Abstract
Purpose of review
In this report we review the evidence that environmental stimuli that perturb naturally selected host-microbe interactions are driving the increasing prevalence of food allergy and examine the mechanisms by which commensal bacteria regulate tolerance to dietary allergens.
Recent findings
Antibiotic use and the consumption of a high fat/low fiber diet have a major, and rapid, impact on gut bacterial populations with long-term consequences for both overall microbial community structure and the regulation of host immunity. Recent work emphasizes the role of mucosa-associated commensal bacteria in eliciting a barrier protective response critical to preventing allergic sensitization to food. Murine model studies are informing the development of novel live biotherapeutic approaches as an adjunctive therapy to enhance antigen specific oral desensitization and promote lasting tolerance in patients with food allergy.
Summary
Strategies based on modulating the composition and/or functionality of the gut microbiome hold promise for the treatment of food allergy.
Keywords: Food allergy, commensal bacteria, epithelial barrier, gut microbiota, probiotics, short chain fatty acids
Introduction
Allergic responses to food are increasingly prevalent in industrialized societies, rising by as much as 20% in a recent ten-year period [1–3]. We have proposed that this dramatic generational upsurge is a consequence of environmentally induced alterations in the composition of the commensal bacteria that normally populate the gastrointestinal tract [4–6]. During the last decade our understanding of the commensal microbiota, the collection of microbes that reside on our skin and mucosal surfaces, has been transformed by the introduction of culture independent methods of analysis. The numbers alone are staggering. Our bodies contain ten times more microbes than eukaryotic cells, which collectively encode 100–1000 times more genetic information, and are referred to as the microbiome [7, 8]. Over the course of millions of years of co-evolution, commensal bacteria have taken on many physiological functions essential to our health, including, but not restricted to, the production of vitamins and the digestion of insoluble dietary fibers [9]. Other constituent microbes including bacteriophage and viruses are at least as numerous but much less well understood or characterized. What is becoming clear, however, is that twenty first century lifestyle practices have shifted the composition of our commensal bacteria away from that which populated our ancestors; this modern-day bacterial community is increasingly correlated with disease [9]. Some environmental influences that can affect the microbiome include pervasive antibiotic use, consumption of a “Western” diet high in fat and sugar and low in fiber, the elimination of previously common enteropathogens, vaccination/reduced exposure to infectious disease, Caesarean birth and formula feeding (reviewed in [6]).
Commensal bacterial community composition varies by anatomic site [8, 9]. We are populated with our founder microbiota at birth which, until comparatively recently, occurred for most humans by natural delivery. Enterobacteria and vaginally derived lactic acid producing bacteria initially predominate; breast milk, which harbors its own microbiome, favors the emergence of Bifidobacteria, which extract nutrients from human milk glycans [10, 11]. Subsequent microbial successions eventually result in a diverse and unique microbiota [12]. Surgical delivery disturbs this process; founder bacterial populations in infants born by Caesarean section are derived from the skin of the mother or caregiver [13]. The neonatal period is a time of great plasticity for the emerging microbiome, which is critically intertwined with the maturation of the immune system of its host. Recent work has highlighted the profound influence of environmental factors on the developing microbiome.
Antibiotic use and diet shape the microbiota and influence susceptibility to food allergy
Beyond the targeted depletion of particular bacterial taxa, the effects of antibiotics on microbial community structure often persist long after cessation of treatment [14]. Medical exposure is only one source; widespread usage of antibiotics in agriculture, especially for their growth promoting properties for livestock, has contributed to their increasing low-residue presence in the food chain [15]. In the United States (and many other developed countries) most infants receive multiple courses of antibiotics during the first two years of life [16]. Murine model studies have demonstrated that early life exposure to orally administered broad-spectrum antibiotics is associated with aberrant immunity to respiratory and dietary antigens [4, 17–19]. Recent work from our laboratory showed that neonatal antibiotic treatment reduced microbial diversity and bacterial load in both fecal and ileal samples and enhanced food allergen sensitization [19]**. Other recent studies support the idea that the neonatal period is particularly critical. Even low dose early life antibiotic exposure can lead to long-lasting effects on metabolic and immune responsiveness [20]**. Data emerging from human studies links the use of anti-microbial agents to the increasing prevalence of food allergy. Maternal use of antibiotics before and during pregnancy, as well as antibiotic courses during the first month of life, are associated with an increased risk of cow’s milk allergy in infants [21]. Higher urinary levels of the common anti-bacterial agent triclosan are detected in children sensitized to food and aeroallergens [22].
Given that its components serve as a source of nutrition for both the host and its microbial inhabitants, it is not surprising that intestinal microbial community structure is strongly influenced by the composition of the diet. The gut microbiota of children consuming a Western style diet of processed food high in sugar and fat and low in fiber content, differs markedly from that of children from a rural African community consuming a low fat, high fiber plant-based diet (potentially resembling that of Neolithic subsistence farmers from 10,000 years ago) [23]. However, the rapidity and extent with which diet can alter human microbial communities has not been well understood; recent work suggests that marked changes can be observed on daily timescales, particularly in response to fiber intake [24].** Moreover, rapid and reproducible alterations in particular gut bacterial taxa can be detected after short term dietary intervention [25]**. Shifts in the abundance of fecal microbial communities were noted in human volunteers after only five days of consumption of a plant based diet (high in fruits, vegetables, grains and legumes) or an animal-based diet of meat, eggs and cheeses [25]. Of particular interest, in individuals consuming an animal-based diet, a reduced abundance of Firmicutes genera that ferment plant polysaccharides was evident even in this short timeframe. These results suggest that the timing of introduction of solid food, and the types of food consumed, may influence the development of food allergy by changing the composition of the intestinal microbiota. A recent study examining the influence of dietary patterns during the first year of life on the development of food allergy at two years of age provides some support for this hypothesis [26]*. Principal component analysis of prospective food diary data in a nested, case-control, within-cohort study showed that non-allergic infants had an ongoing diet that was high in fruits, vegetables, and home-prepared (non-processed) foods when compared to the diet of their challenge-proven food allergic counterparts [26]*.
New work also suggests that microbial transmission patterns have changed with urbanization as humans have become a predominantly indoor species, and with this lifestyle change we, and our children, are being predominantly exposed to our own microbiome [27]**. The controlled indoor environment, which has only arisen in the last 100 years, has fundamentally altered our exposure to the microbial world. These environments are designed, through controlled temperature, humidity and light, to be ‘antimicrobial’ [28]. We spend ~90% of our lives in this ‘clean’ ecosystem. It is likely this significantly reduces our microbial exposure, which may serve to limit the development of our immune system, our associated microbial diversity, and the ability of our commensal microbiota to rebound from composition-altering exposures. Whether the built environment contributes to allergic susceptibility (or might be manipulated to prevent disease) is an idea that is only beginning to be explored.
Alteration of the intestinal microbiota in food allergy
Given the profound influence of multiple environmental stimuli on the composition of the microbiota (and their confounding effects on its analysis) the available data characterizing the microbiota of patients with food allergy is still quite preliminary [29]. The majority of the available data comes from pediatric subjects. 16S rRNA profiling suggests that potentially negative alterations in gut microbiome composition (dysbiosis) may precede the occurrence of allergic manifestations. Azad et al. found that an increased Enterobacteriaceae/Bacteroidaceae ratio and low Ruminococcaceae abundance, in the context of low gut microbiota richness in early infancy, is associated with subsequent food sensitization, suggesting that early gut dysbiosis contributes to subsequent development of food allergy [30]*. Particular bacterial phylotypes, but not the overall gut microbial diversity, were significantly altered in a cohort of Chinese infants with food allergy [31]. When sampled at five months of age, the fecal microbiota of the food allergic infants was characterized by increased relative abundance of Clostridium cluster I and Anaerobacter and a decreased relative abundance of Bacteroides and Clostridium XVIII [31]. We have examined the intestinal microbiota of cow’s milk allergic infants at the time of diagnosis (by double blind oral food challenge) in comparison to age-matched healthy four month old controls [32].** We found that the microbiota of allergic infants in our study was significantly more diverse than that of healthy controls. Strikingly, while the healthy subjects’ microbiota was dominated by Bifidobacteriaceae, Enterobactericeae, and Enterococceae, the microbiota of allergic infants demonstrated a significant increase in the abundance of Ruminococcaceae and Lachnospiraceae, which can predominate in the adult gut [32].
Treatment of food allergy with currently available probiotics
The evidence reviewed thus far suggests that therapeutic modulation of the commensal microbiota may be beneficial for the prevention or treatment of food allergy. Probiotics are typically defined as microorganisms that, when ingested, confer health benefits to the host [33]. Studies examining the efficacy of currently available probiotics in treating food allergy have yielded conflicting results. Differences in study design, populations, probiotic strains and dosages may be responsible for the discrepancies observed [33]. A meta-analysis of clinical trials concluded that administration of probiotics prenatally, or during the period shortly after birth, reduced total IgE levels and the risk of atopic sensitization, but not asthma or wheezing [34]. Recently published guidelines for atopic disease prevention from the World Allergy Organization concluded that there is a likely net benefit in using probiotics for eczema prevention [35]. Administration of Lactobacillus acidophilus was associated with a significantly increased risk of atopic sensitization when compared with other strains, emphasizing the importance of probiotic strain selection [34]. Allen et al demonstrated that high-dose administration of multiple strains of Lactobacilli and Bifidobacteria to mothers during late pregnancy, and to their infant from birth to six months of age, did not prevent eczema or reduce the frequency of asthma in early childhood but did promote a reduced frequency of sensitivity to food antigens [36]. A Japanese study showed that both pre and post-natal supplementation with Bifidobacteria was associated with a significantly reduced risk of eczema/atopic dermatitis during the first 18 months of life [37]. Studies investigating the therapeutic effect of probiotics on challenge confirmed food allergic subjects are scant. In one randomized double blind placebo-controlled study of infants with challenge proven cow’s milk allergy, administration of Lactobacillus casei CRL431 and Bifidobacterium lactis Bb12 for twelve months did not affect the acquisition of tolerance to cow’s milk [38]. In contrast, Berni Canani et al demonstrated in two different prospective clinical trials that an extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG (LGG) accelerated the development of tolerance acquisition in infants with cow’s milk allergy [39,40]. When we compared the fecal microbiota of infants receiving this tolerance-inducing probiotic-supplemented therapy to that obtained from infants receiving an extensively hydrolyzed casein formula (EHCF) alone, we found statistically significant positive correlations between the abundance of genera with the potential for producing butyrate and the concentration of fecal butyrate in the infants that received EHCF plus LGG [32]. Strain level demarcations for butyrate producing genera (including Roseburia, Coprococcus and Blautia) identified in infants that acquired tolerance to cow’s milk suggest that LGG treatment contributes to acquisition of tolerance by altering the strain level community structure of taxa with the potential to produce butyrate [32].
Oral immunotherapy (OIT) has shown promise in eliciting desensitization in patients with food allergy, but its ability to induce long-lasting tolerance in the absence of ongoing allergen administration has not been reliably demonstrated [41]. A double-blind, placebo-controlled randomized trial evaluated the efficacy of 18 months of therapy combining oral peanut desensitization with administration of the probiotic Lactobacillus rhamnosus CGMCC in 1–10 year old children with peanut allergy [42]*. Although “possible sustained unresponsiveness” 2–5 weeks after cessation of OIT was noted in most (82%) of the children receiving the probiotic plus OIT, no comparison was made to treatment groups receiving either OIT or probiotic alone. The effect of daily L. rhamnosus CGMCC treatment on microbial community structure was not evaluated.
Development of novel live biotherapeutics for the prevention and treatment of food allergy
Increasing appreciation of the role of the microbiota in regulating complex immune mediated diseases has led to the emergence of a number of biotechnology companies seeking to commercialize novel live biotherapeutics with microbiome-modulating properties [43]. The efficacy of fecal transplantation in resolving disease in patients with relapsing C. difficile infection has provided proof of principle for therapeutic approaches based on oral administration of constituents of the normal commensal microbiota [44]. Honda and colleagues have identified mucosa-associated Firmicutes in the Clostridia class as the anaerobic component of the indigenous microbiota critical for the induction of regulatory T cells (Treg) in the colonic lamina propria [45]. Both the cell-mediated (Treg) and humoral (IgA) arms of the immune system are dramatically underdeveloped in mice devoid of commensal microbes (germ free) [46]. Atarashi et al showed that Tregs are also induced when spore-forming Clostridia isolated from human feces are transferred into germ free mice [47]**. A Treg-inducing mixture of isolates that can be cultured in vitro has been selected for development as a novel biotherapeutic for the treatment of inflammatory bowel diseases [48].
New work from our laboratory shows that this type of approach may also hold promise for the treatment of food allergy. We found that sensitization to a food allergen is enhanced in germ free mice and in mice that have been treated by neonatal antibiotic administration [19]**. Selective colonization of germ free mice demonstrated that the allergy-protective capacity is contained within the Clostridia class. Moreover, reintroduction of a Clostridia-containing microbiota to antibiotic-treated mice blocked sensitization to a food allergen [19]. Clostridia colonization of germ free mice restored both the Treg and IgA compartments. Microarray analysis of isolated intestinal epithelial cells led to the identification of a novel innate mechanism by which Clostridia protect against sensitization to dietary antigens. Prior work has implicated defects in intestinal permeability to aberrant allergic responses to food [49]. Our discovery that Clostridia colonization induces the production of the barrier protective cytokine IL-22 provided mechanistic insight into how commensal bacteria regulate intestinal barrier permeability. We used a sensitive capture ELISA to demonstrate that IL-22 acts to reduce the concentration of orally administered dietary antigen detectable in the systemic circulation [19]. Our data therefore suggest that Clostridia stimulate both innate and adaptive immune signalling pathways to maintain tolerance to food. This is a paradigm shift since oral tolerance has typically been attributed primarily to an antigen specific Treg response [50,51]. Our work suggests a new model in which tolerance to dietary antigen requires both food antigen specific Tregs and a bacteria-induced barrier protective response [19, 52].
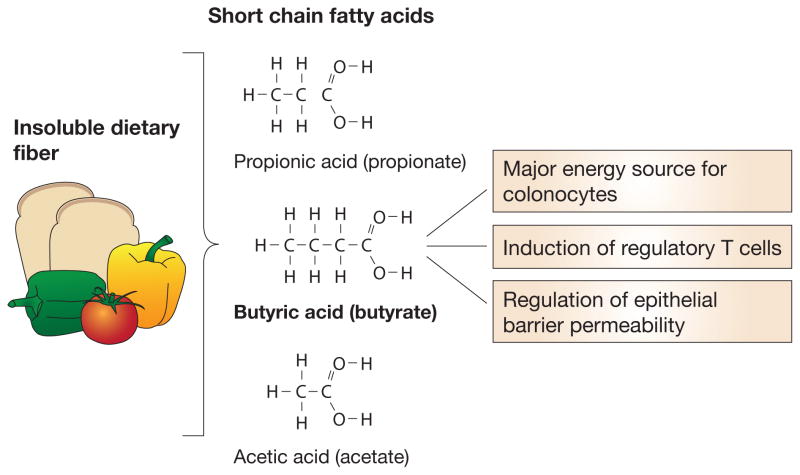
It is not yet clear whether Clostridia modulate host immunity by direct cell-cell contact, secreted metabolites, or both. The ability of particular bacterial taxa to ferment dietary fiber for the production of short chain fatty acids (SCFA) helps to explain the health-promoting role of bacterially produced metabolites [53]. Of the major SCFA, butyrate is the preferred energy source for colonocytes (Figure 1). Its abundant presence in the gut is often considered a sensor of intestinal health [54]. Butyrate-producing bacteria comprise a functional, rather than a taxonomic group, although the Clostridial families Lachnospiraceae and Ruminococceae are among its most prominent producers [55]. Bacteria-produced SCFA have been implicated in the regulation of both the proportions and functional capabilities of colonic Tregs [56, 57] which, in some reports, has been specifically attributed to butyrate production by spore-forming Clostridiales [58]**. Preliminary data from our laboratory also links butyrate, but not other SCFA, to regulation of epithelial barrier permeability (Feehley et al, personal communication). The clinical relevance of this mouse model work is highlighted by a recent report which correlated the severity of atopic disease with intestinal microbial diversity and the abundance of butyrate producing bacteria [59].
Figure 1.
Commensal bacteria ferment insoluble dietary fiber to produce short chain fatty acids (SCFA), the most abundant of which are propionate, butyrate and acetate. Colonocytes metabolize butyrate as their primary energy source. The production of immunoregulatory metabolites is one way that commensal bacteria interact with host immune cells to promote non-responsiveness to innocuous luminal antigens. Recent work suggests that SCFA contribute to mucosal homeostasis through the induction of regulatory T cells and the regulation of epithelial barrier permeability.
Conclusions
Taken together these findings provide a compelling rationale for exploring the administration of novel Clostridia-based biotherapeutics or the administration of butyrate as adjunctive therapies to promote tolerance to food allergens. Protocols which analyze the efficacy of OIT based on multi-parameter flow cytometric analysis of peripheral blood may have limited utility; an elegant recent study has shown that most Teffector/memory cells are tissue resident [60]*. In this regard, evaluation of allergen concentration in the bloodstream in the hours following gavage may provide a more clinically (and immunologically) relevant form of assessment. Moreover, it is tempting to speculate that poor digestibility and the access of undigested protein to the bloodstream with B cell epitopes intact may be a distinguishing feature of food allergens (see refs [61–63]) for peanut, β-lactoglobulin and wheat). Whether this is in fact the case, and whether commensal bacteria regulate the systemic concentration of allergens other than peanut, is now readily testable in pre-clinical murine models. Variations of this assay can be adapted to clinical trials to evaluate the efficacy of administered live biotherapeutic agents/butyrate in modulating allergen concentrations in serum during double blind oral food challenge. Numerous methodological challenges to the understanding of the gut microbiome in food allergy must be considered in future studies and will require multidisciplinary teams of immunologists, clinicians, microbial ecologists, and bioinformaticians (Table 1).
Table 1.
Analysis of the role of the commensal gut microbiome in the regulation of allergic responses to food
| STUDY SUBJECTS | Comparative evaluation of a well characterized patient population with a definitive diagnosis of food allergy (clinical history, positive screening test for immune response against food allergens) prior to any therapeutic intervention to healthy subjects matched for age, gender and exposure to environmental risk factors that can affect the composition of the microbiome including: antibiotic use, birth order, bacterial and viral infections, delivery and feeding modes, degree of social exposure (child care), vaccinations and exposure to animals (endotoxin), smoking and gastric acidity inhibitors. |
| METHODS | Stool samples will be collected from subjects at initial diagnosis and/or in longitudinal studies of samples collected over time for each patient to capture variation in the microbial composition under conditions where food intake and environmental interactions are monitored. These data will then be supplemented with periodic samples and questionnaires collected in an unsupervised setting to improve data density. Bacterial DNA will be extracted from stool using standard techniques outlined by the Earth Microbiome Project (http://www.earthmicrobiome.org/ [64]. The DNA will then be processed for either 16S rRNA amplicons and Illumina sequencing [65] or for shotgun metagenomic analysis with Illumina sequencing [66]. Sequence data will be initially analyzed using published methods (e.g. refs. [27, 65]) followed by a suite of non-parametric statistical tests to identify taxa or genes that show significant differences between cohorts, through time, or with treatment [66]. By leveraging publically available databases of human microbiome data (e.g. hmpdacc.org or microbio.me/americangut/) it will be possible to determine if observed trends show any statistical pattern within a larger population. |
Key Points.
The trillions of bacteria that populate our skin and mucosal surfaces critically regulate key physiological functions.
Environmentally induced changes in commensal bacterial communities have created a dysbiosis that is linked to the increasing prevalence of complex immune-mediated disease.
Understanding how indigenous bacterial communities interact with the innate and adaptive immune system will inform the development of novel live biotherapeutics to prevent or treat food allergy.
Acknowledgments
We thank Taylor Feehley and Sandeep Pawar for critical review of this manuscript.
Financial Support and sponsorship
This work was supported by NIAID AI106302, Food Allergy Research and Education (FARE) and UChicago Digestive Diseases Center Core Grant P30DK42086 (CRN), the Italian Ministry of Health PE-2011-02348447, (RBC) and the U.S. Dept. of Energy under Contract DE-AC02-06CH11357.NS (JAG).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References and Recommended Reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Wang J, Sampson HA. Food allergy. J Clin Invest. 2011;121:827–835. doi: 10.1172/JCI45434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branum AM, Lukacs SL. Food allergy among children in the United States. Pediatrics. 2009;124:1549–1555. doi: 10.1542/peds.2009-1210. [DOI] [PubMed] [Google Scholar]
- 3.Osborne NJ, Koplin JJ, Martin PE, et al. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol. 2011;127:668–676. doi: 10.1016/j.jaci.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 5.Prioult G, Nagler-Anderson C. Mucosal immunity and allergic responses: lack of regulation and/or lack of microbial stimulation? Immunol Rev. 2005;206:204–218. doi: 10.1111/j.0105-2896.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 6.Feehley T, Stefka AT, Cao S, Nagler CR. Microbial regulation of allergic responses to food. Semin Immunopathol. 2012;34:671–688. doi: 10.1007/s00281-012-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin R, Langa S, Reviriego C, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143:754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 11.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108 (Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley LW, Raphael E, Faerstein E. Obesity in the United States - dysbiosis from exposure to low-dose antibiotics? Frontiers in Public Health. 2013;1:69. doi: 10.3389/fpubh.2013.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 17.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Stefka AT, Feehley T, Tripathi P, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. This paper shows that a class of mucosa-associated intestinal bacteria protects against food allergen sensitization by inducing the production of IL-22 in the intestinal lamina propria to regulate the access of allergen to the systemic circulation. The authors suggest that environmentally induced changes in the intestinal bacterial community are driving the increasing prevalence of food allergy and present a new model for the maintenance of tolerance to dietary antigen which requires both an antigen specific Treg response and a bacteria-induced epithelial barrier protective response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. The interesting study used low-dose penicillin (LDP), administered from birth, to examine whether disruption of the developing microbiota has long term consequences. They found that early life exposure to LDP only transiently alters the composition of the microbiota but has lasting effects on the regulation of intestinal immune system gene expression and host metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metsala J, Lundqvist A, Virta LJ, et al. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology. 2013;24:303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 22.Savage JH, Matsui EC, Wood RA, Keet CA. Urinary levels of triclosan and parabens are associated with aeroallergen and food sensitization. J Allergy Clin Immunol. 2012;130:453–460. doi: 10.1016/j.jaci.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.David LA, Materna AC, Friedman J, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biology. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. The authors measured the gut and salivary microbiota of two individuals daily for one year. They found that both communities were generally stable within each individual, but could respond to dietary changes (particularly host fiber intake) with changes in the abundance of 15% of the total community within 24 hrs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. This interesting study examined fecal microbial community structure in 10 volunteers who consumed a diet composed of either entirely animal or entirely plant products (5/group) for five consecutive days. The authors reproducibly demonstrated marked changes in microbial community structure reflective of herbivorous or carnivorous functional profiles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Grimshaw KE, Maskell J, Oliver EM, et al. Diet and food allergy development during infancy: birth cohort study findings using prospective food diary data. J Allergy Clin Immunol. 2014;133:511–519. doi: 10.1016/j.jaci.2013.05.035. When examined at two years of age, children who did not develop food allergies had consumed greater amounts of fruits, vegetables and home prepared foods in later infancy than children with food allergies. [DOI] [PubMed] [Google Scholar]
- 27**.Lax S, Smith DP, Hampton-Marcell J, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. This report found that microbial communities present in the indoor home environment are largely derived from their human and animal occupants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kembel SW, et al. Architectural design drives the biogeography of indoor bacterial communities. PLoS One. 2014;9:e87093. doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrs T, Bruce KD, Logan K, et al. Is there an association between microbial exposure and food allergy? A systematic review Pediatr Allergy Immunol. 2013;24:311–320. doi: 10.1111/pai.12064. [DOI] [PubMed] [Google Scholar]
- 30*.Azad MB, Konya T, Guttman DS, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. 2015 doi: 10.1111/cea.12487. These CHILD study investigators examined the composition of the fecal microbiota in 166 infants at 3 and 12 months of age. They found that a high ratio of Enterobacteriaceae/Bacteroidaceae at 3 months was associated with the development of sensitization to food. [DOI] [PubMed] [Google Scholar]
- 31.Ling Z, Li Z, Liu X, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Berni Canani R, Sangwan N, Stefka AT, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate producing bacterial strains in food allergic infants. ISME J. 2015 doi: 10.1038/ismej.2015.151. in revision. This new study shows that the microbiota of CMA infants at diagnosis is strikingly different from that of age-matched healthy controls. The authors used oligotyping to resolve the analysis of 16S rRNA sequences to the strain level at 100% nucleotide identity. They identified statistically significant positive correlations between the abundance of genera with the potential for butyrate production and the concentration of butyrate in the feces of CMA infants treated with the probiotic-supplemented formula but not the unsupplemented form of the same formula. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill C, Guarner F, Reid G, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 34.Elazab N, Mendy A, Gasana J, et al. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 2013;132:e666–676. doi: 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- 35.Fiocchi A, Pawankar R, Cuello-Garcia J, et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J. 2015;8:4. doi: 10.1186/s40413-015-0055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen SJ, Jordan S, Storey M, et al. Probiotics in the prevention of eczema: a randomised controlled trial. Arch Dis Child. 2014;99:1014–1019. doi: 10.1136/archdischild-2013-305799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enomoto T, Sowa M, Nishimori K, et al. Effects of bifidobacterial supplementation to pregnant women and infants in the prevention of allergy development in infants and on fecal microbiota. Allergology international : official journal of the Japanese Society of Allergology. 2014;63:575–585. doi: 10.2332/allergolint.13-OA-0683. [DOI] [PubMed] [Google Scholar]
- 38.Hol J, van Leer EH, Elink Schuurman BE, et al. The acquisition of tolerance toward cow’s milk through probiotic supplementation: a randomized, controlled trial. J Allergy Clin Immunol. 2008;121:1448–1454. doi: 10.1016/j.jaci.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Berni Canani R, Nocerino R, Terrin G, et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: A randomized trial. J Allergy Clin Immunol. 2012;129:580–582. doi: 10.1016/j.jaci.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Berni Canani R, Nocerino R, Terrin G, et al. Formula selection for managment of children with cow milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. The Journal of Pediatrics. 2013;163:771–777. doi: 10.1016/j.jpeds.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Wood RA, Sampson HA. Oral immunotherapy for the treatment of peanut allergy: is it ready for prime time? The journal of allergy and clinical immunology In practice. 2014;2:97–98. doi: 10.1016/j.jaip.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 42*.Tang ML, Ponsonby AL, Orsini F, et al. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2014.11.034. This randomized placebo-controlled trial showed that possible sustained unresponsiveness was induced in the majority of study participants receiving peanut oral immunotherapy (OIT) together with the probiotic Lactobacillus rhamnosus. However the study did not include a treatment group receiving OIT alone so the relative contributions of the probiotic and OIT to the results observed could not be evaluated. [DOI] [PubMed] [Google Scholar]
- 43.Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31:309–315. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- 44.Youngster I, Russell GH, Pindar C, et al. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312:1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 45.Atarashi K, Tanoue T, Shima T, et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2014 doi: 10.1038/mi.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. The authors used a step-wise approach to isolate a mixture of 17 Clostridia strains from healthy human feces by selecting for their ability to expand and differentiate regulatory T cells in the colonic lamina propria of germ free mice. [DOI] [PubMed] [Google Scholar]
- 48.Narushima S, Sugiura Y, Oshima K, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes. 2014;5:333–339. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menard S, Cerf-Bensussan N, Heyman M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010;3:247–259. doi: 10.1038/mi.2010.5. [DOI] [PubMed] [Google Scholar]
- 50.Hadis U, Wahl B, Schulz O, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao S, Feehley TJ, Nagler CR. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS Letters. 2014;588:4258–4266. doi: 10.1016/j.febslet.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorburn AN, Macia L, Mackay CR. Diet, Metabolites, and “Western-Lifestyle” Inflammatory Diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 54.Miquel S, Martin R, Rossi O, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 55.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5:e00889. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. This report identified the short chain fatty acid butyrate as the microbial metabolite responsible for the induction of regulatory T cells by indigenous commensal spore-forming bacteria. [DOI] [PubMed] [Google Scholar]
- 59*.Nylund L, Nermes M, Isolauri E, et al. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70:241–244. doi: 10.1111/all.12549. These authors correlated fecal microbial community structure with the severity of eczema in infants. They found that disease improvement correlated with an increased abundance of butyrate-producing bacteria. [DOI] [PubMed] [Google Scholar]
- 60**.Thome JJ, Yudanin N, Ohmura Y, et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. This report examines T cell compartmentalization in blood, lymphoid and mucosal tissues collected from 56 individual deceased organ donors between the ages of 3 and 73. The unprecendented body-wide scope of this analysis provides insight in the mechanisms which shape T cell differentiation in humans. Most memory CD4+ T cells are tissue resident and their subset distribution is more complex than suggested by analysis of peripheral blood, where activated and cycling cells are over-represented. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mueller GA, Maleki SJ, Pedersen LC. The molecular basis of peanut allergy. Curr Allergy Asthma Rep. 2014;14:429. doi: 10.1007/s11882-014-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pescuma M, Hebert EM, Haertle T, et al. Lactobacillus delbrueckii subsp. bulgaricus CRL 454 cleaves allergenic peptides of β-lactoglobulin. Food Chemistry. 2015;170:407–414. doi: 10.1016/j.foodchem.2014.08.086. [DOI] [PubMed] [Google Scholar]
- 63.Baar A, Pahr S, Constantin C, et al. Molecular and immunological characterization of Tri a 36, a low molecular weight glutenin, as a novel major wheat food allergen. J Immunol. 2012;189:3018–3025. doi: 10.4049/jimmunol.1200438. [DOI] [PubMed] [Google Scholar]
- 64.Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: successes and aspirations. BMC biology. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knight R, Jansson J, Field D, et al. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]