, CDK4 is activated, or both. Our in vitro experiments with shRNA against CDK4 and overexpression of CDKN2A/p16 gene suggests that a number of therapeutic strategies may become possible including the use of CDK4 inhibitors. Functionally intact pRb signalling is a prerequisite for the effectivity of CDK4 inhibition. Despite LOH of 13q14 has been found in a subset of chondrosarcoma [5, 13, 14], pRb mutations were not found [5]. Moreover, we show deliberate pRb expression in three chondrosarcoma cell lines.
In MCF7 breast cancer cells, CDK inhibitors were effective in treating tumours that overexpress the CDK4-cyclin D1 complex or that have lost INK4a function [37]. Heat shock protein 90 (HSP90) inhibitors could also be potential proliferation blockers in chondrosarcoma, since the stabilization of CDK4, among other proteins, is regulated by HSP90 (reviewed in Whitesell et al., 2005 [38]). Our results indicate that studying the effect of compounds targeting CDK4, as soon as they are made available for (pre-)clinical use, would be the next step to investigate alternative, targeted treatment for high-grade central chondrosarcomas.
Acknowledgments
Dr. R. Hoeben and M. Rabelink, department of Molecular Cell Biology, LUMC, are kindly acknowledged for supplying lentiviral constructs and virus stocks. The authors thank I. Briaire-de Bruin for technical assistance. This study was supported by grants from Netherlands Organisation for Scientific Research [908-02-018 to Y.M.S and 917-76-315 to J.V.M.G.B.] and the Association for International Cancer Research [05-273 to S.L. and A.G.J.] This study was performed within the context of the EuroBoNeT consortium [018814 to Y.M.S., A.M.C.J., P.C.W.H. and J.V.M.G.B.], a European Commission granted Network of Excellence for studying the pathology and genetics of bone tumours.
Supporting Information
Fig. S1 Central chondrosarcoma cell lines SW1353, OUMS27and CH2879 show high levels of CDK4 mRNA in qPCR, comparable to the levels in high-grade chondrosarcoma.
Fig. S2 Central chondrosarcomas, showing gain at 12q13 atarrayCGH, express significantly higher mRNA levels ofMDM2.
Fig. S3 c-MYC mRNA levels are significantly increased in enchondromatosis patients.
Table S1 CDK4 shRNA oligonucleotide sequences.
Table S2 Procedures and details of the primary antibodies used for immuohistochemistry (IHC) and immunoblotting.
Table S3 Primer sequences used for qPCR.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Bertoni F, Bacchini P, Hogendoorn PCW. Chondrosarcoma. In: Fletcher CDM, Unni KK, Mertens F, editors. World Health Organisation classification of tumours. Pathology and genetics of tumours of soft tissue and bone. Lyon: IARC Press; 2002. pp. 247–51. . In: , editors. . [Google Scholar]
- 2.Lucas DR, Bridge JA. Chondromas: enchondroma, periosteal chondroma,and enchondromatosis. In: Fletcher CDM, Unni KK, Mertens F, editors. World Health Organization classification of tumours. Pathology and genetics of tumours of soft tissue and bone. Lyon: IARC Press; 2002. pp. 237–40. . In: , editors. . [Google Scholar]
- 3.Evans HL, Ayala AG, Romsdahl MM. Prognostic factors in chondrosarcoma of bone. A clinicopathologic analysis with emphasis on histologic grading. Cancer. 1977;40:818–31. doi: 10.1002/1097-0142(197708)40:2<818::aid-cncr2820400234>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Brien EW, Mirra JM, Kerr R. Benign and malignant cartilage tumors of bone and joint: their anatomic and theoretical basis with an emphasis on radiology, pathology and clinical biology I. The intramedullary cartilage tumors. Skeletal Radiol. 1997;26:325–53. doi: 10.1007/s002560050246. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi T, Toguchida J, Wadayama B, et al. Loss of heterozygosity and tumor suppressor gene mutations in chondrosarcomas. Anticancer Res. 1996;16:2009–15. [PubMed] [Google Scholar]
- 6.Rozeman LB, Szuhai K, Schrage YM, et al. Array-comparative genomic hybridization of central chondrosarcoma-Identification of ribosomal protein S6 and cyclin-dependent kinase 4 as candidate target genes for genomic aberrations. Cancer. 2006;107:380–8. doi: 10.1002/cncr.22001. [DOI] [PubMed] [Google Scholar]
- 7.Asp J, Sangiorgi L, Inerot SE, et al. Changes of the p16 gene but not the p53 gene in human chondrosarcoma tissues. Int J Cancer. 2000;85:782–6. doi: 10.1002/(sici)1097-0215(20000315)85:6<782::aid-ijc7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 8.Bovée JVMG, Sciot R, Dal Cin P, et al. Chromosome 9 alterations and trisomy 22 in central chondrosarcoma: a cytogenetic and DNA flow cytometric analysis of chondrosarcoma subtypes. Diagn Mol Pathol. 2001;10:228–36. doi: 10.1097/00019606-200112000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Larramendy ML, Tarkkanen M, Valle J, et al. Gains, losses, and amplifications of DNA sequences evaluated by comparative genomic hybridization in chondrosarcomas. Am J Pathol. 1997;150:685–91. [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–30. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, Kinzler KW. p53 Function and dysfunction. Cell. 1992;70:523–6. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–12. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 13.Bovée JVMG, Cleton-Jansen AM, Kuipers-Dijkshoorn N, et al. Loss of heterozygosity and DNA ploidy point to a diverging genetic mechanism in the origin of peripheral and central chondrosarcoma. Genes Chrom Cancer. 1999;26:237–46. [PubMed] [Google Scholar]
- 14.Ropke M, Boltze C, Meyer B, et al. Rb-loss is associated with high malignancy in chondrosarcoma. Oncol Rep. 2006;15:89–95. doi: 10.3892/or.15.1.89. [DOI] [PubMed] [Google Scholar]
- 15.Van Beerendonk HM, Rozeman LB, Taminiau AHM, et al. Molecular analysis of the INK4A/INK4A-ARF gene locus in conventional (central) chondrosarcomas and enchondromas: indication of an important gene for tumour progression. J Pathol. 2004;202:359–66. doi: 10.1002/path.1517. [DOI] [PubMed] [Google Scholar]
- 16.Asp J, Inerot S, Block JA, et al. Alterations in the regulatory pathway involving p16, pRb and cdk4 in human chondrosarcoma. J Orthop Res. 2001;19:149–54. doi: 10.1016/S0736-0266(00)00022-X. [DOI] [PubMed] [Google Scholar]
- 17.Rozeman LB, Hameetman L, Cleton-Jansen AM, et al. Absence of IHH and retention of PTHrP signalling in enchondromas and central chondrosarcomas. J Pathol. 2005;205:476–82. doi: 10.1002/path.1723. [DOI] [PubMed] [Google Scholar]
- 18.Muthusamy V, Hobbs C, Nogueira C, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer. 2006;45:447–54. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- 19.Rozeman LB, Hogendoorn PCW, Bovée JVMG. Diagnosis and prognosis of chondrosarcoma of bone. Expert Rev Mol Diagn. 2002;2:461–72. doi: 10.1586/14737159.2.5.461. [DOI] [PubMed] [Google Scholar]
- 20.Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: liposarcoma. Cancer Genet Cytogenet. 2004;155:1–24. doi: 10.1016/j.cancergencyto.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Morrison C, Radmacher M, Mohammed N, et al. MYC Amplification and polysomy 8 in chondrosarcoma: array comparative genomic hybridization, fluorescent in situ hybridization, and association with outcome. J Clin Oncol. 2005;23:9369–76. doi: 10.1200/JCO.2005.03.7127. [DOI] [PubMed] [Google Scholar]
- 22.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 23.Rozeman LB, Hameetman L, Van Wezel T, et al. cDNA expression profiling of central chondrosarcomas: Ollier disease resembles solitary tumors and alteration in genes coding for energy metabolism with increasing grade. J Pathol. 2005;207:61–71. doi: 10.1002/path.1813. [DOI] [PubMed] [Google Scholar]
- 24.Gil-Benso R, Lopez-Gines C, Lopez-Guerrero JA, et al. Establishment and characterization of a continuous human chondrosarcoma cell line, ch-2879: comparative histologic and genetic studies with its tumor of origin. Lab Invest. 2003;83:877–87. doi: 10.1097/01.lab.0000073131.34648.ea. [DOI] [PubMed] [Google Scholar]
- 25.Kunisada T, Miyazaki M, Mihara K, et al. A new human chondrosarcoma cell line (OUMS-27) that maintains chondrocytic differentiation. Int J Cancer. 1998;77:854–9. doi: 10.1002/(sici)1097-0215(19980911)77:6<854::aid-ijc10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Cleton-Jansen AM, Van Beerendonk HM, Baelde HJ, et al. Estrogen signaling is active in cartilaginous tumors: implications for antiestrogen therapy as treatment option of metastasized or irresectable chondrosarcoma. Clin Cancer Res. 2005;11:8028–35. doi: 10.1158/1078-0432.CCR-05-1253. [DOI] [PubMed] [Google Scholar]
- 27.Oruetxebarria I, Venturini F, Kekarainen T, et al. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoidtumor cells. J Biol Chem. 2004;279:3807–16. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 28.Van De Wetering M, Oving I, Muncan V, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–15. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlotti F, Bazuine M, Kekarainen T, et al. Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol Ther. 2004;9:209–17. doi: 10.1016/j.ymthe.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Baelde HJ, Cleton-Jansen AM, Van Beerendonk H, et al. High quality RNA isolation from tumours with low cellularity and high extracellular matrix component for cDNA microarrays: application to chondrosarcoma. J Clin Pathol. 2001;54:778–82. doi: 10.1136/jcp.54.10.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hameetman L, Rozeman LB, Lombaerts M, et al. Peripheral chondrosarcoma progression is accompanied by decreased Indian Hedgehog (IHH) signalling. J Pathol. 2006;209:501–11. doi: 10.1002/path.2008. [DOI] [PubMed] [Google Scholar]
- 32.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. : research0034.1–.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Driscoll B, Wu L, Buckley S, et al. Cyclin D1 antisense RNA destabilizes pRb and retards lung cancer cell growth. Am J Physiol. 1997;273:941–9. doi: 10.1152/ajplung.1997.273.5.L941. [DOI] [PubMed] [Google Scholar]
- 34.Ouyang P. An in vitro model to study mesenchymal-epithelial transformation. Biochem Biophys Res Commun. 1998;246:771–6. doi: 10.1006/bbrc.1998.8710. [DOI] [PubMed] [Google Scholar]
- 35.Asp J, Brantsing C, Lovstedt K, et al. Evaluation of p16 and Id1 status and endogenous reference genes in human chondrosarcoma by real-time PCR. Int J Oncol. 2005;27:1577–82. [PubMed] [Google Scholar]
- 36.Bovée JVMG, Van den Broek LJCM, Cleton-Jansen AM, et al. Up-regulation of PTHrP and Bcl-2 expression characterizes the progression of osteochondroma towards peripheral chondrosarcoma and is a late event in central chondrosarcoma. Lab Invest. 2000;80:1925–33. doi: 10.1038/labinvest.3780202. [DOI] [PubMed] [Google Scholar]
- 37.Grillo M, Bott MJ, Khandke N, et al. Validation of cyclin D1/CDK4 as an anticancer drug target in MCF-7 breast cancer cells: effect of regulated overexpression of cyclin D1 and siRNA-mediated inhibition of endogenous cyclin D1 and CDK4 expression. Breast Cancer Res Treat. 2006;95:185–94. doi: 10.1007/s10549-005-9066-y. [DOI] [PubMed] [Google Scholar]
- 38.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]


