Abstract
The three-dimensional organization of a genome plays a critical role in regulating gene expression, yet little is known about the machinery and mechanisms that determine higher-order chromosome structure1,2. Here we perform genome-wide chromosome conformation capture analysis, FISH, and RNA-seq to obtain comprehensive 3D maps of the Caenorhabditis elegans genome and to dissect X-chromosome dosage compensation, which balances gene expression between XX hermaphrodites and XO males. The dosage compensation complex (DCC), a condensin complex, binds to both hermaphrodite X chromosomes via sequence-specific recruitment elements on X (rex sites) to reduce chromosome-wide gene expression by half3–7. Most DCC condensin subunits also act in other condensin complexes to control the compaction and resolution of all mitotic and meiotic chromosomes5,6. By comparing chromosome structure in wild-type and DCC-defective embryos, we show that the DCC remodels hermaphrodite X chromosomes into a sex-specific spatial conformation distinct from autosomes. Dosage-compensated X chromosomes consist of self-interacting domains (~1 Mb) resembling mammalian Topologically Associating Domains (TADs)8,9. TADs on X have stronger boundaries and more regular spacing than on autosomes. Many TAD boundaries on X coincide with the highest-affinity rex sites and become diminished or lost in DCC-defective mutants, thereby converting the topology of X to a conformation resembling autosomes. rex sites engage in DCC-dependent long-range interactions, with the most frequent interactions occurring between rex sites at DCC-dependent TAD boundaries. These results imply that the DCC reshapes the topology of X by forming new TAD boundaries and reinforcing weak boundaries through interactions between its highest-affinity binding sites. As this model predicts, deletion of an endogenous rex site at a DCC-dependent TAD boundary using CRISPR/Cas9 greatly diminished the boundary. Thus, the DCC imposes a distinct higher-order structure onto X while regulating gene expression chromosome wide.
To compare the molecular topology of X chromosomes and autosomes in C. elegans, we generated genome-wide chromatin interaction maps from mixed-stage embryos using a modified Hi-C protocol combining conventional 3C with paired-end sequencing10–12 (Fig. 1, Extended Data Fig. 1, and Methods). Interaction data, binned at both 10 kb and 50 kb intervals, revealed features observed in other organisms. Interactions occur most frequently in cis and decay with genomic distance (Extended Data Fig. 1 and Methods). Chromosome compartments comparable to active A and inactive B compartments11,13 are formed (Extended Data Fig. 1, 4–6). Compartments at the left end of X and both ends of autosomes align with binding domains for lamin14, lamin-associated protein LEM-2 (Extended Data Fig. 4–6)15, and the H3K9me3 inactive chromatin mark16, suggesting their similarity to inactive B compartments of mammals.
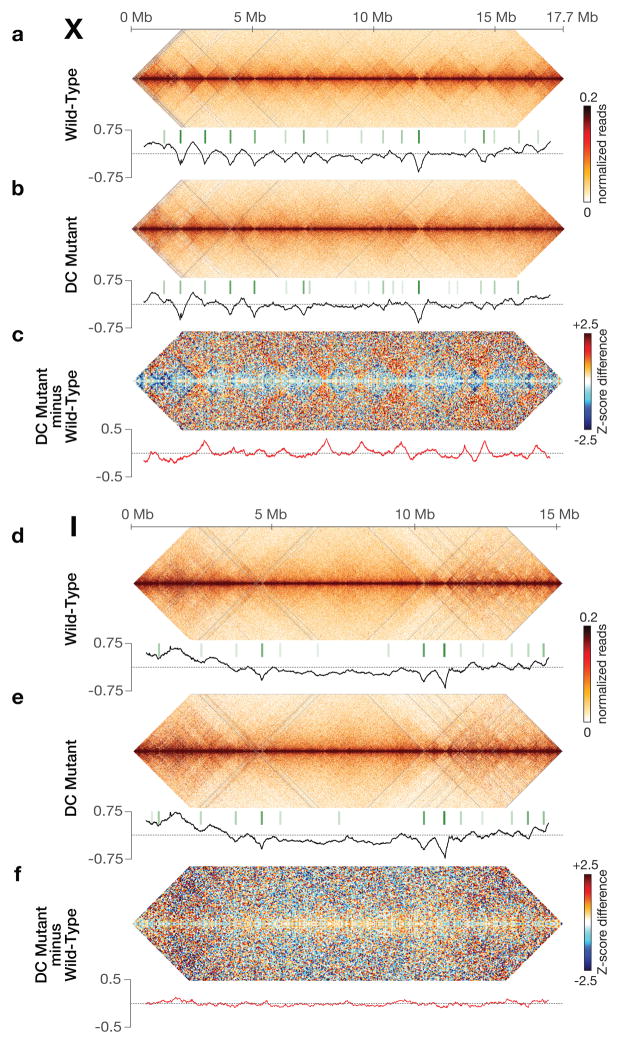
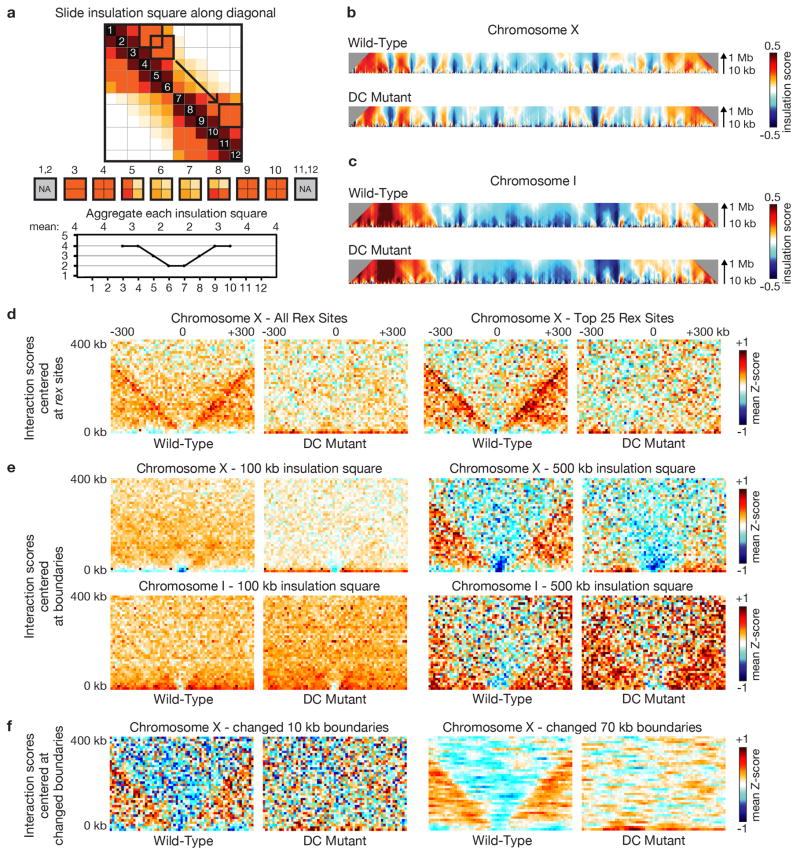
Figure 1. DCC modulates spatial organization of X chromosomes.
a, b, d, e, Chromatin interaction maps binned at 10 kb resolution show interactions 0–4 Mb apart on chromosomes X and I in wild-type and DC mutant embryos. Plots (black) show insulation profiles. Minima (green lines) reflect TAD boundaries. Darker green indicates stronger boundary. c, f, Bluered Z-score difference maps binned at 50 kb resolution for X and I show increased (orange-red) and decreased (blue) chromatin interactions between mutant and wild-type embryos. Differential insulation plots (red) show insulation changes between mutant and wild-type embryos.
Chromatin interaction maps also revealed self-interacting domains (~ 1 Mb), predominantly on X chromosomes. These domains are visible as diamonds along the interaction maps (Fig. 1a, d) and resemble TADs of mammalian and fly chromosomes8,9,12. To quantify TADs, we devised an approach of assigning an “insulation score” to genomic intervals along the chromosome. The score reflects the aggregate of interactions in the interval. Minima of the insulation profile denote areas of high insulation we classified as TAD boundaries (Methods, Fig. 1, Extended Data Fig. 2a, 3a and b).
The insulation profile of X stands out compared to those of autosomes. The insulation signal amplitude is larger on X (Fig. 1a, d; Extended Data Fig. 3d), implying TAD boundaries are stronger. Also, TAD boundaries on X are more abundant and regularly spaced (Extended Data Fig. 3d).
To assess whether the DCC controls the spatial organization of hermaphrodite X chromosomes, we generated chromatin interaction maps for a dosage-compensation-defective mutant (DC mutant; Fig. 1, Extended Fig. 1–6) in which the XX-specific DCC recruitment factor SDC-2 was depleted, severely reducing DCC binding to X3,4,17 (Fig. 2a) and elevating X-chromosome gene expression (see below). The insulation profile of X, but not autosomes, was greatly changed (Fig. 1b, e; Extended Data Fig. 1–6). Of 17 total TAD boundaries on X, five were eliminated and three severely reduced in insulation. TAD boundary strength and spacing on X in DC mutants resembled that of autosomes (Extended Data Fig. 3d).
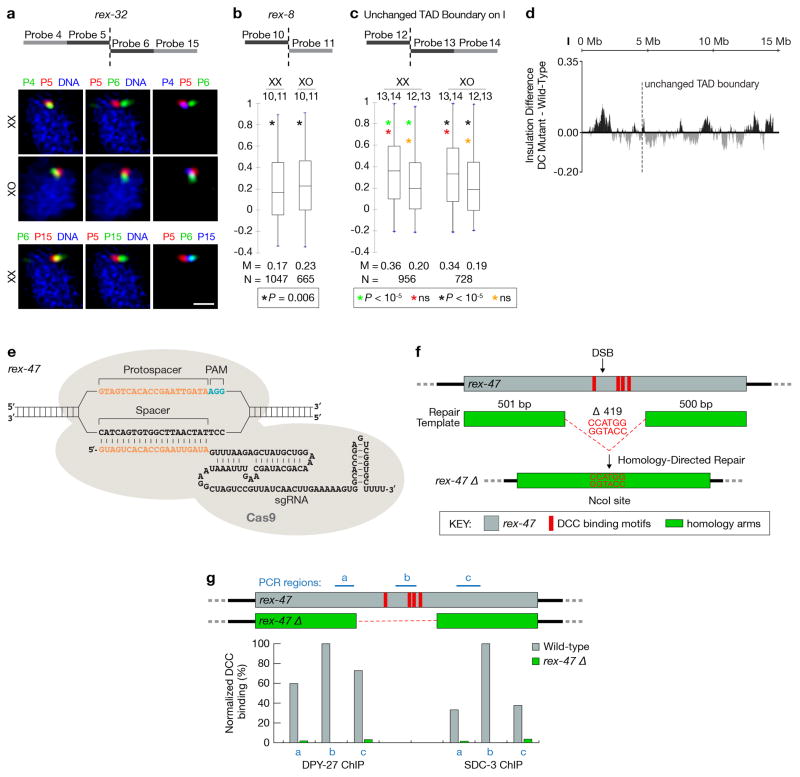
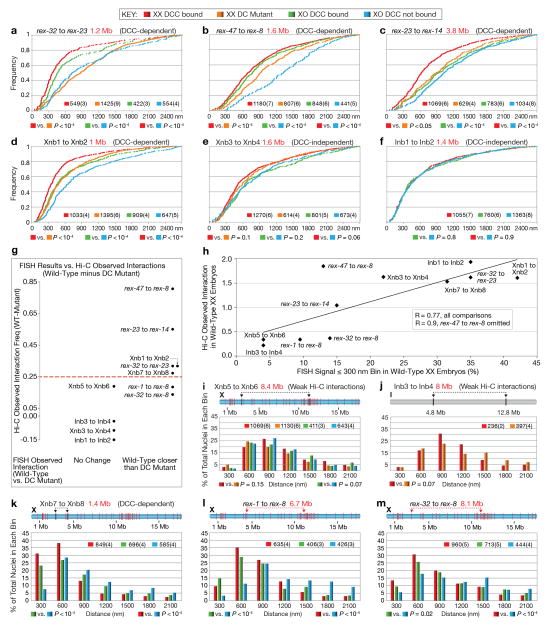
Figure 2. FISH shows DCC-dependent TAD boundaries at high-affinity DCC rex sites.
a, High DCC occupancy correlates with TAD boundaries lost or reduced upon DCC depletion. Top, ChIP-seq profiles of DCC subunit SDC-3 in wild-type (red) and DC mutant (green) embryos. Y-axis, reads per million (RPM) normalized to IgG control. Middle, insulation profiles of wild-type (red) and DC mutant (green) embryos. Bottom, insulation difference plot for wild-type insulation profile subtracted from DC mutant profile. Black lines, TAD boundary locations. Blue dots, boundaries with insulation changes > 0.1 between wild-type and DC mutant embryos. Red lines, locations of 25 highest DCC-occupied rex sites. Cyan bars, sites with the largest insulation loss. b, Confocal images of embryonic nuclei of various genotypes stained with a DNA intercalating dye (blue) and 500 kb FISH probes around the rex-47 TAD boundary. c, d, e, Quantification of FISH probe colocalization confirms DCC-dependent and DCC-independent boundaries found by Hi-C. Box plots, distribution of Pearson’s correlation coefficients between pairwise combinations of FISH probes within (blue) or across (orange) TADs. Boxes, middle 50% of coefficients. Center bars, median (M) coefficients. N, total number of nuclei. Asterisks of same color indicate values compared with one-tailed Mann-Whitney U test. ns, not significant.
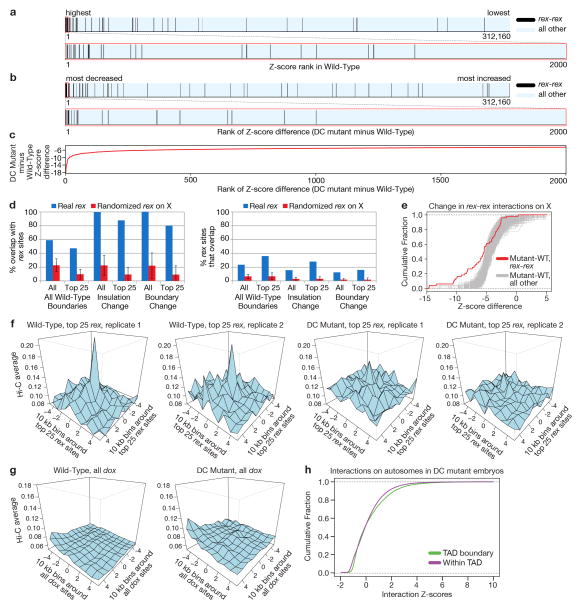
Figure 3. Strong DCC-dependent interactions occur between high-affinity rex sites at TAD boundaries.
a, Cumulative distribution of Hi-C Z-scores for interactions between 10 kb bins with rex sites or with other X interactions in wild-type or DC mutant embryos. Interactions > 4 Mb were excluded from panels (a–e). P-values are corrected for multiple testing. In wild-type embryos, rex-rex interactions are stronger than all other X interactions (P < 2 × 10−16; two-sided KS test) and stronger than rex-rex interactions in DC mutants (P = 1.5 × 10−9; Wilcoxon signed rank test). b, Distributions of Hi-C Z-scores show that rex-rex interactions are stronger than non-rex interactions (P < 2 × 10−16; two-sided KS test) or rex to non-rex interactions (P = 1.7 × 10−14; two-sided KS test). c, Distributions of Z-score differences (DC mutant minus wild-type) show that rex-rex interactions decrease more than any of 1,000 random sets of non-rex interactions of equal number (P < 0.001). d, Average Hi-C interaction profiles (normalized read counts) around pairs of top 25 rex sites or all known rex sites, in wild-type and DC mutants. rex sites are centered at 0. e, Distributions of Hi-C Z-scores for interactions between bins with rex or non-rex sites at TAD boundaries or within TADs of wild-type (left) or DC mutant (middle) embryos. rex sites interact more at TAD boundaries than in TADs (P = 0.0025). These sets of interactions are not different in DC mutants (P = 0.348). Interactions at TAD boundaries or within TADs on autosomes (right). f, Circos plots depict all rex-rex interactions (Z-score >2, colored line) in 50 kb bins in wild-type embryos. Concentric circles show insulation difference plot (black and grey), wild-type TAD boundaries (green boxes), and rex sites (black lines, strongest sites named). g, rex-rex interactions in f that are retained in DC mutants. h, Deletion of rex-47 disrupts TAD boundary. Box plots of Pearson’s correlation coefficients for FISH probe combinations in wild-type, rex-47 Δ, and DC mutant. Probe overlap across TAD boundary increased in rex-47 Δ vs. wild-type (P < 0.01 ANOVA) but was not different in rex-47 Δ vs. DC mutants (P = ns, ANOVA). Probe overlap in TAD was not different in 3 strains (P = 0.075, ANOVA).
To characterize this transformation in conformation, we calculated the difference between chromatin interaction maps of wild-type and DC mutant embryos after converting the interaction data into genomic-distance-normalized Z-scores. In DC mutants, interactions increased across TAD boundaries on X, while interactions within TADs decreased, revealing a DCC-dependent remodeling of X-chromosome structure (Fig. 1c, Extended Data Fig. 1–3 and 5). Weakening of TAD boundaries is expected to cause chromosome-wide changes in chromatin interactions. The largest changes in insulation on X occurred at TAD boundaries. Autosomes appeared unaffected (Fig. 1c and f, Fig. 2a, Extended Data Fig. 1–4 and 6).
TAD boundaries on X are enriched for the highest DCC-occupied rex sites3,4,18 (Fig. 2a, Extended Data Fig. 7d). About 50% of all TAD boundaries and 90% of changed ones overlap the top 25 rex sites, a correlation higher than expected at random (Extended Data Fig. 7d). In DC mutants, the largest insulation losses occurred in regions overlapping the strongest rex sites (Fig. 2a). These results imply the DCC plays a direct role in defining TADs by binding to rex sites to mediate formation of TAD boundaries. In contrast, genomic features such as HOT (Highly Occupied Targets) sites19 do not govern TADs (Supplementary Table 2).
Two TAD boundaries on X that overlap rex sites in the LEM-2 B-like compartment were not greatly reduced in DC mutants (Fig. 1 and 2a; Extended Data Fig. 5e). While the DCC exerts a dominant influence on TAD formation, other forces act on X to form TADs, as on autosomes.
To confirm the DCC-dependent topology of X, we visualized TADs using quantitative 3D fluorescent in situ hybridization (FISH) in wild-type XX embryos and embryos lacking DCC binding on X: male XO and DC mutant XX (Fig. 2b–e). We imaged fluorescent probes that tiled 500 kb regions within TADs or flanking TAD boundaries. Probe overlap was quantified by analyzing the distribution of Pearson’s correlation coefficients between FISH signals from pairwise probe combinations8.
As expected for TADs in wild-type embryos, two adjacent probes within a TAD on either X or autosomes overlapped to a greater extent than two adjacent probes on either side of a TAD boundary (Fig. 2b–e and Extended Data Fig. 8a–d). For DCC-dependent TAD boundaries on X, including rex-47, rex-32, and rex-8, adjacent probes flanking TAD boundaries overlapped and colocalized more in embryos lacking DCC binding than in wild-type XX embryos (Fig. 2c, d and Extended Data Fig. 8b). In contrast, the DCC-independent TAD boundaries on X and autosomes did not change (Fig. 2e and Extended Data Fig. 8c, d). FISH analysis also confirmed that some DCC-dependent TAD boundaries were eliminated (rex-47), and others reduced (rex-32) in DC mutants and XO males (Fig. 2c and d), showing that the DCC alters X structure by strengthening pre-existing TAD boundaries and creating new ones.
Robust correlation between rex sites, DCC-dependent TAD boundaries, and regions of greatest insulation loss in DC mutants (Fig. 2a, Extended Data Fig. 7d; Supplementary Table 2) led us to test whether rex sites interact in a DCC-dependent manner. We found rex-rex interactions to be among the most prominent interactions on X by comparing the ranking (Extended Data Fig. 7a) and cumulative distribution (Fig. 3a, b) of Z-scores for rex interactions with those for all other X interactions. In DC mutants, rex-rex interactions decreased more than any of the 1000 random sets of X interactions (Fig. 3a and c, Extended Data Fig. 7b, c, e). These observations support the hypothesis that DCC binding at rex sites facilitates rex-rex interactions.
The rex-rex interaction frequency was directly related to the level of DCC occupancy at rex sites, as shown by 3D profiles of Hi-C interaction frequencies made for pairwise combinations of 10 kb bins overlapping either the top 25 DCC-occupied rex sites or all 64 rex sites (Fig. 3d, Extended Data Fig. 7f, Fig. 2a and Supplementary Table 2). Interactions for the top 25 rex sites exceeded those for all rex sites.
The correlation between rex-interaction strength and DCC occupancy was reinforced by contrasting results with dependent on X (dox) sites. The DCC spreads to these lower affinity dox sites located in promoters of highly expressed genes once recruited to X by rex sites3,4. dox sites showed no substantial interactions in 3D plots (Extended Data Fig. 7g).
The strongest rex-rex interactions occurred between rex sites at DCC-dependent TAD boundaries on X (Fig. 3e). Weaker rex-rex interactions also occur within TADs. In DC mutants, rex interactions within TADs and between TAD boundaries diminished to the level of non-rex interactions (Fig. 3e). For autosomes, in contrast, interactions between TAD boundaries were not greater than interactions within TADs, and neither set of interactions changed in DC mutants (Fig 3e and Extended Data Fig. 7h). These results suggest that DCC-dependent interactions between rex sites at TAD boundaries contribute more to boundary formation on X than rex interactions within TADs, although DCC-dependent rex interactions within TADs might contribute to TAD integrity.
Visualization of Hi-C interaction data via Circos plots illustrates that almost all rex sites engage in one or multiple strong DCC-dependent interactions with other rex sites, particularly at adjacent TAD boundaries (Fig. 3f, g). Together, our findings reinforce the model that rex sites contribute to TAD formation by recruiting the DCC and facilitating DCC-dependent looping interactions between rex sites at TAD boundaries. In contrast, TAD boundaries on autosomes do not appear to result from looping interactions between boundaries (Fig. 3e right panel and Extended Data Fig. 7h), suggesting that different strategies govern, in part, the formation of DCC-dependent and autosomal TADs.
The model that rex interactions play a critical role in establishing and reinforcing TAD boundaries makes specific predictions. First, rex interactions identified by Hi-C should be evident by FISH in single cells. Second, deletion of a strong rex site from a DCC-dependent TAD boundary should reduce or eliminate the boundary. Both predictions were met.
To confirm DCC-dependent rex-rex interactions and further access X-chromosome topology, we devised a FISH assay using 3–6 kb probes to quantify the spatial separation between two sites (Methods and Fig. 4). We compared distances between loci in XX embryos with (wild-type) and without (DC mutant) DCC binding on X to quantify the level and DCC dependence of interactions. We also compared distances in XO embryos with and without DCC binding on X to quantify DCC-dependent interactions that occur between loci on the same chromosome (Fig. 4 legend). Hi-C analysis did not distinguish between interactions within the same chromosome or across homologous chromosomes.
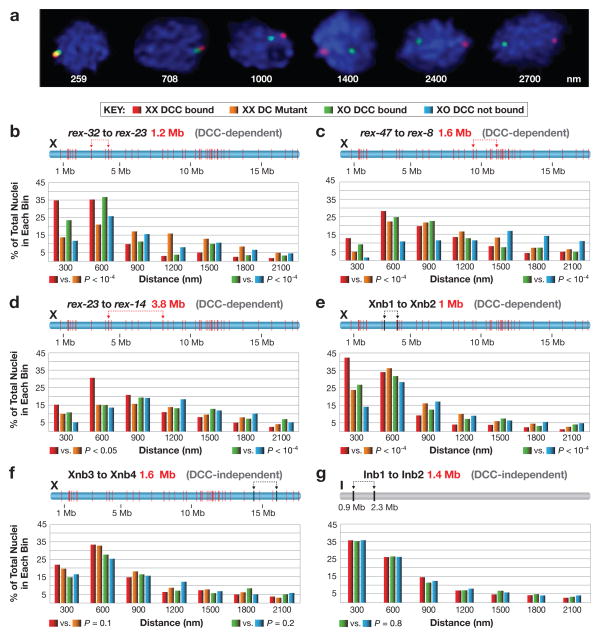
Figure 4. Quantitative FISH shows DCC-dependent association of rex sites in single cells.
a, Representative embryonic nuclei show variability in spacing of FISH probes (red, green) targeting two rex sites. b-g, Quantification of the 3D distance between FISH probes in embryos of different genotypes. DCC binding to the single X of XO embryos was achieved using a xol-1 (XO lethal) mutation, which activates sdc-2, the XX-specific trigger of DCC assembly20. b–d, Pairs of rex sites at DCC-dependent TAD boundaries of varying genomic separation. e, A pair of sites on X that lack DCC binding sites within 100 kb but have DCC-dependent Hi-C interactions. f, g, Loci on chromosomes X and I that lack DCC binding sites within 80–90 kb and display DCC-independent Hi-C interactions. (b–g) Distances between FISH spots were binned in 300 nm intervals and represented in relative frequency histograms. Schematic above each histogram depicts the locations of FISH probes (arrows), their genomic separation (red text), and the location of all rex sites (red bars) or sites lacking DCC binding (black). The DCC dependence or independence of the corresponding Hi-C interactions is indicated above the histogram (grey). P values comparing genotypes were calculated using the chi-square test to compare the 0–300 nm bin with 301–2700 nm bins. The 0–300 nm bin contains FISH probes considered co-localized, because probes < 300 nm apart always overlap visually, while probes 700 nm apart appear only adjacent to each other.
FISH analysis confirmed all categories of interactions shown by Hi-C: (1) strong DCC-dependent interactions between rex sites at DCC-dependent TAD boundaries (rex-32 to rex-23, rex-47 to rex-8, and rex-23 to rex-14); (2) strong DCC-dependent interactions between X loci lacking DCC binding (Xnb1 to Xnb2 and Xnb7 to Xnb8); (3) strong DCC-independent interactions between loci on X (Xnb3 to Xnb4) or I (Inb1 to Inb2) that lacked DCC binding; and (4) weak DCC-independent interactions between distant loci on X (Xnb5 to Xnb6) or I (Inb3 to Inb4) that lack DCC binding (Fig. 4b-g, Extended Data Fig. 9a–f, i–k). FISH and Hi-C results agreed, for both the strength and DCC dependence of interactions (Extended Data Fig. 9g and h).
The only discrepancy occurred for distantly spaced rex loci (rex-1 to rex-8 [6.7 Mb]; rex-32 to rex-8 [8.1 Mb]), which showed greater DCC-dependent spatial proximity by FISH analysis than predicted by Hi-C (Extended Data Fig. 9l and m). Loss of sensitivity in our Hi-C data for sites separated by >5 Mb likely accounts for the difference.
Both FISH and Hi-C experiments showed that the DCC-dependent topology of X brings many distant, non-rex sites into close proximity. If the DCC compacted X uniformly, pairs of non-rex loci separated by similar distances should exhibit comparable levels of DCC-dependent interactions. They did not. For example, two pairs of non-rex loci (Xnb1 and Xnb2 [1 Mb]; Xnb7 and Xnb8 [1.4 Mb]) showed strong DCC-dependent interactions (Fig. 4e; Extended Data Fig. 9g, h, k), but the non-rex loci Xnb3 and Xnb4 (1.6 Mb) showed strong DCC-independent interactions (Fig. 4f). Thus, the DCC affects the overall topology of X but does not cause uniform compaction across X.
To test directly whether DCC-dependent interactions between rex sites create TAD boundaries, we deleted the endogenous rex-47 site from a DCC-dependent TAD boundary using genome editing with CRISPR/Cas9 (Extended Data Fig. 8e, f) and assayed TAD structure with FISH (Fig. 3h). ChIP-qPCR showed the deleted rex locus (rex-47 Δ) lacked DCC binding (Extended Data Fig. 8g). The TAD boundary was greatly diminished, as predicted (Fig. 3h). For FISH probes on either side of the rex-47 TAD boundary, overlap was increased in rex-47 Δ and DC mutant embryos over that in wild-type embryos. In contrast, overlap was not statistically different between rex-47 Δ and DC mutant embryos. Thus, the DCC plays a key role in inducing and reinforcing X TAD boundaries by mediating long-range interactions between its highest-affinity rex sites.
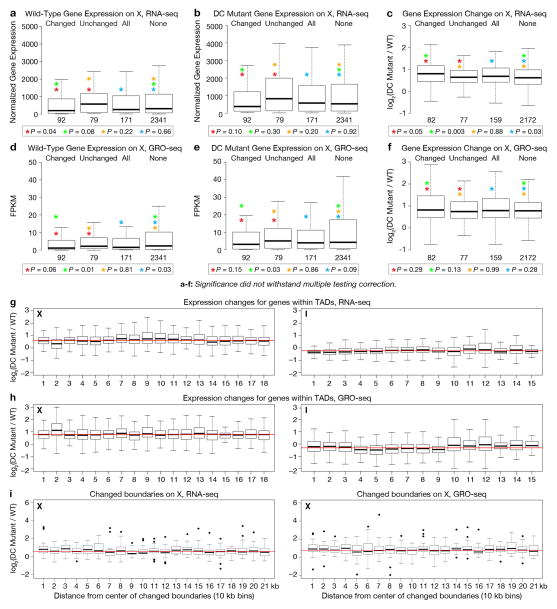
We explored the relationship between TAD structure and gene expression. Our prior work showed that the DCC acts at a distance to repress gene expression,3,4,20 suggesting that a unique, DCC-dependent X-chromosome structure might mediate chromosome-wide gene repression, as supported by our Hi-C and FISH data. We assessed whether the structure of individual TADs affects gene expression locally or whether the chromosome-wide topology created from TADs regulates gene expression globally. Both RNA-seq data derived from the embryo preparations used for Hi-C analysis and GRO-seq data from independent embryo preparations support the latter hypothesis for the following reasons.
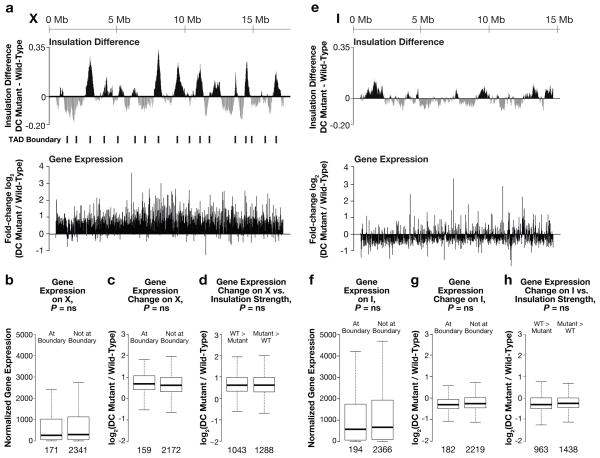
First, in wild-type embryos, genes at TAD boundaries were not expressed at significantly different levels from genes within TADs, for either X (Fig. 5b, Extended Data Fig. 10a, d) or chromosome I (Fig. 5f). Second, although X is organized into DCC-dependent TADs in wild-type animals, no similarly coordinated block of genes exhibited elevated expression in DC mutants (Fig. 5a). That is, the changes in expression were not significantly different for X-linked genes within TADs, at all TAD boundaries, at changed TAD boundaries, or within regions of changed insulation (Fig. 5c, d, Extended Data Fig. 10b, c, e–i). Similarly, DC mutations did not alter gene expression on chromosome I in any discernible pattern (Fig. 5e, g–h, Extended Data Fig. 10g–i).
Figure 5. DCC-dependent TADs influence global rather than local gene expression.
a, Insulation changes and TAD boundaries are compared to median fold-changes in expression (10 kb bins across X) between wild-type and DC mutant embryos. a–h, No discernible pattern was detected between mutant-induced changes in expression and gene locations relative to TADs or TAD boundaries. b, Box plots show comparison of expression levels for X genes within or outside TAD boundaries in wild-type embryos. Expression levels, normalized read number per kilobase of gene length. c, Box plots show expression changes for X genes within or outside TAD boundaries. d, Comparison of expression changes (DC mutant/wild-type) for X gene sets with greater insulation scores in wild-type embryos (grey domains in a) versus in DC mutants (black domains in a). e–h, Same as a–d but for genes on chromosome I. P-values for b–d and f–h, Mann-Whitney U Test; no significant P-values withstood multiple testing correction. ns, not significant. (a, c, d, e, g, h), Lowest-expressed genes (bottom 10%) were removed from analyses.
Our results support the model that TAD structure on X mediated by DCC binding to rex sites creates a 3D topology that acts chromosome wide to repress gene expression. Given that changes in TAD boundaries occur locally, while changes in gene expression occur chromosome wide, a parsimonious model posits that DCC-dependent changes in X structure imposed by rex-rex interactions drive the chromosome-wide reduction in gene expression. Potential DCC-dependent nuclear positioning of X might also affect gene expression, as speculated by others21.
In summary, DCC-induced formation of TAD structure on X demonstrates a dramatic remodeling of chromosome topology that reveals a central role for condensin in shaping the 3D landscape of interphase chromosomes. Not only does condensin compact and resolve mitotic and meiotic chromosomes, it acts as a key structural element to regulate gene expression. No other molecular complex or set of DNA binding sites is yet known to cause comparably strong effects on Mb-scale TAD structure in higher eukaryotes22–24. Our new understanding of the topology of dosage-compensated chromosomes provides fertile ground to decipher the detailed mechanistic relationship between higher-order chromosome structure and chromosome-wide regulation of gene expression.
Methods
Nematode strains
The strains that follow were used in our study.
Wild-type: TY125, N2 Bristol, XX
Dosage Compensation mutants:
sdc-2 (y93, RNAi) X (XX strain used in all experiments requiring a DC mutant strain, except those listed below using TY2222 or TY1996)
TY1996, szT1/sdc-2(y74) unc-3(e151) X (XX DC mutant in Fig. 2b–e, Fig. 3h, Fig. 4b–f and Extended Data Fig. 9a–e, i)
TY2222, her-1(hvly101) V; xol-1(y9) sdc-2(y74) unc-9(e101) X (XX DC mutant used only in Extended Data Fig. 9j)
TY0810, sdc-2(y93) X (XX strain used to create sdc-2 (y93, RNAi) XX embryos)
TY0525, him-8(e1489) IV; xol-1(9) X (used for XX and XO DCC bound)
CB1489, him-8(e1489) IV (used for XO DCC not bound)
Nematode culture
ChIP-seq, RNA-seq and chromosome conformation capture
To obtain wild-type control embryos, wild-type N2 worms were grown at 20°C on NG agar plates with concentrated HB101 bacteria. For DC mutant embryos, 10 μl of packed synchronous sdc-2(y93) L1 worms were placed onto 10 cm RNAi plates (NG agar with 1 mM IPTG and 100 μg/ml Carbenicillin) seeded with 2–3 ml of concentrated HT115 (DE3) bacteria carrying the Ahringer feeding library plasmid25 expressing the coding region of sdc-2. The RNAi plates were incubated at 25°C overnight before L1s were added.
Immunofluorescence and FISH analysis
Animals were grown at 20°C on NG agar plates seeded with OP50 grown in Luria Broth (LB). The worms were grown at 20°C until gravid adults, then dissected for their embryos and stained as described below.
Antibodies
Rat polyclonal SDC-3 (PEM4A) antibodies were made against amino acids 1067-1340 of SDC-3 fused to GST. Rabbit polyclonal antibodies against DPY-27 (rb699) and SDC-3 (rb1079) were as described previously18,26. Mouse monoclonal Mab414 antibody (1 mg/ml) was obtained from Abcam (ab24609). Normal rabbit IgG (400 μg/ml) was from Santa Cruz Biotechnology (sc-2027). Rabbit polyclonal LMN-1 antibody (500 mg/ml) was from SDIX (3853.00.02)
ChIP-seq library creation and analysis
Libraries were made and analyzed from one batch of wild-type embryos (data consistent with all previously wild-type published ChIP-seq data20) and two biological replicates of sdc-2(y93, RNAi) embryos as described previously20.
Modified Hi-C embryo isolation and crosslinking
Worms of appropriate genotype, either wild-type (two biological replicates) or sdc-2(y93, RNAi) (two biological replicates), were grown until gravid adults. The worms were harvested and bleached to release the embryos and remove the carcasses. Following bleaching, embryos were centrifuged for ~45 sec at 1500–1800 rpm and washed 3 times in 1X M9 buffer to remove bleach solution. An equal volume of 1X M9 was added to the embryos and they were frozen in 1 ml aliquots and stored at −80°C. The frozen embryos were thawed on ice and supplemented with 1 mM PMSF and 5 mM DTT. The embryos were then washed once in 50 ml formaldehyde solution (1X M9 solution with 2% (v/v) formaldehyde, Polysciences 18814-20). Embryos were cross-linked in 50 ml of formaldehyde solution for 30 min at room temperature while shaking. Following crosslinking embryos were washed once with 50 ml of 100 mM Tris-HCl, pH 7.5, followed by two 50 ml washes of 1X M9. The embryos were then washed once in lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl and 0.2% (v/v) Igepal CA-630 (Sigma I8896)) supplemented with 5 mM DTT, 1 mM PMSF, 0.1% (v/v) protease inhibitors (EMD 539134) and 0.5 mM EGTA. To obtain extract, embryos were dounced 10 times using the large pestle (Kontes 2 ml glass dounce, Spectrum 985-44182; clearance 0.076-0.127 mm), and then 10 times using the small pestle (clearance 0.01-0.069 mm). All douncing steps were performed on ice. The dounced extract was spun for 5 min at 100xg at 4°C, and the supernatant was saved. The pellet was resuspended in 750 μl of supplemented lysis buffer and dounced again. This procedure was repeated 7–10 times. After each spin, a 9 μl aliquot was taken from the supernatant, mixed with 1 μl of 10 ng/ml DAPI and visualized under a microscope. All supernatants containing only nuclei, and not broken carcasses, were combined. An aliquot of the combined supernatant was stained with DAPI and the nuclei were counted using a hemocytometer, and then spun down for 5 min at 2000xg at 4°C. The nuclei were resuspended in the appropriate volume of 1.25X DpnII buffer (NEB B0543S) to create a Hi-C library as described below.
Modified Hi-C library preparation
The 3C libraries were made as described below. The protocol is based on the published protocols, with some modifications12,27,28. Approximately 1.5 × 108 C. elegans nuclei were pipetted into 5–10 1.7 ml tubes and resuspended in 300 μl of 1.25X DpnII buffer. 38 μl of 1% (w/v) SDS was added per tube and the tubes were incubated at 65°C for 10 min. After the addition of 34 μl of 20% (v/v) Triton X-100, the tubes were incubated at 37°C for 1 hr, shaking at 1000 rpm. 30 μl (1500 U) of DpnII (NEB R0543M) were added to each tube, and they were incubated overnight at 37°C while rocking. 26 μl of 20% (w/v) SDS was added to each tube and they were incubated at 65°C for 20 min, shaking at 1000 rpm. The reaction was then added to 7.6 ml of ligation master mix (745 μl of 10% Triton X-100, 745 μl of 10X T4 ligation buffer (500 mM Tris-HCl, pH 7.5, 100 mM MgCl2, 100 mM DTT), 80 μl of 10 mg/ml BSA, 80 μl of 100 mM ATP, and 5.96 ml water). 100 μl (100 U) T4 DNA ligase (Invitrogen 15224-025) was added and the reactions were incubated for 4 hrs at 16°C. After incubation 50 μl of 10 mg/ml Proteinase K was added and the tubes were further incubated at 65°C overnight. The next day 50 μl of 10 mg/ml Proteinase K was added to the reactions, and they were incubated at 65°C for an additional 2 hrs. 2 μl of RNaseA (1 mg/ml) was added to each sample and incubated for 30 min at 37°C. The ligated DNA was then phenol-chloroform extracted and ethanol precipitated overnight. DNA was pelleted at 14,000xg for 30 min at 4°C, and then washed twice with 70% ethanol and air-dried. The DNA pellets from all Hi-C reactions were combined and dissolved in a total of 500 μl of 1X TE buffer, pH 8.0. Excess salt was removed from the samples via centrifugation using a filter unit (AMICON® Ultra Centrifugal Filter Unit – 0.5 ml 30 KDa) following the manufacture’s instruction. Briefly, the samples were spun at 18,000xg for 10 min to reduce the volume to 40–50 μl. Flow through was discarded and 450 μl of 1X TE, pH 8.0 buffer was added to each unit and spun as before. This wash step was repeated at least 5 times. The volume of the eluate was adjusted to 100 μl with water. The concentration of DNA was determined and 10 μg of the Hi-C library was resuspended in 100 μl of water. AMPure beads, supplied as a suspension of magnetic beads in a PEG solution (Beckman Coulter, A63880), were used to remove large DNA fragments (> 10 kb), following the protocol provided by the manufacturer. Specifically, for the first DNA selection, 35 μl of AMPure beads were added to the 100 μl of DNA. The supernatant was kept and the beads, which bind only large DNA molecules under these PEG conditions, were discarded. To then remove smaller fragments, 65 μl of AMPure beads were added to the supernatant and the beads, which bind all DNA molecules greater than 100 bp due to the greater PEG concentration, were kept and washed with 70% ethanol. The DNA was eluted from the beads in 100 μl of 10 mM Tris-HCl, pH 8.5. The eluted DNA was then adjusted to 125 μl with 1X TE, pH 8.0 and sheared to 500–1000 bp using a Covaris S2 (Covaris, 520045) in micro tubes with the following settings: duty cycle, 5%; intensity, 3; cycles/burst, 200; time, 65 sec. The sheared DNA was then size selected for fragments larger than ~100 bp using AMPure beads and eluted in 34 μl of water. The DNA was quantified and 500 ng was used to make a paired-end Illumina sequencing library following the standard protocol (PE-930-1001), with the exception that we size selected 500–600 bp at the gel excision step prior to adding adapters for sequencing. The library was sequenced using 100 bp paired end reads with a HiSeq2500s machine.
Read Mapping/Binning/ICE correction
Iterative mapping and error correction of the chromatin interaction data were performed as previously described29. Supplementary Table 1 summarizes the mapping results and lists the different categories of DNA molecules encountered in the libraries. We obtained around 70 million valid pairs that represent chromatin interactions per replicate. The frequency of redundant read pairs, due to PCR amplification, were found to be below ~5%. Redundant read pairs were removed. The number of Hi-C interactions mapped to sequences belonging to homologous chromosomes (both intra-chromosomal [cis] and inter-homolog [trans] interactions) was much higher than the interactions mapped to non-homologous chromosomes (inter-chromosomal [trans] interactions). Assuming that inter-homolog interactions (trans) are as frequent as non-homologous inter-chromosomal interactions (trans), we estimate that 80–90% of interactions mapped to the same chromosomes are intra-chromosomal (cis) interactions, with DC mutant (90%) higher than wild-type (> 85%). Whether this difference reflects a biological phenomenon or is due to technical differences is currently not known. Conversion of interaction data into Z-scores eliminates this difference (see below).
The data were binned at both 10 kb and 50 kb non-overlapping genomic intervals. Binned data were normalized for intrinsic biases such as differences in number of restriction fragments within bins using the previously developed ICE method29. To normalize for differences in read depth of different datasets we summed the entire genome-wide binned ICE-corrected interaction matrix, excluding the diagonal (x = y) bins. We then transformed each interaction into a fraction of the matrix sum (minus diagonal x = y bins). Each fraction was then multiplied by 106. Biological replicates were highly correlated (Pearson’s correlation coefficients > 0.98 for 50 kb binned data excluding short-range interactions up to 50 kb). The correlations between biological replicates were higher than those between wild-type and DC mutant. Overall these numbers indicate that the modified Hi-C procedure was reproducible and performed as expected. For most analyses sequence reads obtained for biological replicates were pooled and ICE-corrected as described above to create a combined replicate dataset.
At 10 kb resolution, very long-range interactions are not sampled deeply enough to provide robust and reliable data. Therefore, we truncated the 10 kb binned data to include only cis interaction pairs separated by 4 Mb or less in linear genomic distance. This distance cutoff was chosen based on the observation that beyond this point, both wild-type and DC mutant datasets have no observed reads in more than 50% of bin-bin interactions. In addition to limiting the dynamic range of interaction counts at these large distances, this high frequency of un-sampled interactions beyond 4 Mb causes a dramatic collapse in the standard deviation of the overall chromatin interaction decay over distance, making the LOWESS expected and Z-score calculations beyond 4 Mb unreliable. For 50 kb bins, all distances were included in analyses, because the coverage of cis interaction pairs never dropped below 50% for any distance at this resolution.
TAD Calling (Insulation Square Analysis)
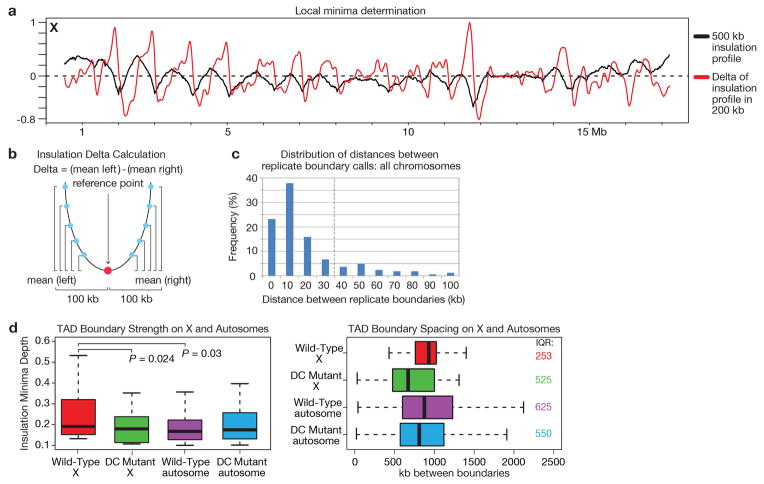
To calculate the ‘insulation’ score of each bin in the 10 kb binned Hi-C data, we calculated the average number of interactions that occurred across each bin. This can be visualized by sliding a 500 kb × 500 kb (50 bins × 50 bins) (Extended Data Fig. 2 and 3) square along the matrix diagonal, and aggregating all signal within the square. The mean signal within the square was then assigned to the 10 kb diagonal bin and this procedure was then repeated for all 10 kb diagonal bins. For any bins within 500 kb of the matrix start/end, an insulation score was not assigned, as the 500 kb × 500 kb insulation square would extend beyond the matrix bounds. The insulation score was then normalized relative to all of the insulation scores across each chromosome by calculating the log2 ratio of each bin’s insulation score and the mean of all insulation scores. Valleys/Minima along the normalized insulation score vector represent loci of reduced Hi-C interactions that occur across the bin. These valleys/minima are interpreted as TAD boundaries or areas of high local insulation. The valleys/minima were detected as follows: First, a delta vector was calculated to approximate the slope of the normalized insulation vector. The delta vector is defined as the difference between the amount of insulation change 100 kb to the left of the central bin and 100 kb to the right of the central bin (relative to the central bin) (Extended Data Fig. 3b and c). The delta vector crosses the horizontal 0 at all peaks and all valleys. All bins where the delta vector crosses 0 were extracted. Zero-crossings occurring at peaks were removed, and the remaining zero-crossings, all occurring at potential valleys were passed through a boundary strength filter. The boundary strength was defined as the difference in the delta vector between the local maximum to the left and local minimum to the right of the boundary bin. All boundaries with a boundary strength < 0.1 were removed. This method in practice is very similar to the widely used zero-derivative method for detecting peaks/valleys in various data vectors.
The precision with which we could define a boundary was determined by comparing boundary calls across biological replicates (Extended Data Fig 3c). The final boundary zones were defined as +/− 30 kb around the pooled replicate insulation minima bins (70 kb total) because most (>80%) replicate boundary calls overlapped within this window. Wild-type and DC mutant insulation profiles were compared by subtracting the wild-type insulation profile from the DC mutant insulation profile. We compared the insulation profiles and boundary calls resulting from a full range of alternative insulation square sizes (Extended Data Fig 2b, c). We find that a 500 kb square size captures best the major robust boundaries that change in the DC mutant. In contrast, boundaries detected by a 100 kb insulation square, for example, only affect interactions within a few bins of the boundary rather than insulating larger genomic regions from one another and do not change in the DC mutant (Extended Data Fig. 2e).
Z-Score Calculation
We modeled the overall chromatin interaction decay with distance using a modified LOWESS method (alpha = 0.5%, ignore zeros, IQR filter), as described previously30. LOWESS calculates the weighted-average and weighted-standard deviation for every genomic distance by pooling all data genome-wide. We transformed interaction data into a Z-score by calculating: [(observed signal – lowess-average)/lowess-stdev]. Observed signals with a count of 0 were excluded from the Z-score transformation. By expressing interaction data as Z-scores, we corrected for minor differences in the overall decay with genomic distance that can vary slightly between samples.
To calculate the difference between the wild-type and DC mutant Hi-C data, we calculated the difference between the combined replicate DC mutant Z-score data and the combined replicate Wild-Type Z-score data (DC Mutant Z-score minus Wild-Type Z-score). (Fig. 1c, f; Extended Data Fig. 1, 4–6).
Compartment analysis and comparison to LEM-2 associated domains
The presence and locations of A/B-compartments can be quantified using principle component analysis, where the largest eigenvector typically represents the compartment profile11,13,29. Applying this approach to 50 kb binned interaction data, we determined the positions of such preferentially associating compartments along each C. elegans chromosome (Extended Data Fig. 4e, 5e, 6c, g, k, o). Compartment positions quantified in this manner closely align with the large sub-chromosomal domains that are visible in the chromatin interaction maps.
LEM-2 binding data15 (log2 ratio of ChIP signal over input) were lifted from the ce4 genome assembly to the ce10 assembly, and data were averaged in 50 kb bins. These bins correspond exactly to the coordinates of the binned chromatin interaction data. Binned LEM-2 binding data were then plotted along each chromosome, and compared to the compartment profiles (Extended Data Fig. 4e; 5e; 6c, g, k, o).
3D plots
To test for elevated levels of interaction between certain classes of sites in the genome, we constructed 3D plots. For each plot, a list was first made of all 10 kb bins meeting desired criteria: containing any rex or Prex site (Fig. 3d), containing a rex or Prex site in the top 25 by ChIP-seq signal (Fig. 3d and Extended Data Fig. 7f), or containing any dox site (Extended Data Fig. 7g). Prex sites are defined as those with very strong ChIP-seq signal that was greatly diminished in sdc-2 mutants. Unlike rex sites, which also have these properties, Prex sites have not been tested for autonomous DCC recruitment in vivo through an array assay4. Next, sub-matrices of wild-type or DC mutant interactions were prepared for all possible pairs of bins in this list, extending 50 kb away from the central bin in all directions. Pairs of bins that were separated by less than 100 kb were excluded so that no sub-matrices would overlap the whole-chromosome interaction matrix diagonal (interactions within the same bin). All pairwise sub-matrices were then averaged together and the values plotted in 3D. If sub-matrices stretched past the end of the chromosome or overlapped bins with no data (unmappable sequence, etc.), only the part of the sub-matrix containing data was included in the average.
Cumulative Plot Randomization
To assess the significance of the decrease in Z-scores observed for the set of rex-rex interactions, we selected 1000 random sets of 785 interactions (Fig. 3c, Extended Data Fig. 7e). These random interaction sets were thus the same size as the rex-rex interaction set. The p-value represents the fraction of the 1000 randomized interaction sets that changed more from wild-type to DC mutant than the rex-rex set (according to the KS test statistic).
Circos plots
Plots were generated using the Circos package to highlight the strength of various sets of rex-rex interactions in wild-type and DC mutant at 50 kb resolution. A Z-score threshold of 2 was selected and interactions were colored and given line thickness proportional to their Z-score. Z-scores greater than 8 were determined to correspond to “singleton” outlier interactions and were excluded.
TAD FISH
Preparation of FISH probes
FISH probes covering 400–500 kb genomic regions were prepared using pooled fosmids (BioScience LifeSciences), as described previously8. 1 μg DNA was labeled with Alexa-488, Alexa-594, Alexa-555 or Alexa-647 using FISH Tag DNA Kit (Invitrogen). The genomic locations of tested regions are listed below:
| Probe1 | Chr. X | 9.05–9.45 Mb |
| Probe2 | Chr. X | 9.5–9.9 Mb |
| Probe3 | Chr. X | 9.95–10.35 Mb |
| Probe4 | Chr. X | 2.0–2.5 Mb |
| Probe5 | Chr. X | 2.5–3.0 Mb |
| Probe6 | Chr. X | 3.0–3.5 Mb |
| Probe7 | Chr. X | 11.2–11.7 Mb |
| Probe8 | Chr. X | 11.7–12.3 Mb |
| Probe9 | Chr. X | 12.3–12.8 Mb |
| Probe10 | Chr. X | 10.6–11.1 Mb |
| Probe11 | Chr. X | 11.1–11.6 Mb |
| Probe12 | Chr. I | 4.1–4.6 Mb |
| Probe13 | Chr. I | 4.6–5.1 Mb |
| Probe14 | Chr. I | 5.1–5.6 Mb |
| Probe15 | Chr. X | 3.5–4.1 Mb |
FISH procedure
C. elegans embryos were obtained by dissecting gravid N2, him-8(e1489) or szT1/sdc-2(y74) unc-3(e151) adults in 13 μl of water on poly-lysine coated slides. A coverslip was added on top of the dissected worms, and the slides were then frozen in liquid nitrogen for at least 1 min. Coverslips were cracked off, and the samples were dehydrated in 95% ethanol for at least 10 min. 35 μl of fix (2% (v/v) paraformaldehyde in egg buffer (25 mM HEPES, pH 7.3, 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl=) was added and slides were incubated in a humid chamber for 5.5 min. Slides were washed 3 times 10 min with 1X PBS-T (0.5% Triton X-100 in 1X PBS) at room temperature. Excess 1X PBS-T was then removed and 15 μl of hybridization solution (30% (v/v) formamide, 3X SSC, 10% dextran sulfate) containing approximately 50 ng of each FISH probe was added. Hybridization was performed in a temperature-controlled slide chamber (Bio-Rad ALD0211 Alpha Unit Block Assembly). The following FISH program was typically run overnight: 90°C for 5 min, 0.5°C/second to 50°C, 50°C for 1 min, 0.5°C/sec to 45°C, 45°C for 1 min, 0.5°C/sec to 40°C, 40°C for 1 min, 0.5°C/sec to 38°C, 38°C for 1 min, 0.5°C/sec to 37°C, 37°C overnight. Slides were then washed at 39°C as follows: 3 times 10 min with 30% (v/v) formamide in 2X SSC, 3 times 10 min with 20% (v/v) formamide in 2X SSC, 3 times 5 min with 10% (v/v) formamide in 2X SSC, 3 times 5 min with 2X SSC, and 3 times 1 min with in 1X SSC. Slides were then washed 3 times 10 min in 1X PBS-T. For N2 embryos, the slides were mounted in Prolong Gold antifade reagent (Invitrogen, P36934) containing DAPI (1 ng/ml). For him-8(e1489) and sdc-2(y74) embryos, immunostaining with SDC-3 antibody was performed following FISH to determine the sex and/or genotype of embryos as described below.
Immunofluorescence
Excess 1X PBS-T was removed and 35 μl of primary antibody (rat anti-SDC-3 antibody, 1:400) were added. Samples were incubated in a humid chamber for 6 hrs to overnight. Slides were washed 3 times 10 min with 1X PBS-T at room temperature and then incubated in secondary antibody (Alexa-Fluor-647 goat anti-rat antibody (Invitrogen), 1:250) for 6 hrs to overnight. Slides were then washed 3 times 10 min with 1X PBS-T at room temperature and then mounted.
Microscopy and co-localization analysis
Embryos were imaged on a Leica TCS SP8 microscope using 63X, 1.4 NA objective lenses. The scanning settings for SP8 were: 1024 × 1024 pixels frame size, 51.5 nm pixel size, 3.5 zoom factor, 400 Hz scanning speed and 83.9 nm step size for Z-sections. Image deconvolution was performed using Huygens Professional Software.
After deconvolution, the homozygous sdc-2(y74) unc-3(e151) XX embryos were determined based on the lack of SDC-3 staining on the X chromosomes and their sex was further confirmed by examining the number of X-chromosome FISH signals. For all genotypes, embryos between 200 and 400-cell stages which match the developmental stage of Hi-C samples were selected for further analysis.
The deconvolved image stacks of embryos were manually segmented based on DAPI staining using Priism software31. FISH signals in individual embryos were thresholded to make the total signals from each probe occupy equal volume. The center-of-mass coordinates for the FISH signals from the probe in the middle of the probe set were determined using a built-in Find Points function in Priism. Regions of equal volume were then created around the FISH signals to encompass the entire sets of FISH signals on the same chromosomes using a Python script. Pearson’s correlation coefficients between pairs of FISH probes were then calculated: the more the two probes overlap, the higher the correlation coefficient.
3D quantitative FISH for measuring the interaction frequency between genomic loci
FISH experimental design
To examine the DCC dependence of interactions between genomic loci, and to distinguish between inter-homolog (trans) and intra-chromosomal (cis) interactions, we performed the 3D FISH analysis in both XX and the XO embryos in which the DCC was bound or not bound to X chromosomes. For these experiments, we acquired confocal images of embryos hybridized with FISH probes to two genomic loci and also stained with lamin (LMN-1) antibody and DAPI to help segment the nuclei. Newly developed software was used to measure the 3D distance between FISH probes automatically.
To assay XO embryos having DCC binding on X, we performed the experiments using xol-1(y9); him-8(e1489) animals. These animals carried a deletion of the master switch gene (xol-1) that inhibits DCC binding to X chromosomes of XO embryos. DCC association with X kills XO animals by the L1 larval stage. To enrich for XO male embryos in our experiments, we used mutation in him-8 (high incidence of males), which elevated the frequency of male progeny in a hermaphrodite brood from 0.02% to 37%. The XX embryos deficient in DCC binding were obtained from szT1/sdc-2(y74) unc-3(e151) animals, as described above.
To measure the distance between FISH foci in Z stacks of confocal images, we developed software (Mets and Meyer, unpublished) that identified foci automatically, assigned foci to appropriate nuclei, and quantified the distance between foci in 3D space, thereby permitting the unbiased quantification of probe-interaction frequency. The quantification involved several steps. Each FISH spot was center fitted, and its location was recorded in X, Y, and Z. For all nuclei, distances between all combinations of red and green FISH spots were calculated using a distance quantification algorithm that employs LMN-1 and DAPI co-staining to segment the nuclei. In XX embryos, four FISH spots (two red and two green) were generally apparent for X-linked probes in each nucleus, corresponding to the hybridization of both probes to their target sites on both homologous chromosomes. To eliminate the bias in our calculations for interactions caused by the inclusion of distances between probes on different chromosomes, we utilized only the shortest of the four possible distances between red and green probes in each nucleus for X-linked loci in XX embryos and for autosomal loci in all embryos.
We segmented the distances into 300 nm bins and plotted the relative contribution of each bin to the total number of measured distances. The limit of resolution of the confocal microscope is ~200 nm in X and Y, making 300 nm a reasonable choice for the smallest bin. Furthermore, probes spaced < 260 nm apart appear overlapping by visual inspection, and probes spaced ~700 nm apart appear adjacent, indicating that the smallest bin size (300 nm) represents a degree of overlap that would be consider co-localized. Chi-square tests comparing the number of FISH pairs within 0–300 nm to those within 301–2700 nm were used to assess the similarity of data sets from different classes of embryos. The unbinned data were also represented in cumulative plots (Extended Data Fig. 9a–f).
Preparation of FISH probes
Primers were created to amplify 3–6 kb sequences of DNA corresponding to each site. 1 μg of the probe DNA was labeled using the FISH tag DNA Red Kit (Molecular Probes, F32949) or the FISH tag DNA Green Kit (Molecular Probes, F32947) according to the manufacturer’s protocol, with the following exceptions: the DNaseI was diluted 1:1000, and the labeled probes were eluted in 10 μl but then diluted 1:10 for use in staining. Primers to make the probes are listed below:
| rex-23 F | rex-23 | gcccattcaacccattgtcc |
| rex-23 R | rex-23 | gcactcgcatattccaaaacg |
| rex-32 F | rex-32 | cgcagctggccgttaaatg |
| rex-32 R | rex-32 | cattgcaggtgcgttcacaac |
| rex-47 F | rex-47 | ccgaaacacaacaacaatgc |
| rex-47 R | rex-47 | agactggcgaagaggaacaa |
| rex-8 F | rex-8 | tgtgatgcaagccagagttgg |
| rex-8 R | rex-8 | cattgagccgaatttccaaagg |
| rex-14 F | rex-14 | ttgcagttgcgaaagaaatg |
| rex-14 R | rex-14 | tttttgaggagatcgggatg |
| rex-1 F | rex-1 | ctcaagagctgcgaagtgc |
| rex-1 R | rex-1 | aaagttcaacgaccagaatgc |
| Xnb1 F | Xnb1 | tcgaatgacctcaagcactg |
| Xnb1 R | Xnb1 | tcaccactgaaatcggcata |
| Xnb2 F | Xnb2 | aaaacgcggtgaaacgatac |
| Xnb2 R | Xnb2 | gttttcctctccccaacaca |
| Xnb3 F | Xnb3 | gtatgcacacgcctcaaaaa |
| Xnb3 R | Xnb3 | ttggaatctctcaccggagt |
| Xnb4 F | Xnb4 | atggtaggacgttccgtttg |
| Xnb4 R | Xnb4 | aatccagccctctggttttc |
| Xnb5 F | Xnb5 | atttgcttgggcattaaacg |
| Xnb5 R | Xnb5 | ttcaatgaagagacgcgatg |
| Xnb6 F | Xnb6 | ccgtttttggcaatgaactt |
| Xnb6 R | Xnb6 | agaggatggtttggacgttg |
| Xnb7 F | Xnb7 | gagcgacgattctgtcttcc |
| Xnb7 R | Xnb7 | cgtcatgtccattttgcttg |
| Xnb8 F | Xnb8 | atcgtgccaagacctattcg |
| Xnb8 R | Xnb8 | ttttcgcatttcctgcttct |
| Inb1 F | Inb1 | aaaggaccctccccctaact |
| Inb1 R | Inb1 | tccatgcctacttgcctacc |
| Inb2 F | Inb2 | caggcgagcattctaccact |
| Inb2 R | Inb2 | ccggaaagagcattgattgt |
| Inb3 F | Inb3 | gcactgcaattgccaaccag |
| Inb3 R | Inb3 | ttcaaagacactcctcccatcc |
| Inb4 F | Inb4 | attgccgctaacccaagtgc |
| Inb4 R | Inb4 | tccaacgccaacaaaactcc |
Combined FISH and immunofluorescence procedure
FISH followed by immunofluorescence was performed as described in the previous section. 5–10 ng (0.5–1 μl of 1:10 dilution) of each FISH probe was used for hybridization. For immunofluorescence, primary antibodies were applied at the following dilutions in 1X PBS-T: rat anti-SDC-3, 1:400; rabbit LMN-1, 1:400. Secondary Alexa-Fluor-555 donkey anti-rabbit and Alexa-Fluor-647 donkey anti-rat antibodies (Invitrogen) were used at a 1:200 dilution.
Microscopy and image analysis
Embryos were imaged on a Leica TCS SP2 AOBS confocal microscope or a Leica TCS SP8 microscope using 63X, 1.4 NA objective lenses. The scanning settings for SP2 were: 1024 × 1024 pixels frame size, 46.5 nm pixel size, 5.0 zoom factor, 400 Hz scanning speed and 81 nm step size for Z-sections. The scanning settings for SP8 were as described in the previous section. The images were then deconvolved using Huygens Professional with the appropriate settings. The images were visualized and processed in Priism. The embryos were first cut out from the background using the Edit Polygon and Cut Mask function. Then the DAPI and LMN-1 channels were blurred using the 3-D Filter Function to make the nuclear signal continuous and thus allow for the nuclei to be accurately segmented. This protocol permits each nucleus to be counted as one spot by the Find Points function. A new processed image was made by discarding the Z-sections in the top and bottom 10% of the image, and by substituting the new blurred channels for those in the original image. The Find Points function was then used to count and record the local center of mass (LCOM) of each nucleus and each FISH spot in X, Y and Z using user-defined threshold values. The data for the location of the nuclei and the FISH, along with the processed image are processed using the software described in FISH experimental design section above.
rex-47 deletion
Plasmid construction
Expression vectors for both codon-optimized Cas9 and sgRNA (Peft-3::cas9-SV40_NLS::tbb-2 3′UTR and PU6::unc-119_sgRNA32 were obtained from Addgene. To enhance the expression and assembly of sgRNA, the sgRNA vector was modified by introducing an A-U flip in the sgRNA stem loop and extending the Cas9 binding hairpin33. To clone the protospacer sequence for the sgRNA targeting rex-47 (5′-GTAGTCACACCGAATTGATA -3′), the modified sgRNA vector was PCR amplified using primers GTAGTCACACCGAATTGATAGTTTAAGAGCTATGCTGGAAACAGCATAG and AACAGCTATGACCATGATTACGCCAAGCTTCACAGCCGACTATGTTTGGCGTCGAG or GACGTTGTAAAACGACGGCCAGTGAATTCCTCCAAGAACTCGTACAAAAATGCTCTGAAG and TATCAATTCGGTGTGACTACAAACATTTAGATTTGCAATTCAATTATATAG to generate two fragments with overlapping protospacer sequences. The two PCR products were then inserted into the sgRNA vector backbone generated by EcoRI/HindIII digestion using a previously described Gibson Assembly protocol34. To clone the repair template for making the 419 bp rex-47 deletion, two 500 bp homology arms flanking the target region were PCR amplified from C. elegans genomic DNA using primers ACGACGTTGTAAAACGACGGCCAGTGAATTCGACGTGTCGAAATTTTCAG and TTGAATTATTGACCATGGCAGACAGAGCGTAACGAGTAAT or ACGCTCTGTCTGCCATGGTCAATAATTCAATGCAATGAAG and CTATGACCATGATTACGCCAAGCTTAATAATAAACTTCCATAAGA. The homology arms and the sgRNA vector backbone were assembled using Gibson Assembly. The resulting repair template contains an NcoI restriction site between the homology arms, which facilitates the identification of desired mutations.
Cas9-mediated mutagenesis and mutant screening
To generate Cas9-mediated heritable rex-47 deletion, DNA microinjection was performed according to standard protocols. The Cas9 expression vector, sgRNA expression vector, repair template and two co-injection markers: pCFJ90 (Pmyo-2::mCherry) and pCFJ104 (Pmyo-3::mCherry) were mixed and injected into the germline of 34 N2 young adults at the following concentrations: Cas9 (50 ng/μl), sgRNA (200 ng/μl), repair template (50 ng/μl), pCFJ90 (2.5 ng/μl) and pCFJ104 (5 ng/μl). Three days post-injection, 269 F1s expressing both Pmyo-2::mCherry and Pmyo-3::mCherry markers were cloned into liquid culture in 96-well plates and propagated at 20°C as described previously35. Worms from each well were lysed and PCR amplified using primers CCGAAACACAACAACAATGC and TGGTAGCCGTATGCACAGTT. We identified 8 deletion mutants from the 269 F1s (3%) based on the size of PCR products. These deletions were further verified by NcoI digestion of the PCR fragments. The progeny of the F1s carrying the rex-47 deletion alleles were then cloned into a new set of wells for the identification of homozygote mutants. PCR products from the homozygote mutants were sequenced to verify the precision of the deletions.
ChIP-qPCR
Wild-type and rex-47 deletion embryos were obtained as described earlier. Input and ChIP samples using rabbit anti-DPY-27 or rabbit anti-SDC-3 antibody were prepared according to previously published protocols20. Three pairs of qPCR primers (ACTTTGCAAGAGTATGTAGTGAA/ACGAGTAATACTTTGAGCATACTT, TACGGCTACCAATCTTGTAA/TCTGTATCTCTAATCCCTAATAGT and TGTGACTACTTGCCCAATAAA/TATCTCTCCCTTCGCCTAAA) were used to amplify three ~100 bp regions located upstream, downstream or within the rex-47 deletion region, respectively. qPCR was performed using iQ SYBR Green Supermix (Bio-Rad,170-8880) on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad).
FISH analysis of rex-47 deletion strain
The legend for Fig. 3h provides the quantification for three-way comparisons of FISH probe colocalization among wild-type, DC mutant, and rex-47 deletion strains. For two-way comparisons using the one-tailed Mann Whitney U test, the rex-47 deletion strain differed significantly from the wild-type strain (P < 10−5) for probes on each side of the TAD boundary, and the rex-47 deletion strain was not statistically different from the DC mutant strain (P = ns), as expected.
RNA-Seq library creation
Embryos of appropriate genotype, four total wild-type biological replicates (two from the Hi-C biological replicates) and three total sdc-2 (y93, RNAi) biological replicates (two from the Hi-C biological replicates), were isolated following the procedures above and frozen at −80°C in 1X M9 buffer. RNA was extracted using a protocol described previously36, except that 10 μl of a 20 mg/ml glycogen solution was used as a carrier. Libraries were prepared from 10 μg of total RNA. PolyA RNA was purified using the Dynabeads mRNA purification kit (Ambion) and fragmented using Fragmentation Reagent (Ambion). First strand cDNA was synthesized from polyA RNA using the SuperScript III Reverse Transcriptase Kit with random primers (Life Technologies). Second strand cDNA synthesis was performed using Second Strand Synthesis buffer, DNA Pol I, and RNase H (Life Technologies). cDNA libraries were prepared for sequencing using the mRNA TruSeq protocol (Illumina).
Gene expression analysis
Libraries were sequenced with Illumina’s HiSeq2000 platform. Reads were required to have passed the CASAVA 1.8 quality filtering to be considered further. To remove and trim reads containing the sequencing barcodes, we used cutadapt version 0.9.5 (http://code.google.com/p/cutadapt/). Reads were aligned to the transcriptome using GSNAP37 version 2012-01-11. Uniquely mapping reads were assigned to genes using HTSeq version 0.5.4p3 using the union mode. Gene expression levels and changes in gene expression were determined by analysis with DESeq38. Gene expression analysis were conducted both with these RNA-seq data sets and published GRO-seq data sets20. For each chromosome, scatter plots analyzed the log2 of the median fold-change in gene expression (DC-mutant expression/wild-type expression) calculated for each 10 kb bin along the chromosome versus the change in insulation score for that bin in wild-type versus DC mutant embryos. No significant correlation was found between the change in gene expression and the change in insulation score: for chromosomes I, II, and X, R = 0.04; for chromosome III and IV, R = 0.00; for chromosome V, R = 0.03.
Extended Data
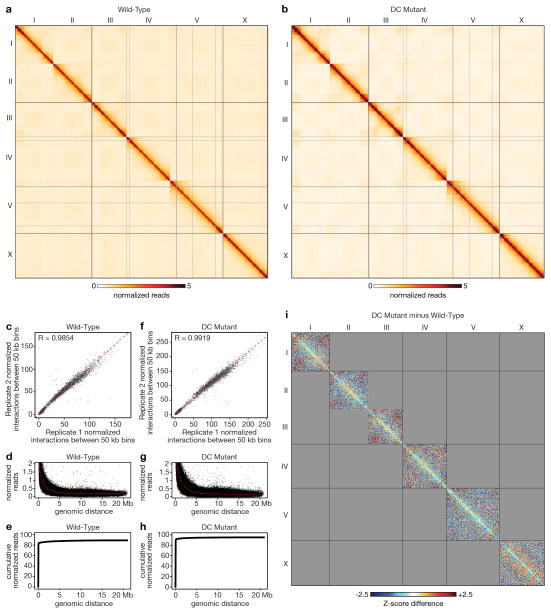
Extended Data Figure 1. Genome-wide chromatin interaction maps for wild-type or DC mutant embryos and genome-wide difference chromatin interaction map.
a, b Genome-wide chromatin interaction maps for wild-type embryos (a) and DC mutant embryos (b) from Hi-C data of combined replicates binned at 50 kb and corrected with ICE. c, f Scatter plots comparing normalized interactions between pairs of 50 kb bins in the two replicates from wild-type embryos (c) or DC mutant embryos (f) (both excluding x = y diagonal). A strong correlation between biological replicates is shown for wild-type embryos (Pearson’s correlation coefficient = 0.9854) and for DC mutant embryos (Pearson’s correlation coefficient = 0.9919). d, g Overall interaction frequency decays with increasing genomic distance in wild-type embryos (d) and in DC mutant embryos (g). e, h Cumulative reads versus linear genomic distance in wild-type embryos (e) and in DC mutant embryos (h). i, Genome-wide difference chromatin interaction map. Shown is the 50 kb binned heatmap depicting the Z-score difference between wild-type and DC mutant embryos (see Methods for Z-score difference calculation). The most apparent differences are on the X chromosome: blue signal within TADs (loss of intra-TAD interactions) and red signal between TADs (gain of inter-TAD interactions).
Extended Data Figure 2. Insulation profile calculation parameters and boundary calling.
a, Cartoon shows approach for calculating the insulation profile. A square is slid along each diagonal bin of the interaction matrix to aggregate the amount of interactions that occur across each bin (up to a specified distance up- and downstream of the bin). Bins with a high insulation effect (e.g. at a TAD boundary) have a low insulation score (as measured by the insulation square). Bins with low insulation or boundary activity (e.g. in the middle of a TAD) have a high insulation score. Minima along the insulation profile are potential TAD boundaries. b, c, Heatmaps of X and I represent the insulation profiles calculated using insulation square sizes ranging from 10 kb to 1 Mb. At the 100 kb scale, weak boundaries are observed on X and autosomes, but they are generally not changed in DC mutants. These boundaries cannot be detected at larger scales, meaning they do not insulate over distances beyond ~100 kb (see e). These smaller scale structures may represent sub-TAD domains not correlated with dosage compensation. Boundaries called using a 500 kb insulation square represent TAD boundaries that define domains observed in chromosome-wide interaction maps of X at 10 kb resolution. These boundaries are used in this paper (Fig. 1) and insulate over the larger distances defining the Mb-sized TADs. Boundaries on X are the strongest and are DC dependent. d, e, f, Pile up plots depict aggregate (mean) Hi-C 10 kb Z-score data centered on specified ‘anchors’ (e.g. rex sites, boundaries, changed boundaries). d, Pile up plots centered on all rex sites or top 25 rex sites in wild-type and DC mutant. e, Pile up plots centered on all boundaries called using insulation squares of 100 kb (left) or 500 kb (right) for X and I in wild-type and DC mutant. f, Pile up plots using boundaries called with a 500 kb insulation square, centered (left) on the single 10 kb bin at the midpoint of all 8 changed boundaries or (right) on all seven 10 kb bins within changed boundaries.
Extended Data Figure 3. TAD boundary analysis.
a, Insulation/Delta plot of the 10 kb binned wild-type combined replicate chromosome X Hi-C data calculated using a 500 kb insulation square size. The insulation profile is depicted in black. In red, the ‘delta’ vector is depicted. It is derived from the insulation vector using a 200 kb delta window (see insulation methods). The ‘delta’ vector is used to facilitate the detection of the valleys/minima along the insulation profile. b, Cartoon example showing how the delta vector is calculated from the insulation data vector. For each bin (reference point) the average insulation differences are calculated between all points up to 100 kb left of the reference point relative to the reference point. The same is repeated for all points up to 100 kb right of the reference point. The delta value is then defined as the difference between the mean (left difference) and mean (right difference). c, Bar plot shows the distribution of distances between boundary calls obtained with biological replicate Hi-C data across all chromosomes. Dotted vertical line indicates that +/− 30 kb was chosen for boundary definition, as it was the window in which the majority of replicate boundary calls (> 80%) overlap. d, Boxplots compare boundary strength (left) and spacing (right) in wild-type vs. DC mutant embryos. Wild-type boundary strength on X (defined as the distance from the insulation minimum to the largest neighboring maximum in the insulation profile) is higher than the DC mutant chromosome X boundary strength (P = 0.024) and higher than the boundary strength on wild-type autosomes (P = 0.03). TAD boundary strength on autosomes does not change in the DC mutant compared to wild-type (P = 0.979). Boundaries on X have less variance in spacing (interquartile range (IQR) = 253 kb) compared to the DC mutant (IQR = 525 kb) embryos. DC mutant X boundary spacing is more similar to the boundary spacing on the autosomes in wild-type (IQR = 625 kb) and DC mutant embryos (IQR = 550 kb).
Extended Data Figure 4. Compartment and insulation analysis for chromosome I in wild-type and DC mutant embryos. (see next page.).
a, ICE corrected chromatin interaction maps are shown for wild-type and DC mutant embryos for both 10 kb binned and 50 kb binned data across replicate 1, replicate 2, and the combined replicates. b, Insulation profiles are shown for each biological replicate (replicate 1, orange line; replicate 2, blue line) for 50 kb and 10 kb binned data in wild-type and DC mutant embryos. Insulation profiles are calculated using a 500 kb × 500 kb insulation square (10 bins × 10 bins for the 50 kb binned Hi-C data, and 50 bins × 50 bins for the 10 kb binned Hi-C data). The insulation profiles are consistent across replicates. Green tick marks, TAD boundaries identified using combined replicate data. c, Differential insulation plots derived from the insulation profiles calculated above (50 kb binned and 10 kb binned Hi-C data). d, 50 kb binned heatmap depicting the difference in chromatin interactions expressed as the difference in Z-scores between wild-type and DC mutant. e, Plot showing the compartment analysis calculated using the 50 kb binned wild-type Hi-C data. A/B compartment profile was determined by principle component analysis. First Eigen Vector value representing compartments (black) is plotted along the chromosome, revealing three zones for each autosome: two outer sections and the middle third of the chromosome. Positive Eigen1 signals represent the B (inactive compartment) and negative Eigen1 signals represent the A (active compartment). The compartments at chromosome ends display increased interactions with each other, both in cis and in trans (see Extended Data Fig. 1a). Also shown is the average binding of the lamin-associated protein LEM-2 along the chromosomes (grey). Overall compartmentalization correlates with LEM-2 binding, showing that compartments at both ends of chromosome I are located near the nuclear periphery.
Extended Data Figure 5. Compartment and insulation analysis for chromosome X in wild-type and DC mutant embryos.
a–e, See legend to Extended Data Fig. 4. In e, only two compartments are observed for X, compared to three for chromosome I. Overall compartmentalization correlates with LEM-2 binding, showing that the compartment at the left end of X is located near the nuclear periphery.
Extended Data Figure 6. Compartment and insulation analysis for chromosomes II, III, IV, and V in wild-type and DC mutant embryos.
a–d, Chromosome II. e–h, Chromosome III. i–l, Chromosome IV. m–p, Chromosome V. a, e, i, m, Insulation profiles for each biological replicate (replicate 1, orange line; replicate 2, blue line) for 50 kb or 10 kb binned Hi-C data in wild-type and DC mutant embryos. Green lines, TAD boundaries identified from combined replicate data. b, f, j, n, Differential insulation plots made from insulation profiles (50 kb binned or 10 kb binned Hi-C data). c, g, k, o, Plots show chromosome compartment analysis calculated with 50 kb binned data. Average binding of the lamin-associated protein LEM-2 is shown along the chromosomes (grey). Compartmentalization correlates with LEM-2 binding; compartments at both ends of autosomes are near the nuclear periphery. d, h, l, p, Heatmaps (50 kb bins) show differences in chromatin interactions as the differences in Z-scores (DC mutant minus wild-type).
Extended Data Figure 7. rex sites are enriched at TAD boundaries and in top Hi-C interactions.
a, Tick plots rank the interaction Z-scores for the top 25 highest-affinity rex sites (black) among all other 10 kb bin Hi-C interactions on X (light blue). Bottom plot amplifies top 2000 interactions. Density of black ticks (left) shows strong enrichment of rex-rex interactions among the most significant X interactions. b, Tick plots rank the Z-score differences (DC mutant minus wild-type) for interactions between the top 25 rex sites among all other differences on X. Bottom plot amplifies top 2000 changes. c, Quantification of Z-score differences for top 2000 changes in (b). d, Bar graphs depict overlap between X TAD boundaries and rex sites. Three sets of TAD boundaries are shown: all 17 boundaries; 8 boundaries with an insulation change (DC mutant minus wild-type) > 0.1; 5 boundaries present in wild-type embryos but absent in DC mutants. Overlap is calculated for the entire set of rex sites or just the top 25 rex sites. (left), Percent of boundaries that overlap rex sites. (right), Percent of rex sites that overlap each set of boundaries. Red bars, same sets of overlaps were calculated with 1000 random sets of rex-site positions along X. Average overlap and standard deviation are shown. No randomized set had as much overlap as the true rex set (P < 0.001). e, Cumulative comparison of Z-score differences for rex interactions and for 1000 randomized sets of non-rex interactions (same number as in rex set). These rex or non-rex interactions had Z-scores > 4 in wild-type embryos. rex interactions are reduced more in DC mutants than other similarly strong X interactions (P = 0.037; rex-interaction differences were significantly more reduced [KS test] than random interaction sets for 963 of 1000 cases). f, 3D plots of Hi-C interaction profiles (normalized read counts) around top 25 rex sites for 2 Hi-C replicates of wild-type or DC mutants. g, 3D plots of interactions between dox sites in wild-type and DC mutants show no interaction peak. h, Cumulative plots show no difference in DC mutants for the distribution of autosomal Hi-C interaction Z-scores (10 kb bins) in TADs or at boundaries.
Extended Data Figure 8. Visualization and disruption of TAD boundaries.
a–d, Visualization of DCC-dependent TAD boundaries in single cells confirms Hi-C analysis. a, Representative confocal images of embryonic nuclei of different genotypes stained with a DNA intercalating dye (blue) and FISH probes surrounding rex-32. Scale bar, 1 μm. b, Quantification of colocalization between FISH probes flanking rex-8 (see Fig. 2a) in XX and XO embryos confirms the DCC-dependent boundary identified by Hi-C. Because TADs on either side of rex-8 are small, we could only use one 500 kb FISH probe for each TAD. c, Quantification of colocalization between FISH probes for a TAD boundary on chromosome I (dashed line in d) in XX and XO embryos confirms the DCC-independent boundary identified by Hi-C. b–c, Box plots show the distribution of Pearson’s correlation coefficients between pairwise combinations of FISH probes. Boxes represent the middle 50% of coefficients, and the central bar within indicates the median coefficients (M). N, total number of nuclei. P-values derived using the one-tailed Mann-Whitney U Test are shown below each graph. ns, not significant. d, Insulation difference plot of chromosome I for DC mutant insulation profile minus wild-type insulation profile. e–g, Deletion of endogenous rex-47 by Cas9 disrupts DCC binding and TAD boundary formation. e, Schematic illustration of the sgRNA-Cas9 complex interacting with the rex-47 target sequence. f, Cas9-mediated deletion of rex-47. Top, diagram showing the location of DCC binding motifs within rex-47 (red bars) and Cas9-induced double strand break (arrow). Middle, diagram of the double-stranded repair template containing two ~500 bp homology arms and an NcoI restriction site. Bottom, after precise homology-directed repair, a 419 bp region containing all DCC binding motifs was deleted and replaced with NcoI. g, Loss of DCC binding at endogenous locus carrying the rex-47 deletion. DCC binding at three ~100 bp regions located upstream (a), within (b) or downstream (c) of the 419 bp deletion was examined using ChIP-qPCR. Histogram shows the ChIP-qPCR signal for DCC components DPY-27 or SDC-3 at target regions relative to the level at region b in wild-type embryos.
Extended Data Figure 9. Quantitative FISH shows that rex sites colocalize more frequently if the DCC is bound to X.
a–f, Data from histograms in Fig. 4b–g shown as cumulative plots. Number of nuclei and embryos (parentheses) assayed are shown (also for i–m). Distance between loci (red) and DCC dependence or independence of Hi-C interactions (black) are shown. P-values (chi-squared test) compare values in the 0–300 nm bin to those in 301–2700 nm bins. Same statistical analysis for (i–m). g, Correlation between DCC-dependent Hi-C interactions and DCC-dependent FISH colocalization. Y-axis, difference between wild-type and DC mutant Hi-C observed interaction frequency at 50 kb resolution. Higher number shows greater DCC-dependence. X-axis shows two categories defined by FISH: sites with unchanged colocalization frequency in DC mutant (DCC-independent) (left); sites with less frequent colocalization in a DC mutant (DCC-dependent) (right). Red dotted line, cutoff for calling a Hi-C interaction “changed” between wild-type and DC mutant. h, Scatter plot shows correlation between Hi-C and FISH data. Y-axis, Hi-C observed interaction frequency in 50 kb bins. X-axis, % colocalization (i.e. 300 nm bin) by FISH. R = 0.77 for all comparisons; R = 0.9 if the rex-47-rex-8 interaction is omitted. i–m, Histograms show quantification of 3D distances between two FISH probes. i, j, Distant loci on X or I with weak Hi-C interactions. k, DCC-dependent interaction between X sites lacking DCC binding. l–m, DCC-dependent interactions between distant rex sites.
Extended Data Figure 10. DCC-dependent TADs influence global rather than local gene expression.
Gene expression analysis was assayed using RNA-seq or GRO-seq, as indicated. a–b, Boxplots depict expression levels for wild-type or DC mutant embryos assayed by RNA-seq for X genes at changed TAD boundaries, unchanged TAD boundaries, all TAD boundaries or genes not at TAD boundaries. Expression levels are given as normalized read number per kilobase of gene length. c, Boxplots depict the fold change in expression assayed by RNA-seq between wild-type and DC mutant embryos for genes at changed TAD boundaries, unchanged TAD boundaries, all TAD boundaries or genes not at boundaries. The lowest-expressing genes (bottom 10%) were removed from analysis. d–f, As in a–c, but assayed by GRO-seq with gene expression levels given as fragments per kilobase of transcript per million mapped reads (FPKM). For a–f, P-values were calculated using the Mann-Whitney U Test; significance did not withstand multiple testing correction. g–h, Boxplots depict the fold change in the gene expression between wild-type and DC mutant embryos based on RNA-seq or GRO-seq for X and I. Each box has genes from one TAD on X (left) or I (right). Lowest-expressing genes (bottom 10%) were removed from analysis. No discernible pattern was evident for expression changes versus gene location. i, Boxplots depict the fold change in X gene expression between wild-type and DC mutant embryos relative to the distance from the TAD boundary. Each box contains genes in 10 kb bins radiating out from the center of each TAD boundary. The lowest-expressing genes (bottom 10%) were removed from analysis. No discernible pattern to the gene expression changes exists, as assayed by RNA-seq (left) or GRO-seq (right). Weak significance and lack of concordance between RNA-seq and GRO-seq data suggest no biologically relevant correlation between TAD boundaries and local regulation of gene expression.
Supplementary Material
Acknowledgments
We thank D. Mets for the computer script to analyze FISH data, A. Michel for initial conformation experiments, D. Stalford for figures, Vincent J. Coates Genomics Sequencing Laboratory (NIH S10RR029668), and K. Brejc, T. Cline, and A. Freund for manuscript comments. Research was supported in part by NIGMS grant R01 GM030702 to B.J.M. and NHGRI grant R01 HG003143 to J.D. B.J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
E.C. conducted Hi-C, ChIP-seq, and FISH experiments. Q.B. conducted FISH and rex-47 deletion experiments. R.P.M. conducted statistical and long-range interaction analyses. B.R.L. analysed HiC data and mapped TADs. B.S.W. conducted RNA-seq studies. E.J.R., S.U., and all authors analysed data and edited the manuscript, J.D. guided and performed Hi-C analysis and wrote manuscript sections. B.J.M. guided the study and wrote the manuscript.
Hi-C, ChIP-seq, and RNA-seq data are available at the NCBI Gene Expression Omnibus (GEO) repository, accession number GSE59716.
The authors declare no competing financial interests.
References
- 1.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 3.Jans J, et al. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009;23:602–618. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csankovszki G, et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol. 2009;19:9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 11.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 15.Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawes HE, et al. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- 18.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–618. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen RA, et al. Extreme HOT regions are CpG-dense promoters in C. elegans and humans. Genome Res. 2014;24:1138–1146. doi: 10.1101/gr.161992.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma R, et al. Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev. 2014;28:2591–2596. doi: 10.1101/gad.248864.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seitan VC, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 26.Chuang PT, Albertson DG, Meyer BJ. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 27.van Berkum NL, Dekker J. Determining spatial chromatin organization of large genomic regions using 5C technology. Methods Mol Biol. 2009;567:189–213. doi: 10.1007/978-1-60327-414-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struc Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 32.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Met. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Met. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 35.Lo TW, et al. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 37.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.