Abstract
Sooty mangabeys (Cercocebus atys) are natural SIV hosts and the presumed source of HIV-2 and SIVmac, which makes them a valuable model for HIV/SIV research. However, like other African primates, little is known about their major histocompatibility complex (MHC) genetics. In this study, we used Roche/454 and Illumina MiSeq deep sequencing in order to determine the MHC class I transcripts in a cohort of 165 sooty mangabeys from the Yerkes National Primate Research Center (YNPRC). We have characterized 121 functionally full-length classical (Ceat-A and Ceat-B) and non-classical (Ceat-F and Ceat-I) alleles and have also identified 22 Ceat-A/Ceat-B haplotype chromosomal combinations. We correlated these Ceat-A/Ceat-B haplotype combinations to recently described microsatellite haplotypes from the YNPRC colony. These newly identified alleles and haplotypes establish a resource for studying cellular immunity in sooty mangabeys and provide a framework for rapidly cataloging MHC class I sequences in an understudied, yet important, nonhuman primate species.
Keywords: MHC class I, sooty mangabey, SIV natural host, haplotyping, deep sequencing
Introduction
Sooty mangabeys (Cercocebus atys) are an important model for HIV/AIDS research because they are a natural SIV host and the source of HIV-2 and SIVmac (Apetrei et al. 2005b; Locatelli et al. 2014; Sharp and Hahn 2011). Sooty mangabeys are native to West Africa, ranging from Guinea to Ghana. Two subspecies are recognized: C. a. atys, ranging from Guinea to Cote d’Ivoire, is the subspecies in which SIV is more prevalent, though the subspecies found in Cote d’Ivoire and Ghana, C. a. lumunatus, can also harbor the virus (Santiago et al. 2005).
SIV is prevalent in wild sooty mangabeys, with current estimates of infection between 59–63% (Apetrei et al. 2005a; Santiago et al. 2005). Interestingly, these animals do not appear to develop AIDS, despite persistently high viral loads (Chahroudi et al. 2012; Rey-Cuillé et al. 1998). These high viral loads engender SIV-specific CD8+ T cell responses that resemble those observed in non-natural hosts such as rhesus macaques (Silvestri 2005; Wang et al. 2006). These responses also include cytokine secretion that is measureable ex vivo in the peripheral blood (Wang et al. 2006). Research continues to elucidate the mechanism of non-pathogenic infection in natural SIV hosts; two of the main hypotheses are limited immune activation and low CCR5 expression (Estes et al. 2008; Paiardini et al. 2011; Silvestri et al. 2007). Only 7 SIV-specific CTL epitopes have been identified in sooty mangabeys (Kaur et al. 2000), at least in part due to the currently limited knowledge of the major histocompatibility complex (MHC) molecules that bind SIV epitopes in these animals.
Most of what is known about Old World monkey genetics is inferred from the intensively studied Asian macaques. Rhesus, cynomolgus, and pig-tailed macaques have MHC class I loci that are both polymorphic and polyallelic. MHC class I A and B loci have undergone gene duplication, such that individual animals can have upwards of 20 of these genes on a single haplotype (Daza-Vamenta et al. 2004) A subset of these are transcriptionally abundant in peripheral blood mononuclear cells (PBMC), whilst others are transcribed at low levels, or, in some cases, not at all (Budde et al. 2011). Because of their importance in binding and presenting peptides to T cells, the classical MHC-A and MHC-B genes have received the most attention. In addition to these classical MHC class I genes, however, it should be noted that macaques also have non-classical MHC-E, MHC-F, and MHC–I loci that may be involved in specialized immune processes such as regulation of cytolytic T cells (Adams and Parham 2001).
The organization of MHC genes in African monkeys is largely unknown. Five functional MHC-I sequences (2 MHC-A, 2 MHC-B, and 1 MHC-E) have been identified from a single yellow baboon; these alleles have greater than 90% nucleotide similarity to rhesus macaques (Sidebottom et al. 2001). The baboon MHC-I appears to have high intralocus variability, mainly within the peptide-binding pocket, similar to MHC loci of Asian macaques (Sidebottom et al. 2001). A recent study of African green monkeys, which are also a SIV natural host, identified duplicated MHC class I A and B alleles (Cao et al. 2014). Sooty mangabeys are most closely related to baboons, followed by macaques and African green monkeys (Rogers and Gibbs 2014). Therefore, we would expect the organization of the sooty mangabey MHC-I region to be similar to these populations.
In this study, we used deep sequencing to define MHC class I allele repertoires of 165 sooty mangabeys. We demonstrate an expansion of MHC-A and MHC–B genes relative to humans, as has been previously described in macaques (Wiseman et al. 2013), baboons (Sidebottom et al. 2001), and African green monkeys (Cao et al. 2014), and provide the first description of MHC class I sequences in this important population of African monkeys.
Materials and Methods
Animals
PBMC were obtained from 165 sooty mangabeys housed at Yerkes National Primate Research Center (YNPRC). The YNPRC sooty mangabey colony was founded by 22 animals in 1968 (Sharma et al. 2014). The YNPRC is fully accredited by AAALAC, International and all animals are cared for under procedures approved by the Emory University Institutional Animals Care and Use committee.
cDNA Synthesis and PCR Amplification for Roche/454 Sequencing
RNA was isolated from frozen PBMC using the Roche MagNA Pure instrument and high performance RNA kit (Roche, Indianapolis, IN, USA) according to manufacturer’s protocols. cDNA was synthesized using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) and each sample was normalized to 10 ng/μL. PCR was performed to amplify a 530bp amplicon, spanning exons 2–4 of MHC-I, using Phusion High Fidelity DNA Polymerase (New Wngland biolabs, Ipswich, MA, USA) and the previously described SBT568F and SBT568R primers (Fernandez et al. 2011; Karl et al. 2013; Wiseman et al. 2013). All primers contained a Roche/454 sample-specific MID tag. Thermocycler conditions were: denaturation at 98°C for 3 min, 23–30 elongation cycles of 98°C for 5 s, 60°C for 10 s, and 72°C for 20 s, and a final extension at 72°C for 5 min. Amplicons were confirmed using the FlashGel System (Lonza Group Ltd., Basel, Switzerland). Amplified samples were purified twice with AMPure XP SPRI beads (Agencourt Bioscience Corporation, Beverly, MA, USA) at a 1:1 bead to sample volume ratio. All samples were then quantified using the Quant-iT dsDNA HS Assay Kit (Life Technologies, Grand Island, NY, USA) and the DTX800 Multimode Detector (Beckman-Coulter, Brea, CA, USA).
Roche/454 Sequencing
Purified PCR products from individual animals were pooled together at equimolar ratios. According to Roche/454 protocols, emulsion PCR (emPCR) was then performed on the pool with thermocycler conditions: 94°C for 4 min and then 50 cycles of 94°C for 30 sec, 58°C for 4.5 min, 68°C for 30 sec. emPCR was followed by breaking, enriching, and sequencing on either the Roche/454 GS FLX or GS Junior instrument using Titanium technology.
Roche/454 Data Analysis
The Roche/454 data for the first 83 samples were analyzed to create a database of sooty mangabey exon 2–4 sequences essentially as previously described (Karl et al. 2013). In brief, small form factor (SFF) files were imported into Genious Pro (v5.4) (Biomatters Limited, Aukland, New Zealand) to create uni-directional contigs of sequences by assembling at 100% identity. These contigs were then imported into CodonCode Aligner (v3.5) (CodonCode Corporation, Centerville, MA, USA) and bi-directional contigs were created by interrogating the overlap region between the uni-directional contigs for unambiguous assembly; for example, if a forward contig matched two or more reverse contigs, those three contigs were excluded from the database. All unique, unambiguously assembled contigs represented by at least 3 independent reads were used to create a database of 530bp sooty mangabey sequences.
Once a database was established, the data from all 165 sooty mangabey samples were processed through a custom script to convert Roche/454 SFF files into FASTQ sequencing reads using sff-extract (v0.3.0) (available as part of seq.crumbs at bioinf.comav.upv.es/seq-crumbs/download.html). Reads corresponding to each MID index were binned. Primer and sequencing adapter sequences were removed using FLEXBAR v2.34 (Dodt et al. 2012). Trimmed reads were mapped to the database of 530bp sooty mangabey sequences using Bowtie2 (v2.1.0) (Langmead et al. 2012). Only reads that were a perfect match to a database sequence were reported by the script, and the number of reads supporting each allele assignment for each MID tagged sample were aggregated and used for haplotype assessment.
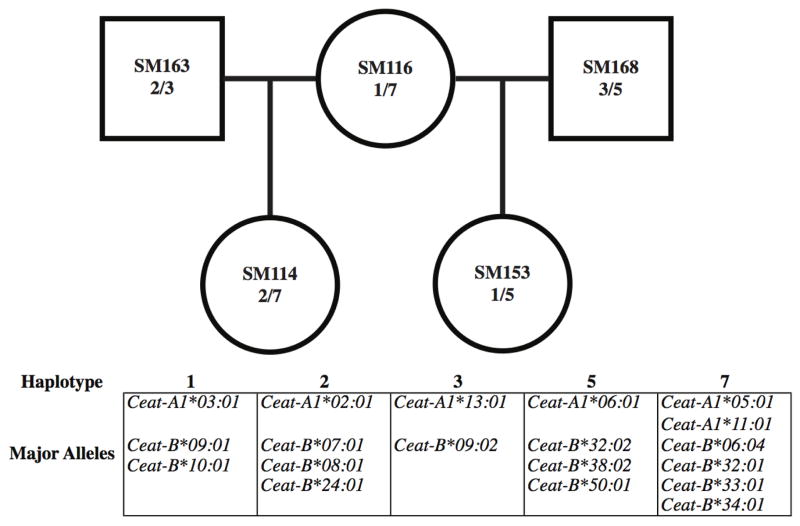
Animals were organized into dam/sire/progeny sets using familial information from the YNPRC colony. This pedigree data was used to deduce Ceat-A and Ceat-B Roche/454 amplicon haplotypes. Haplotypes are defined as a group of alleles that are inherited together, presumably on the same chromosome. First, we looked at the dam, sire, and progeny sets and determined haplotypes via patterns of allele sharing. For trio sets where both parents and their progeny were all Roche/454 genotyped, the parental chromosomes inherited by the progeny were easily determined (Figure 1). If only one parent was genotyped, after using direct inheritance to assign alleles to one haplotype of the progeny, we inferred that the remaining alleles belonged to the other haplotype in each animal. For the animals where neither parent was genotyped, we used our previously determined haplotype combinations to deduce the segregation of alleles. Alleles were also classified as “major” or “minor” in terms of transcriptional abundance, with an allele considered major if it was represented by an average of at least 4% of the total reads per animal and was present in the majority of animals with the particular haplotype (Karl et al. 2013). After individual A and B haplotypes were determined, we determined chromosomal Ceat-A/Ceat-B combinations prevalent in this cohort.
Fig 1.
Using pedigrees to determine haplotypes. SM116 is the dam of both progeny; SM114 inherited haplotype 7 and SM153 inherited haplotype 1. SM114 therefore inherited haplotype 2 from her sire, SM163. Likewise, SM153 inherited haplotype 5 from her sire, SM168. Haplotype 3 of each of the sires was not passed on to these progeny
Animal Selection for Full-Length Allele Discovery
Using the haplotypes deduced for all 165 animals, 32 animals were selected for further full-length MHC class I interrogation. All haplotypes present in this 32 animal cohort were represented at least twice.
cDNA Synthesis, PCR Amplification, and Illumina MiSeq Sequencing
Sample preparation from cDNA to Illumina MiSeq sequencing was performed as previously described (Dudley et al. 2014). Fragmented pools of full-length MHC amplicons ranged in average length from 471 to 762bp, determined using the Agilent High Sensitivity Bioanalyzer Kit (Agilent Technologies, Santa Clara, CA, USA). Concentrations ranged from 0.189ng/μL to 3.72ng/μL (via Quant-iT HS assay kit and Qubit fluorometer (Life Technologies)). The samples were then pooled, denatured, and diluted to 12 pM according to Illumina MiSeq sample preparation protocols before being run on the Illumina MiSeq using the 600 cycle MiSeq Reagent Kit v2.
MiSeq Data Analysis
Full-length sequence analysis was performed using Geneious Pro Software (v7.1.5) with a semi-automated pipeline. FASTQ files were imported into Geneious and parsed based on Illumina indices. For each animal, forward and reverse reads were paired. Duplicate reads were then determined and assigned a pseudoquality score based on number of reads. In this analysis, we focused on alleles with high numbers of duplicate reads representing high-confidence alleles present in our animals. Unique reads were binned for later use in the pipeline. The top 2000 pseudoquality-scored sequences were de novo assembled, with a minimum overlap of 150bp at 100% stringency, allowing gaps, and 5% mismatches. Consensus sequences were then generated from contigs 1000bp or longer, which represent functionally full-length MHC class I sequences. The unique reads were mapped against these consensus sequences, allowing 1% mismatches. This process was repeated 5 times, de novo assembling the 2000 next highest-scoring sequences and mapping the remaining unique reads to the consensus sequence. The original paired reads were then mapped to the consensus sequences to extend each sequence as far as possible, generating extension contigs that were subject to further analysis. These extension contigs were then compared against the exon 2–4 database from the Roche/454 data analysis using the Blast Local Alignment Search Tool (BLAST), and validated for correct reading frame and to look for apparent PCR recombination chimerism. Sequences were submitted to GenBank (KP166446-KP176471, KP176494-KP176596) and to the Immuno Polymorphism Database (IPD) for official naming.
Results and Discussion
Allele Discovery
In this study we characterized 121 functionally full-length (from start codon or leader peptide through stop codon) alleles: 26 Ceat-A, 83 Ceat-B, 4 Ceat-F, and 8 Ceat-I (Table 1). The distribution across the loci is similar to the MHC class I of other nonhuman primates with a significantly higher number of alleles in the B region than the A, F, or I loci (Daza-Vamenta et al. 2004). There is also a significant founder effect due to the limited overall variability provided by the 22 animals that established the YNPRC sooty mangabey colony (Smith et al. 2015). While this limited diversity is likely not representative of the wild sooty mangabey population, it could inform studies using sooty mangabeys from the YNPRC. The diversity observed in this colony is intermediate between the inbred Mauritian cynomolgus macaques and the more diverse Indian or Chinese cynomolgus macaque colonies (Karl et al. 2013; Wiseman et al. 2007).
Table I.
Sooty mangabey alleles
| Allele | GenBank Accession No. | No. Animals |
|---|---|---|
| Ceat-A1*01:01 | KP176446 | 2 |
| Ceat-A1*02:01 | KP176447 | 2 |
| Ceat-A1*03:01 | KP176452 | 8 |
| Ceat-A1*04:01 | KP176454 | 3 |
| Ceat-A1*05:01 | KP176455 | 2 |
| Ceat-A1*06:01 | KP176456 | 12 |
| Ceat-A1*08:01 | KP176463 | 3 |
| Ceat-A1*09:01 | KP176459 | 2 |
| Ceat-A1*10:01 | KP176460 | 2 |
| Ceat-A1*11:01 | KP176464 | 2 |
| Ceat-A1*11:02 | KP176562 | 5 |
| Ceat-A1*12:01 | KP176465 | 5 |
| Ceat-A1*13:01 | KP176466 | 5 |
| Ceat-A1*14:01 | KP176467 | 1 |
| Ceat-A1*15:01 | KP176468 | 1 |
| Ceat-A1*16:01 | KP176469 | 2 |
| Ceat-A1*17:01 | KP176470 | 2 |
| Ceat-Aw1*01:01 | KP176448 | 4 |
| Ceat-Aw1*01:02 | KP176449 | 1 |
| Ceat-Aw1*01:03 | KP176450 | 3 |
| Ceat-Aw1*01:08 | KP176471 | 3 |
| Ceat-Aw2*01:01 | KP176453 | 1 |
| Ceat-Aw2*02:01 | KP176458 | 2 |
| Ceat-Aw2*02:02 | KP176461 | 2 |
| Ceat-Aw2*02:03 | KP176462 | 2 |
| Ceat-Aw2*03:01 | KP176457 | 1 |
| Ceat-B*01:01Sp | KP176583 | 1 |
| Ceat-B*03:01 | KP176498 | 3 |
| Ceat-B*04:02 | KP176500 | 3 |
| Ceat-B*05:01 | KP176499 | 3 |
| Ceat-B*05:02 | KP176533 | 3 |
| Ceat-B*06:01 | KP176501 | 3 |
| Ceat-B*06:02 | KP176502 | 7 |
| Ceat-B*06:03 | KP176564 | 2 |
| Ceat-B*06:04 | KP176571 | 8 |
| Ceat-B*07:01 | KP176503 | 2 |
| Ceat-B*08:01 | KP176504 | 2 |
| Ceat-B*09:01 | KP176505 | 7 |
| Ceat-B*09:02 | KP176506 | 10 |
| Ceat-B*10:01 | KP176507 | 7 |
| Ceat-B*10:02 | KP176508 | 6 |
| Ceat-B*10:03 | KP176509 | 7 |
| Ceat-B*11:01 | KP176510 | 7 |
| Ceat-B*11:02 | KP176511 | 10 |
| Ceat-B*12:01 | KP176512 | 6 |
| Ceat-B*13:01N | KP176513 | 5 |
| Ceat-B*13:02N | KP176515 | 2 |
| Ceat-B*14:01N | KP176514 | 3 |
| Ceat-B*15:01 | KP176516 | 4 |
| Ceat-B*15:02 | KP176552 | 2 |
| Ceat-B*15:03 | KP176563 | 3 |
| Ceat-B*16:01 | KP176517 | 4 |
| Ceat-B*16:02 | KP176518 | 8 |
| Ceat-B*17:01:01 | KP176520 | 2 |
| Ceat-B*17:01:02 | KP176522 | 2 |
| Ceat-B*17:02 | KP176519 | 3 |
| Ceat-B*17:03 | KP176521 | 1 |
| Ceat-B*18:02 | KP176523 | 3 |
| Ceat-B*20:01N | KP176524 | 3 |
| Ceat-B*21:01 | KP176595 | 1 |
| Ceat-B*22:01 | KP176596 | 1 |
| Ceat-B*23:01 | KP176525 | 1 |
| Ceat-B*23:02 | KP176526 | 5 |
| Ceat-B*24:01 | KP176527 | 2 |
| Ceat-B*25:01 | KP176528 | 1 |
| Ceat-B*26:01 | KP176529 | 3 |
| Ceat-B*27:03 | KP176530 | 1 |
| Ceat-B*28:01N | KP176531 | 1 |
| Ceat-B*29:01 | KP176532 | 4 |
| Ceat-B*30:01 | KP176534 | 2 |
| Ceat-B*31:02 | KP176535 | 2 |
| Ceat-B*31:03 | KP176536 | 3 |
| Ceat-B*32:01 | KP176537 | 6 |
| Ceat-B*32:02 | KP176538 | 7 |
| Ceat-B*33:01 | KP176539 | 8 |
| Ceat-B*34:01 | KP176540 | 8 |
| Ceat-B*35:01 | KP176541 | 1 |
| Ceat-B*35:03 | KP176542 | 1 |
| Ceat-B*37:01 | KP176543 | 5 |
| Ceat-B*38:01 | KP176544 | 4 |
| Ceat-B*38:02 | KP176545 | 9 |
| Ceat-B*39:01 | KP176546 | 4 |
| Ceat-B*40:01 | KP176547 | 4 |
| Ceat-B*40:02 | KP176548 | 8 |
| Ceat-B*40:03 | KP176574 | 1 |
| Ceat-B*41:01 | KP176549 | 3 |
| Ceat-B*42:01 | KP176550 | 3 |
| Ceat-B*43:01Sp | KP176551 | 3 |
| Ceat-B*44:01N | KP176553 | 1 |
| Ceat-B*45:01N | KP176554 | 3 |
| Ceat-B*45:02 | KP176579 | 3 |
| Ceat-B*46:01 | KP176555 | 7 |
| Ceat-B*47:01 | KP176556 | 2 |
| Ceat-B*48:01 | KP176557 | 3 |
| Ceat-B*49:01 | KP176558 | 2 |
| Ceat-B*49:02 | KP176568 | 1 |
| Ceat-B*50:01 | KP176559 | 9 |
| Ceat-B*51:01 | KP176560 | 6 |
| Ceat-B*51:02 | KP176573 | 7 |
| Ceat-B*52:01 | KP176561 | 5 |
| Ceat-B*53:01 | KP176565 | 3 |
| Ceat-B*54:01 | KP176566 | 1 |
| Ceat-B*55:01 | KP176567 | 1 |
| Ceat-B*56:01 | KP176569 | 1 |
| Ceat-B*57:01 | KP176570 | 4 |
| Ceat-B*58:01 | KP176572 | 4 |
| Ceat-B*59:01 | KP176575 | 2 |
| Ceat-B*60:01 | KP176576 | 4 |
| Ceat-B*61:01 | KP176592 | 2 |
| Ceat-F*01:01 | KP176494 | 11 |
| Ceat-F*01:02 | KP176495 | 5 |
| Ceat-F*01:03 | KP176496 | 2 |
| Ceat-F*01:04 | KP176497 | 1 |
| Ceat-I*01:01 | KP176584 | 6 |
| Ceat-I*01:02 | KP176585 | 2 |
| Ceat-I*01:03 | KP176586 | 3 |
| Ceat-I*02:01 | KP176587 | 2 |
| Ceat-I*02:02 | KP176588 | 1 |
| Ceat-I*02:03 | KP176589 | 7 |
| Ceat-I*02:04 | KP176590 | 2 |
| Ceat-I*03:01 | KP176591 | 3 |
Official IPD names and GenBank accession numbers of all full-length alleles and the number of animals in which they were observed
Haplotype and Chromosomal Combinations Characterization
While Ceat-A, Ceat-B, Ceat-F, and Ceat-I alleles were identified, our further analysis focused solely on the classical A and B loci, because their peptide presentation functions are better characterized and they are the focus of most immunological research performed in nonhuman primates. The use of colony breeding data allowed us to identify 17 trios and 44 dam/sire and offspring duos (Supplemental Table 1). This pedigree information allowed us to deduce haplotypes for this colony. We were able to identify 22 chromosomal combinations of Ceat-A and Ceat-B haplotypes (Table 2). Each haplotype is composed of 1–2 major and 0–2 minor Ceat-A alleles and 1–4 major and 0–11 minor Ceat-B alleles (Supplemental Table 2). The haplotypes for which no minor alleles were identified may prove to contain very lowly expressed minor alleles below the limit of detection of the current study.
Table 2.
Sooty mangabey haplotypes
| Haplotype | No. Chromosomes | A major 1 | A major 2 | B major 1 | B major 2 | B major 3 | B major 4 |
|---|---|---|---|---|---|---|---|
| 1 | 50 | Ceat-A1*03:01 | Ceat-B*09:01 | Ceat-B*10:01 | |||
| 2 | 38 | Ceat-A1*02:01 | Ceat-B*07:01 | Ceat-B*08:01 | Ceat-B*24:01 | ||
| 3 | 36 | Ceat-A1*13:01 | Ceat-B*09:02 | ||||
| 4 | 35 | Ceat-A1*04:01 | Ceat-A1*08:01 | Ceat-B*05:02 | Ceat-B*29:01 | ||
| 5 | 32 | Ceat-A1*06:01 | Ceat-B*32:02 | Ceat-B*38:02 | Ceat-B*50:01 | ||
| 6 | 24 | Ceat-A1*12:01 | Ceat-B*14:01N | Ceat-B*41:01 | |||
| 7 | 17 | Ceat-A1*05:01 | Ceat-A1*11:01 | Ceat-B*06:04 | Ceat-B*32:01 | Ceat-B*33:01 | Ceat-B*34:01 |
| 8 | 14 | Ceat-A1*11:02 | Ceat-B*06:03 | Ceat-B*52:01 | |||
| 9 | 14 | Ceat-A1*17:01 | Ceat-B*06:04 | Ceat-B*32:01 | Ceat-B*33:01 | Ceat-B*34:01 | |
| 10 | 13 | Ceat-A1*01:01 | Ceat-B*03:01 | Ceat-B*04:02 | Ceat-B*05:02 | Ceat-B*06:01 | |
| 11 | 11 | Ceat-A1*16:01 | Ceat-B*06:02 | Ceat-B*09:02 | |||
| 12 | 10 | Ceat-A1*14:01 | Ceat-B*06:03 | Ceat-B*52:01 | |||
| 13 | 8 | Ceat-A1*06:01 | Ceat-B*06:02 | Ceat-B*37:01 | Ceat-B*38:01 | Ceat-B*39:01 | |
| 14 | 7 | Ceat-A1*09:01 | Ceat-B*06:04 | Ceat-B*32:01 | Ceat-B*33:01 | Ceat-B*34:01 | |
| 15 | 6 | Ceat-A1*10:01 | Ceat-B*09:02 | ||||
| 16 | 5 | Ceat-A1*03:01 | Ceat-B*37:01 | Ceat-B*38:01 | |||
| 17 | 4 | Ceat-A1*07:01 | Ceat-B*02:01 | Ceat-B*03:02 | Ceat-B*04:01 | ||
| 18 | 2 | Ceat-A1*12:01 | Ceat-B*06:04 | Ceat-B*32:01 | Ceat-B*33:01 | Ceat-B*34:01 | |
| 19 | 1 | Ceat-A1*15:01 | Ceat-B*54:01 | Ceat-B*55:01 | |||
| 20 | 1 | Ceat-A1*03:01 | Ceat-B*03:01 | Ceat-B*04:02 | Ceat-B*05:01 | Ceat-B*06:01 | |
| 21 | 1 | Ceat-A1*02:01 | Ceat-B*06:03 | Ceat-B*52:01 | |||
| 22 | 1 | Ceat-A1*09:01 | Ceat-B*09:01 | Ceat-B*10:02 |
The major alleles of all 22 A/B haplotypes found in this cohort and the number of chromosomes representing each combination
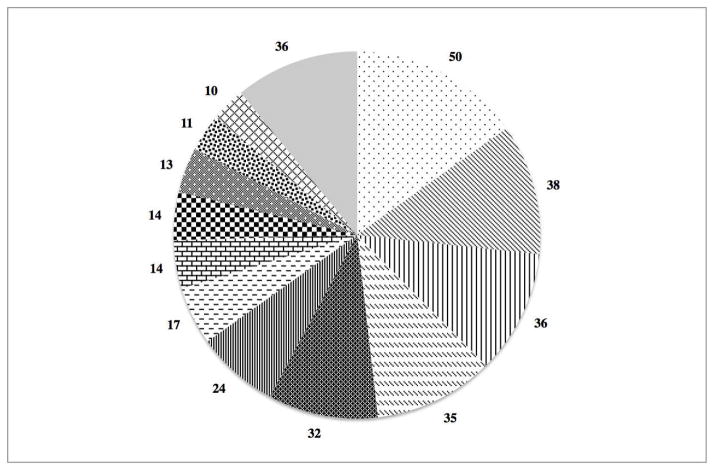
The 12 most common haplotypes were all observed at least 10 times amongst our analysis of the 165 animals, again showing the limited diversity of this cohort (Figure 2). The five most common haplotypes encompass 191 of the total 330 chromosomes in our cohort (58%).
Fig 2.
Diversity of the YNPRC sooty mangabey colony. The 5 most common haplotypes encompass 58% of the total chromosomes found in this cohort (191/330). The number of chromosomes for the top 12 haplotypes are shown, haplotypes 13–22 have been combined
Noteworthy, the allele Ceat-B*12:01, a minor allele associated with haplotypes 1 and 22 (Supplemental Table 2), has been found to present a SIV-Nef epitope LRARGETY (Wang manuscript in preparation). This allele was present in 15.4% of the chromosomes in our cohort. This information may be useful for studies interested in characterizing T cell responses of the YNPRC sooty mangabey colony, especially Nef-restricting responses.
Interestingly, we have identified several singleton haplotypes found in only one animal. For example, the alleles present in haplotype 19 were only observed in one animal (which had no ancestors included in our cohort), but had extremely strong representation in that animal. Haplotype 10, an abundant haplotype in the colony, was also present in this animal, supporting the assignment of remaining alleles to an alternative haplotype. This singleton haplotype is likely a rare haplotype in the YNPRC colony, and thus animals with this haplotype were not sampled more than once in our cohort. There are some indications of recombination events in the MHC class I region, even though this colony has only been breeding for approximately 10 generations to date. Haplotype 20 appears to be a recombination of haplotype 1 and haplotype 10. We verified the haplotype 20 recombination event by comparing the animal with this haplotype to 3 siblings via the dam and 17 siblings via the sire; the dam, though not genotyped, can be inferred through her progeny to carry haplotype 1 and haplotype 10. These singleton haplotypes may inform selection of animals from this colony for further studies; if MHC-I genetics are a desired control, animals with these rare haplotypes should not be selected.
Another recent study of this sooty mangabey colony, exploring a different subset of animals from the YNPRC, characterized haplotypes using microsatellite analysis instead of next generation sequencing (Smith et al. 2015). 109 of the same animals were included in both our deep sequencing and the Smith microsatellite cohort, providing further confirmation of our haplotypes and allowing correlation between both haplotype definitions (Table 3). To do this we looked at the animals present in both cohorts and compared the microsatellite repeat lengths within the MHC-I region. We found that animals that we had assigned to the same deep sequencing haplotype had the same repeat lengths profiles across the MHC-I region, corresponding to certain microsatellite haplotypes. Most of the deep sequencing haplotypes are associated with more than one microsatellite haplotype because the microsatellite haplotypes included markers spanning the entire ~5 Mb genomic MHC region, including MHC class II and class III regions. Thus, multiple microsatellite haplotypes often shared the same MHC class I region. Microsatellite repeat sizes for the common haplotypes defined by Smith et al. were associated with 11 of the 22 deep sequencing haplotypes (Table 3). Our correlation between deep sequencing and microsatellite haplotypes allows for more efficient and cost-effective haplotyping of the YNPRC sooty mangabey colony and other understudied sooty mangabey populations in the future. Deep sequencing techniques will provide more confident, replicable, and informative results containing the functional alleles present in animals, rather than nonfunctional microsatellite marker repeat lengths.
Table III.
Comparison of deep sequencing (DS) and microsatellite (MS) haplotypes
| Condensed MHC-I Microsatellite Haplotype | Deep Sequencing Haplotype | No. Chromosomes (Both DS and MS Typed) | MHC-I Microsatellite Markers | Microsatellite Haplotypes (Entire MHC region) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D6S1571 | D6S1621 | MML4SJ3 | MML4SJ5 | MML4SJ16 | STRMICA | ||||||||||||
| MS1 | DS1 | 38 | 126 | 280 | 204 | 167 | 180 | 224 | 4 | 11 | 13 | 17 | 18 | 25 | 71 | 52 | |
| MS2 | DS3 | 29 | 126 | 280 | 212 | 146 | 173 | 224 | 8 | 19 | 28 | 65 | 14 | ||||
| MS3 | DS5 | 24 | 137 | 280 | 212 | 156 | 171 | 207 | 7 | 27 | 37 | 42 | 67 | 23 | 30 | ||
| MS4 | DS2 | 22 | 114 | 282 | 212 | 146, 165 | 176, 180 | 207 | 1 | 34 | 15 | 44 | 57 | 58 | 64 | 36 | 29 |
| MS5 | DS4 | 21 | 127 | 280 | 214 | 167, 184 | 176 | 224 | 16 | 20 | 22 | 38 | 45 | 49 | 55 | ||
| MS6 | DS6 | 20 | 139 | 280 | 210 | 149 | 180 | 224 | 5 | 61 | |||||||
| MS7 | DS7 | 12 | 114 | 280 | 204 | 154 | 161 | 224 | 26 | 68 | |||||||
| MS8 | DS9 | 10 | 139 | 280 | 204 | 154 | 161 | 224 | 9 | 24 | 47 | ||||||
| MS9 | DS11 | 10 | 126 | 280 | 206 | 146 | 173 | 224 | 6 | 72 | 69 | ||||||
| MS10 | DS8 | 6 | 124 | 280 | 214 | 143 | 161 | 224 | 21 | 31 | |||||||
| MS11 | DS12 | 6 | 126 | 280 | 214 | 146 | 173 | 224 | 33 | 66 | |||||||
The correlation between deep sequencing haplotypes and consolidated MHC-I microsatellite haplotypes is shown. Condensation of the multiple microsatellite haplotypes into one haplotype based on markers solely within the MHC class I region, showing the specific microsatellite repeat lengths associated with the haplotypes defined by Smith et al. 2015
This study of MHC class I alleles in sooty mangabeys increases our knowledge of MHC genetics for this important natural host of SIV. We have shown the MHC class I genetics of sooty mangabeys resemble other nonhuman primates, such as Asian macaques, as all of these populations have multiply duplicated MHC class I B genes. Knowing the common haplotypes present in this breeding colony may aid design of studies in captive sooty mangabeys. Even though few new studies are initiated because these animals are endangered, these animals are still used to understand SIV pathogenesis and diabetes (Jones et al. 2014). Moreover, the characterization of MHC class I sequences from African primates can contribute to the understanding of host:pathogen interactions in free-living animals whose viral and bacterial infections are known. For example, combining non-invasively collected MHC genetic data with SIV sequence data could probe the role of T cell selection on viral evolution in naturally infected sooty mangabeys. In addition, our full-length sequences can be used to generate tetramer reagents to study specific MHC alleles for transplant or disease-related biomedical research.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (HHSN272201100013C), the National Center for Research Resources (R24 RR021745), and the Wisconsin National Primate Research Center Base Grant from the National Center for Research Resources (P51 RR000167) and the Office of Research Infrastructure Programs (P51 OD011106) of the National Institutes of Health. This research was conducted at a facility constructed with support from the Research Facilities Improvement Project (RR15450-01, RR020141-01). This work was also supported by funding by National Institutes of Health grants P51 OD0111103 to NEPRC, and P51 OD011132 to YNPRC. We gratefully acknowledge the help of Tracy Meeker, Stephanie Ehnert and Kay Lee Summerville at YNPRC for assistance with collection and shipping of samples from mangabeys.
References
- Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunological Reviews. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- Aetrei C, Metzger MJ, Richardson D, Ling B, Telfer PT, Reed P, Robertson DL, Marx PA. Detection and partial characterization of simian immunodeficiency virus SIVsm strains from bush meat samples from rural Sierra Leone. J Virol. 2005a;79(4):2631–2636. doi: 10.1128/JVI.79.4.2631-2636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J Virol. 2005b;79(14):8991–9005. doi: 10.1128/JVI.79.14.8991-9005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde ML, Lhost JJ, Burwitz BJ, Becker EA, Burns CM, O’Connor SL, Karl JA, Wiseman RW, Bimber BN, Zhang GL, Hildebrand W, Brusic V, O’Connor DH. Transcriptionally abundant major histocompatibility complex class I alleles are fundamental to nonhuman primate simian immunodeficiency virus-specific CD8+ T cell responses. J Virol. 2011;85(7):3250–3261. doi: 10.1128/JVI.02355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li A, Li L, Yan X, Fa Y, Zeng L, Fan J, Liu B, Sun Z. Identification of 32 major histocompatibility complex class I alleles in African green monkeys. Tissue Antigens. 2014;84(3):304–307. doi: 10.1111/tan.12389. [DOI] [PubMed] [Google Scholar]
- Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science. 2012;335(6073):1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty D. Genetic Divergence of the Rhesus Macaque Major Histocompatibility Complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 2012;1(3):895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DM, Karl JA, Creager HM, Bohn PS, Wiseman RW, O’Connor DH, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, O’Connor DH. Full-length novel MHC class I allele discovery by next generation seqencing: two platforms are better than one. Immunogenetics. 2014;66(1):15–24. doi: 10.1007/s00251-013-0744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes JD, Gordon SN, Zeng M, Chahroud AM, Haase AT. Early Resolution of Acute Immune Activation and Induction of PD-1 in SIV-Infected Sooty Mangabeys Distinguishes Nonpathogenic from Pathogenic Infection in Rhesus Macaques. J Immunol. 2008;18(10):6798–6807. doi: 10.4049/jimmunol.180.10.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CS, Reece JC, Saepuloh U, De Rose R, Ishkandriati D, O’Connor DH, Wiseman RW, Kent SJ. Screening and confirmatory testing of MHC class I alleles in pig-tailed macaques. Immunogenetics. 2011;63(8):511–521. doi: 10.1007/s00251-011-0529-5. [DOI] [PubMed] [Google Scholar]
- Jones A, Herndon J, Courtney C, Collura L, Cohen J. Clinicopathologic Characteristics, Prevalence, and Risk Factors of Spontaneous Diabetes in Sooty Mangabeys (Cercocebus atys) Comparative Medicine. 2014;64(3):200–210. [PMC free article] [PubMed] [Google Scholar]
- Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, O’Connor DH. Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3 (Bethesda) 2013;3(7):1195–1201. doi: 10.1534/g3.113.006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A, Yang J, Hempel D, Gritz L, Mazzara GP, McClure H, Johnson RP. Identification of Multiple Simian Immunodeficiency Virus (SIV)-Specific CTL Epitopes in Sooty Mangabeys with Natural and Experimentally Acquired SIV Infection. The Journal of Immunology. 2000;164(2):934–943. doi: 10.4049/jimmunol.164.2.934. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzber S, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noe R, Peeters M, Brookfield JF, Shaw GM, Sharp PM, Hahn BH. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, McKean KA, Sesink Clee PR, Gonder MK. The Evolution of Resistance to Simian Immunodeficiency Virus (SIV): A Review. Int J Primatol. 2014;35(2):349–375. [Google Scholar]
- Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nat Med. 2011;17(7):830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey-Cuillé M-A, Berthier J, Bomsel-Demontoy M, Chaduc Y, Montagnier L, Hovanessian A, Chakrabarti L. Simian Immunodeficiency Virus Replicates to High Levels in Sooty Mangabeys without Inducing Disease. Journal of Virology. 1998;72(5):3872–3886. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Gibbs RA. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet. 2014;15(5):347–359. doi: 10.1038/nrg3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noe R, Peeters M, Brookfield JF, Shaw GM, Sharp PM, Hahn BH. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d’Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79(19):12515–12527. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Chennareddi L, Greene-Hartsfield EZ, Villinger F, Cohen JK, Herndon JG. Hematology and serum chemistry values of sooty mangabeys (Cercocebus atys): comparison with rhesus monkeys. J Med Primatol. 2014;43(2):78–88. doi: 10.1111/jmp.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1(1) doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidebottom D, Kennedy R, Hildebrand WH. Class I MHC Expression in the Yellow Baboon. The Journal of Immunology. 2001;166(6):3983–3993. doi: 10.4049/jimmunol.166.6.3983. [DOI] [PubMed] [Google Scholar]
- Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. Understanding the benign nature of SIV infection in natural hosts. J Clin Invest. 2007;117(11):3148–3154. doi: 10.1172/JCI33034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G. Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS? J Med Primatol. 2005;34:243–252. doi: 10.1111/j.1600-0684.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- Smith GR, Bauer L, Crane MM, Johnson ZP. Immunogenetic characterization of a captive colony of sooty mangabeys (Cercocebus atys) used for SIV research. J Med Primatol. 2015 doi: 10.1111/jmp.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Metcalf B, Ribeiro RM, McClure H, Kaur A. Th-1-type cytotoxic CD8+ T-lymphocyte responses to simian immunodeficiency virus (SIV) are a consistent feature of natural SIV infection in sooty mangabeys. J Virol. 2006;80(6):2771–2783. doi: 10.1128/JVI.80.6.2771-2783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O’Connor SL, O’Connor DH. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81(1):349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O’Connor DH. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J. 2013;54(2):196–210. doi: 10.1093/ilar/ilt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.