Abstract
Stem cell technologies have facilitated the development of human cellular disease models that can be used to study pathogenesis and test therapeutic candidates. These models hold promise for complex neurological diseases such as Alzheimer’s disease (AD) because existing animal models have been unable to fully recapitulate all aspects of pathology. We recently reported the characterization of a novel three-dimensional (3D) culture system that exhibits key events in AD pathogenesis, including extracellular aggregation of β-amyloid and accumulation of hyperphosphorylated tau. Here we provide instructions for the generation and analysis of 3D human neural cell cultures, including the production of genetically modified human neural progenitor cells (hNPCs) with familial AD mutations, the differentiation of the hNPCs in a 3D matrix, and the analysis of AD pathogenesis. The 3D culture generation takes 1–2 days. The aggregation of β-amyloid is observed after 6-weeks of differentiation followed by robust tau pathology after 10–14 weeks.
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of age-related dementia and is characterized by progressive memory loss and cognitive impairment. Familial, early-onset (<60 years), autosomal-dominant forms of AD (FAD) can be caused by mutations in the genes amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2)1. Sporadic AD typically presents at a later onset and is due to multifactorial genetic and environmental risk factors. The two pathological hallmarks of AD are extracellular amyloid plaques, mainly composed of amyloid β (Aβ) peptides derived from APP via serial cleavage by β- and γ-secretase, and intracellular neurofibrillary tangles, composed of filamentous aggregation of hyperphosphorylated tau (p-tau) proteins1. It is hypothesized that the accumulation and aggregation of Aβ causes the formation of neurofibrillary tangles2–5, but this has not been experimentally proven in any animal models. Only the models that overexpress frontotemporal dementia (FTD)-associated P301L MAPT (tau) mutant forms in addition to human APP and/or PSEN1 with FAD mutations develop both plaques and tangles, albeit in a disconnected manner2–4. Fundamental species-specific differences in genome and protein composition between humans and mice, such as the difference in number of tau isoforms, have precluded an accurate recapitulation of AD pathology.
Advances in the field of stem cell generation have further advanced the prospect of in vitro systems that model AD in human neurons, including the generation of induced pluripotent stem cells (iPSCs) from FAD patient fibroblasts5–10. However, it has been challenging to replicate the aged AD brain environment in the presence of high levels of soluble and insoluble toxic Aβ species and thereby to realize full AD pathology11–13. Recently, we reported that genetically engineered human neural stem cells overexpressing FAD genes combined with a three-dimensional (3D) culture condition induced robust AD pathogenesis, including extracellular aggregation of Aβ and accumulation of hyperphosphorylated/aggregated tau as neurofibrillary tangles14. In this article, we describe our protocol for the generation of these 3D human neural culture models of AD, detail their technical background, describe the techniques we applied for their analysis, and discuss their use and application.
Development of 3D human neural cell culture models of AD
We designed our AD model around two central technologies: human neural progenitor cells (hNPCs) that produce high concentrations of pathogenic Aβ species and a Matrigel-based 3D culture system that provides an environment that favors Aβ deposition.
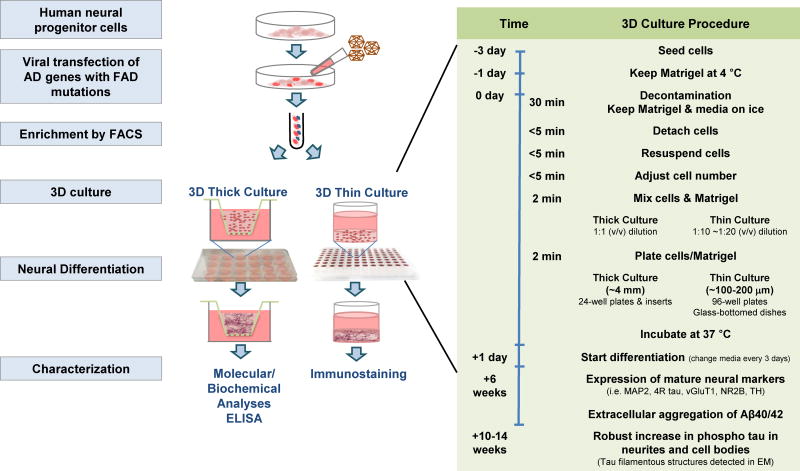
First, we engineered a FAD cell line that could exhibit significant amyloid pathology, be easily maintained, and survive through multiple passages. We chose the immortalized hNPC cell line ReNcell VM (ReN) as a base for our platform because the cells can be maintained for more than 45 passages, are commercially available, and can differentiate into neurons and glial cells with simple growth-factor deprivation15–27. The ReN cells were then transfected with IRES-mediated polycistronic lentiviral vectors containing FAD genes encoding human APP with both K670N/M671L (Swedish) and V717I (London) mutations (APPSL), PSEN1 with ΔE9 mutation (PSEN1(ΔE9)), and APPSL/PSEN1(ΔE9) with GFP or mCherry as a reporter for viral infection (Fig. 1 and 2). Fluorescence-activated cell sorting (FACS) was then employed to enrich the population of cells with the highest expression levels (Fig. 2 and 3).
Figure 1. Polycistronic lentiviral vectors used in this study.
Diagrams showing lentiviral internal ribosome entry sites (IRES) constructs that were designed to express human APP with Swedish/London FAD mutations (APPSL) along with coexpressed green fluorescent protein (GFP), mCherry, PSEN1 with E9 FAD mutation (PSEN1 E9) mCherry tagged, or both APPSL and PSEN1 E9 FAD mutations together with mCherry. CMV, cytomegalovirus promotor.
Figure 2. Overview of ReN VM cell 3D culture protocol.
The experimental procedure begins with the generation of human neural progenitor cell lines virally transfected with APP and/or PSEN1 FAD mutations and enriched based on GFP and/or mCherry signals by fluorescence-activated cell sorting (FACS). The right-hand column shows in detail the timing for each step of the 3D culture method. Day 0 indicates the day on which cells are mixed with Matrigel. MAP2, microtubule associated protein 2; 4R tau, 4-repeat tau isoform; vGluT1, Vesicular glutamate transporter 1; NR2B, N-methyl D- aspartate receptor subtype 2B; TH, Tyrosine Hydroxylase.
Figure 3. FACS (Fluorescence-activated cell sorting) enrichment of ReN-G and -GA cells for higher expressions of APP with FAD mutations.
a. The transfected ReN cells stably expressing GFP alone (ReN-G) were enriched by FACS based on the GFP signal intensity. Rectangular boxes indicate the cell populations that were selected/enriched for the experiments. ReN-G50 or 50, ReN-G cells with the top 50% GFP signal intensity; ReN-G10 or 10, ReN-G cells with the top 10%; ReN-G2 or 2, ReN-G cells with the top 2%; fsc, forward scatter which is roughly size/diameter; ssc, side scatter which is granularity. b. The transfected ReN cells stably expressing APPSL/GFP (ReN-GA) were enriched by FACS based on the GFP signal intensity. ReN-GA50 or 50, ReN-GA cells with the top 50% GFP signal intensity; ReN-GA10 or 10, ReN-GA cells with the top 10%; ReN-GA2 or 2, ReN-GA cells with the top 2%. c. Representative confocal fluorescence images showing ReN-G and ReN-GA cells with the top 2% (Ren-G2 and Ren-GA2), 10% (Ren-GA10) and 50% (ReN-GA50) GFP signal intensity. Green, GFP; Scale bar, 25 μm. d. The confocal fluorescence microscope images of enriched ReN-G2, -GA2 cells that were differentiated by growth-factor deprivation for 4 weeks (Green, GFP; Scale bar, 25 μm).
Second, we differentiated and maintained the FACS-sorted ReN cells expressing high levels of FAD genes in a 3D Matrigel culture system to promote extracellular deposition of Aβ (Fig. 4 and Supplementary Fig. 1). We posited that, in two-dimensional models, secreted Aβ may diffuse into the cell culture medium, disrupting aggregation, whereas the 3D Matrigel may prevent this diffusion of Aβ, allowing for high local concentrations that are sufficient to initiate aggregation. We chose Matrigel specifically as a 3D culture matrix because it can be easily solidified with ReN cells through moderate thermal change and because it provides a brain-like environment rich in structural proteins such as laminin, entactin, collagen, and heparan sulfate proteoglycans28. We confirmed that the 3D Matrigel serves as an excellent 3D seeding structure for Aβ aggregation once the aggregation is initiated although it is still permeable to Aβ species. Moreover, previous studies suggested that 3D conditions that more closely mimic in vivo environments can accelerate neuronal differentiation and neural network formation23,29–34. Indeed, we found that 3D differentiated FAD-gene ReN cells expressed higher levels of specific neural markers and elevated 4-repeat adult tau (4R tau) isoforms versus 2D14.
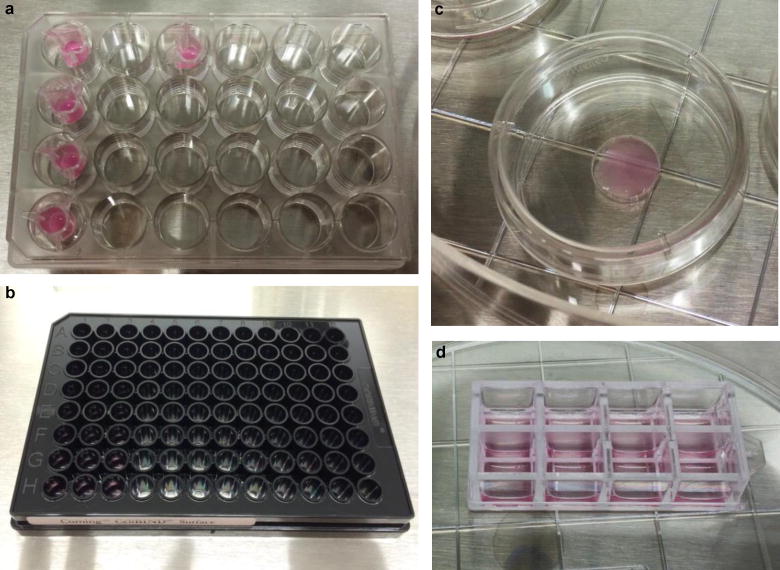
Figure 4. Various 3D culture formats.
a. A 24-well plate with inserts yields the thick-layer cultures ideal for molecular, biochemical, and ELISA analyses. b–d. For thin culture, the 96-well plate (b), 35 mm dishes (c), and 8- or 16-chambered cover glass compartments (d) offer glass-bottom options that facilitate high-resolution immunofluorescence staining.
While the characteristics of the 3D culture system are essential for fully recapitulating AD, the same properties cause several technical difficulties in classical biochemical and imaging analyses. For immunofluorescence (IF) and fluorescence/confocal microscopy, we developed thin layer cultures (~100–300 μm) that could be imaged at high magnification. For biochemical analyses that required higher concentrations of biomolecules, we differentiated cells into thick layer cultures (~4 mm) (Fig. 2). We confirmed that both thin layer and thick layer 3D cultures robustly differentiate into neurons and glia14.
Comparison with other AD iPSC models
Most iPSC-derived human neurons with either APP or PSEN1 FAD mutations exhibit significant increases in Aβ42 levels as compared to the control cells, consistent with previous findings in other model systems5–10. Interestingly, increases in p-tau levels were reported in some of these models. Israel et al. showed a ~2-fold increases of p-tau/total tau ratio in AD neurons6. Recently, Muratore et al. showed that AD neurons harboring APPV717I mutation also showed increases in p-tau and total tau levels and furthermore, demonstrated that abnormal elevation in tau levels could be decreased by treating with anti-Aβ antibodies10. Another recent study determined that differentiated human neurons from Down syndrome (DS) patients generated high levels of Aβ40 and 42 due to an extra copy of the APP gene7. Interestingly, DS neuronal cultures revealed intracellular and extracellular aggregates of Aβ42 as well as increases of p-tau levels, even in 2D culture conditions after extended differentiation periods (>90 days). However, these studies could not recapitulate either robust extracellular aggregation of Aβ or the aggregated p-tau pathology that are evident in our 3D culture study. Limitations of 2D culture protocols and/or low Aβ42 levels may be responsible for the lack of robust AD pathology in the previously described studies using stem cell-derived neuronal cultures.
Applications and limitations of the protocol
As this model is the first human cellular model to comprehensively show Aβ-driven tauopathy this protocol can now be used to examine many other central parts of AD pathogenesis in vitro, including the molecular mechanisms underlying the production of high concentrations of Aβ, the accumulation of extracellular Aβ, the deposition of Aβ aggregates, the hyperphosphorylation of tau, and p-tau aggregation. These paths may lead to new diagnostic and prognostic biomarkers of AD. The model can also be used to test other genetic or environmental factors associated with AD, either in conjunction with the FAD-causing mutations used in this study or in place of them. The flexible scalability of the system also makes it ideal for use in larger scale testing and drug screening. More generally, the following protocols may also be applicable to other neurodegenerative diseases with genetic variations and can be especially suitable for diseases with abnormal aggregation of misfolded proteins.
However, it is important to note that while the 3D-differentiated ReN cells with FAD mutations have reproduced key events in the pathogenic cascade of AD including Aβ deposition and p-tau accumulation/aggregation, they are not surrogate human brain systems. First, our 3D culture model lacks human microglia cells (data not shown) that play a crucial role in Aβ clearance, brain inflammation, and synaptic and neuronal damage. Our 3D differentiation protocol is also not designed to model the specific brain regions that are most affected in AD, i.e. the hippocampus and specific cortices. Indeed, we found that 3D-differentiated ReN cells are composed of a heterogeneous cell population, expressing markers for GABAergic, glutamatergic and dopaminergic neurons as well as astrocytes and oligodendrocytes14. The overexpression of AD genes with multiple FAD mutations may generally restrict the use of our model for the study of the early stages of the disease, in which Aβ gradually accumulates without robust increases in tau phosphorylation. Moreover, our model also characterizes the late stages of the disease, where hyperphosphorylated tau aggregation develops.
Regardless, we believe that the model and techniques described in this protocol offer an exciting model for the AD field, especially given that plaques and tangles formation are the two major hallmarks of this disease. Moreover, the model provides a methodology for the potential development of human neural cell cultures for the study of other neurodegenerative diseases and a robust platform for finding new AD disease targets and therapeutics.
Although the 3D culture described in here is optimized for ReN cells, we have also confirmed that the same 3D culture condition can be applied for differentiating human iPSC-derived neural stem cells including It-NES cells35,36 (data not shown).
Experimental design
Establishment of the ReN cells expressing high levels of Aβ (Step 1–16)
This protocol is optimized for a commercially available human neural progenitor cell line (ReNcell VM, EMD Millipore), which is immortalized, well characterized, and genetically stable after prolonged passaging (>45 passages)15–27. ReN cells were transfected with either CSCW-APP-GFP, CSCW-PSEN1(ΔE9)-mCherry or CSCW-APP-IRES-PSEN1(ΔE9)-IRES-mCherry alone or together (Fig. 1–4). The FAD mutations APP (K670N/M671L (Swedish) and V717I (London)) and PSEN1(ΔE9) were chosen specifically because they cause high production of Aβ42 and Aβ40 and a high ratio of Aβ42/Aβ40. ReN cells with CSCW-IRES-GFP or CSCW-IRES-mCherry alone were also generated as a control. Other FAD mutations may be selected for further characterization or comparison. Other gene editing and overexpression strategies including electroporation, Zinc-finger nucleases (ZFNs), transcription activator like effector nucleases (TALENs), or clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas based RNA-guided DNA endonucleases could also be utilized37–39.
Differentiation of control and FAD ReN cells in 3D Matrigel (Step 17–34)
We describe how to develop thin-layer (~100~300 μm) and thick-layer (~4 mm) 3D cultures by mixing different concentrations of Matrigel with cells. Thin-layer 3D cultures can be used for immunocytochemistry and thick-layer 3D cultures can be used for molecular, biochemical, and ELISA analyses (Fig. 2, 5–9). Also differentiate cells transfected with the GFP or mCherry control lentiviral particles. The presence of Aβ aggregates and p-tau aggregates described herein may depend on the hNPC line, the passage number, the type of FAD mutations, the number of cells seeded, and the proportion of enriched cells by FACS. Generally, we observed extracellular Aβ aggregates starting at 6 weeks of differentiation and robust p-tau accumulation after 10 weeks. We also observed that both Aβ aggregates and p-tau pathology were further elevated through 17 weeks, depending on the cell lines (Fig. 7–9).
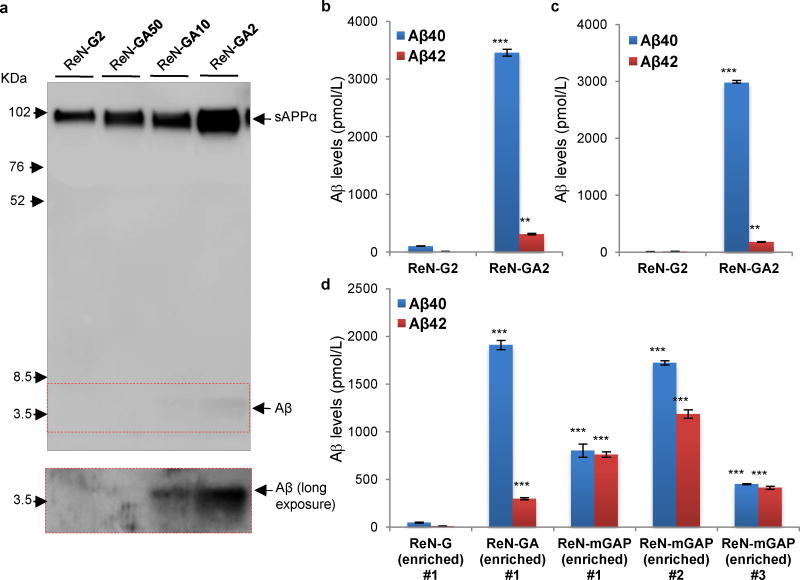
Figure 5. Analysis of soluble Aβ levels in the enriched FAD ReN cells by Western Blot and Aβ ELISA.
a. Western Blot of Aβ levels in the conditioned medium collected from undifferentiated enriched ReN-G and -GA cells. The conditioned medium was collected from T25 flask after culturing the cell for 2 days. 6E10 anti-Aβ detected ~4 kDa Aβ monomer bands in the media collected from the enriched FAD ReN cells (ReN-GA10, ReN-GA2). The 6E10 antibody also detected sAPPα (~100 kDa), which would represent the amount of full-length APPs in these cell lines. b. Increased Aβ40 and 42 levels in undifferentiated FAD ReN-GA2 cells, detected through Aβ ELISA (***, p<0.001; Student t-test; n=3 per each sample). c. Increased Aβ40 and 42 levels were also detected in a conditioned media collected from 1-week old thick-layer 3D cultures of ReN-GA2 cells (***, p<0.001; Student t-test; n=3 per each sample). The soluble Aβ levels cannot be directly compared to the levels shown in (b) since the total cell numbers of thick-layer cultures are higher than 2D undifferentiated cultures used in (b). d. Soluble Aβ levels of the various enriched FAD ReN cells. The FAD cells with PSEN1ΔE9 showed elevated Aβ42/40 ratio as compared to the enriched ReN-GA cells.
Figure 9. Detection of aggregated p-tau in the enriched ReN-G and HReN-mGAP cells.
a. Western blot analyses of p-tau and total tau levels in 1% Sarkosyl-soluble and -insoluble fractions prepared from 3D cultures of 8-week 3D-differentiated ReN-mGAP (enriched) cells and control ReN-G cells. ReN-mGAP (enriched) cell line is one of the FACS-enriched cell lines that stably express both GFP:APPSL and mCherry:PSEN1ΔE9 constructs. b. Dot blot analyses of p-tau and total tau in Sarkosyl-insoluble fractions prepared from ReN-G, ReN-mGAP and ReN-mGAP (enriched) cells (8-week 3D differentiated).
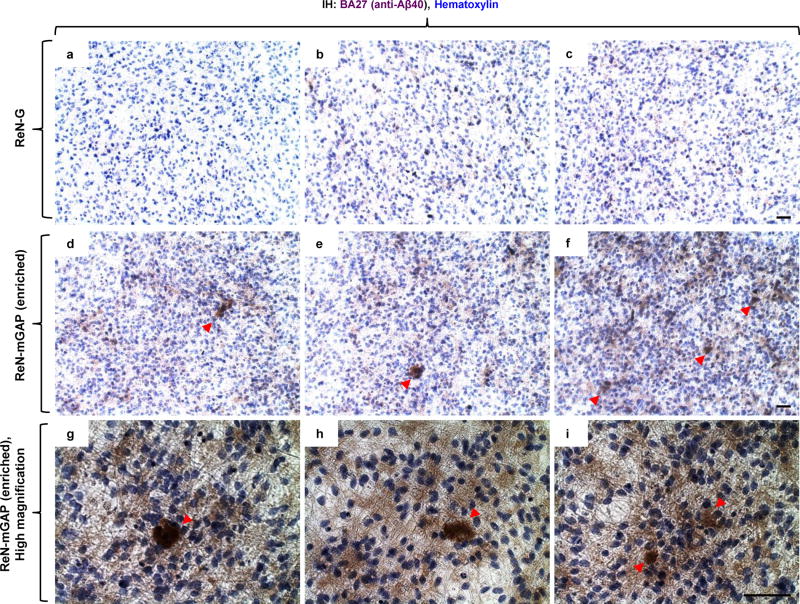
Figure 7. Reconstitution of amyloid-β aggregates in 3D-cultured FAD ReN cells.
a–f. IHC of amyloid-β deposition, detected by anti-Aβ40 BA27 antibodies, in control (ReN-G cells, a–c) and FAD ReN (ReN-mGAP (enriched), d–f) cells. Arrowheads indicate amyloid-β aggregates. g–i. Enlarged images of amyloid-β deposition in ReN-mGAP cell cultures. Scale bars: 50 μm.
Endpoint analysis of Aβ and p-tau pathologies (Step 35–36)
For the thin-layer 3D culture, we describe how to analyze Aβ and abnormal p-tau accumulation using immunofluorescence staining and immunohistochemistry (Fig. 7–8), as previously shown14. Since most of our FAD ReN cells express high levels of GFP and mCherry, the UV-compatible amyloid dye Amylo-Glo can be used for detecting β-amyloid aggregates in the thin layer 3D culture (Supplementary Fig. 2). For thick-layer 3D culture, we describe biochemical extraction to show accumulation of Aβ and p-tau species in both soluble and insoluble fractions. Specifically, for detecting Aβ aggregates, we recommend using TBS/2%SDS/Formic acid serial extraction, while for detecting tau aggregation, we recommend using a 1% Sarkosyl extraction method, as shown in Fig. 9. Accumulation of SDS-resistant Aβ oligomers, including dimers, trimers and tetramers, can also be used as indirect markers for Aβ aggregation. To monitor the proper 3D differentiation, analyze expression of adult neural markers, including 4-repeat tau, by using RT-PCR, qRT-PCR14 and immunofluorescence staining (as shown Fig. 6). LDH levels in 3D culture media should also be measured routinely to detect the cell death rate in these cultures.
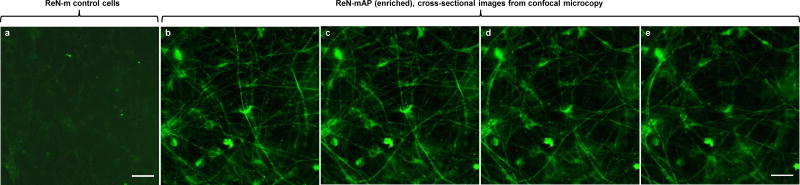
Figure 8. p-Tau accumulation in cell body and neurites of the enriched ReN-mAP cells.
a–e. Immunofluorescence of p-tau, detected by PHF-1 antibody, in the ReN-m control (a) and the enriched ReN-mAP cells (b–e) after 3D differentiation. b–e. Select confocal Z-stack images of p-tau in the soma and neuritis of ReN-mAP cells in 3D cultures. Z-sections were capture in 0.4 μm intervals. The ReN-mAP (enriched) and control ReN-m cells were 3D-differentiated for 8 weeks. Scale bars: 25 μm.
Figure 6. Images of ReN-G2-differentiated neural cell populations in 3D cultures after 2- and 4-week differentiation.
Representative images of 2-week (upper panels) or 4-week (lower panels) differentiated ReN-G2 cells (a and d) that are co-labeled with MAP2 (b and e). The overlay images (c and f) demonstrate that most ReN-G2 cells differentiated in 3D cultures show neuronal properties. Scale bar: 25 μm.
MATERIALS
REAGENTS
Production of FAD cells
Lentiviral particles. See Reagent set up.
KnockOut Serum Replacement (Life Technologies, cat. no. 10828-028)
Propidium Iodide (Life Technologies, cat. no. P1304MP)
3D Cell Culture
ReNcell VM cells (EMD Millipore, SCC008)
DMEM/F12 medium (Gibco/Life Technologies, cat. no. 11320-033)
Heparin (Stemcell Technology, cat. no. 07980)
B27 (Gibco/Life Technologies, cat. no. 17504-044)
bFGF (Stemgent, cat. no. 03-0002)
EGF (Sigma-Aldrich, cat. no. E9644)
Penicillin/Streptomycin/Amphotericin B (Lonza, cat. no. 17-745E)
ReNcell Neural Stem Cell Freezing Medium (EMD Millipore, cat. no. SCM007)
Accutase solution (Life Technologies, cat. no. A11105-01)
Matrigel (Corning, cat. no. 356230; cat. no. 356234 also works)
D-PBS (Lonza, cat. no. 17-512Q)
RNA extraction, RT-PCR and qRT-PCR
RNaseZap® (Life Technologies, cat. no. AM9780)
D-PBS (Lonza, cat no. 17-512Q) ▲CRITICAL We have not tested DEPC-treated D-PBS for this protocol but opened a fresh bottle keeping it aliquoted only for RNA work.
Buffer RLT (Qiagen, cat. no. 79216)
2-Mercaptoethanol (Sigma-Aldrich, cat. no. M3148) ▲CRITICAL It should be dedicated for RNA work only.
HPLC-grade H2O (Fisher Scientific, cat. no. W5-1)
QIAshredder (Qiagen, cat. no. 79656)
RNeasy Mini Kit (Qiagen, cat. no. 74104 or 74106, depending on size)
Ethanol 200 proof (absolute) (Sigma-Aldrich, cat. no. E7023)
Buffer RW1 (Qiagen, cat. no. 1053394)
Buffer RPE (Qiagen, cat. no. 1018013)
RNase-free H2O (Qiagen, cat. no. 129112)
Oligo(dT)20 Primer (Life Technologies, cat. no. 18418020)
10 mM dNTP Mix (Life Technologies, cat. no. 18427013, 18427088, depending on size)
10x RT Buffer (Life Technologies, cat. no. 18067-017)
25 mM MgCl2 (Life Technologies, cat. no. 18080-051)
0.1 M DTT (Life Technologies, cat. no. 18080-051)
RNaseOUT™ (40U/μl) (Life Technologies, cat. no. 10777-019)
SuperScript® III RT (200U/μl) (Life Technologies, cat. no. 18080-093)
RNase-H (Life Technologies, cat. no. 18021-071)
SYBR® Select Master Mix (Life Technologies, cat. no. 4472908)
3R4RTau Forward Primer: 5′-AAGTCGCCGTCTTCCGCCAAG-3′40 (synthesized in the MGH DNA core facility)
3R4RTau Reverse Primer: 5′-GTCCAGGGACCCAATCTTCGA-3′40 (synthesized in the MGH DNA core facility)
Agarose (Apex, cat. no. 20–102)
Quick-Load® 2-Log DNA ladder (0.1–10 kb) (New England Biolabs, cat. no. N0469S)
DNA Loading Dye (6x) (Thermo Scientific, cat. no. R0611)
Ethidium Bromide (Sigma-Aldrich, cat. no. E1510)
3R Tau forward primer (AGGCGGGAAGGTGCAAATAG)41 (synthesized in the MGH DNA core facility)
4R Tau forward primer (GAAGCTGGATCTTAGCAACG)41 (synthesized in the MGH DNA core facility)
3R Tau reverse primer (TCCTGGTTTATGATGGATGTT)41 (synthesized in the MGH DNA core facility)
4R Tau reverse primer (GACGTGTTTGATATTATCCT)41 (synthesized in the MGH DNA core facility)
Immunofluorescence and Immunohistochemistry
Paraformaldehyde (Electron Microscopy Sciences, cat. no. 15710)
Tris (Fischer Scientific, cat. no. BP152)
Tween-20 (Acros Organics, cat. no. 23360051)
Bovine Serum Albumin (Sigma-Aldrich, cat. no. A2153)
Gelatin from Bovine Skin, Type B (Sigma-Aldrich cat. no. G9391)
Glycine (American Bioanalytical, cat. no. AB00730)
Donkey serum (Sigma-Aldrich, cat. no. D9663)
Anti-fade gold (Life Technologies, cat. no. P36930)
H2O2 (Sigma-Aldrich, cat. no. 1001582615)
ImmPRESS Polymer Detection Kit (Vector Laboratories, cat. no. MP-7402 and MP-7401)
DAB Peroxidase (HRP) Substrate Kit, 3,3′-diaminobenzidine (Vector Laboratories, cat. no. SK-4100)
ImmPACT DAB Peroxidase Substrate kit (Vector Laboratories, cat. no. SK-4105)
Amyloid β-Protein Immunohistostain Kit (Wako, cat. No. 299-56701)
Chromopure goat IgG (Jackson Immuno Research, cat. No. 005-000-003)
Amylo-Glo 100x (Biosensis, cat. no. TR-300-AG)
0.9% saline (B. Braun Medical Inc., cat. no. L8001)
1% Triton X-100 (Fischer Scientific, cat no. 11332481001)
A summary of antibodies we used is shown in table 1
Table 1.
Antibodies used for immunofluorescence and immunohistochemistry.
| Useful markers for | Antibody | Host | Supplier | Cat. No. | Dilution (I.F.) | Dilution (I.H.C.) |
|---|---|---|---|---|---|---|
| Neuron (synaptic) | anti-NR2B | Mouse | NeuroMab | N/59/36 | 1:100 | |
| Neuron (synaptic) | anti-GluR2 | Mouse | NeuroMab | L21/32 | 1:100 | |
| Neuron (neuritic) | anti-beta-tubulin III | Rabbit | Abcam | ab24629 | 1:200 | 1:100 |
| Neuron (dendritic) | anti-MAP2 | Rabbit | Millipore | 8717 | 1:400 | |
| Neuron (dendritic) | anti-MAP2 | Rabbit | Cell Signaling Technology | 4542 | 1:200 | |
| Neuron (dendritic) | anti-MAP2 | Chicken | EMD Millipore | AB5543 | 1:500 | |
| Astrocyte | anti-GFAP | Mouse | Antibodies Incorporated | N206A/8 | 1:200 | |
| Astrocyte | anti-GFAP | Rabbit | DAKO | Z0334 | 1:2000 | |
| Neuronal | anti-GABAR2 | Rabbit | Cell Signaling Technology | 4819 | 1:100 | |
| Neuron (dopaminergic) | anti-Tyrosine Hydroxylase | Rabbit | Cell Signaling Technology | 2792 | 1:100 | |
| p-Tau | AT8 | Mouse | Thermo Scientific | MN1020 | 1:30 | 1:20 |
| p-Tau | PHF1 | Mouse | A gift from Dr. Davies | N/A | 1:2000 | 1:1500 |
| Total tau | tau46 | Mouse | Cell Signaling Technology | 4019 | 1:200 | 1:200 |
| Aβ | 3D6 | Mouse | A gift from Lilly | N/A | 1:400 | |
| Aβ 42 | β-Amyloid (1–42 Specific) | Rabbit | Cell Signaling Technology | D9A3A | 1:1600 | |
| Aβ 42 | BC05 | Mouse | Wako Chemicals USA | 299-56701 (A part of Aβ immuno staining kit) | 1:1 | |
| Aβ 40 | BA27 | Mouse | Wako Chemicals USA | 299-56701 (A part of Aβ immuno staining kit) | 1:1 | |
| Cy5 secondary antibody | anti-mouse | Goat | Jackson Immunoresearch | 715-175-150 | 1:300 | |
| AlexaFluor350,405, 488, 568 secondary antibodies | anti- mouse,rabbit, chicken | Goat | Life technologies | A-21049/A-21140/A-31553/A-11001/A-11008/A-11004/A-11011/A-11041 | 1:200-400 |
Biochemical Extraction, Western Blot and Dot Blot
HPLC-grade H2O (Fisher Scientific, cat. no. W5-1)
4x LDS sample loading buffer (Life Technologies, cat. no. NP0007)
Invitrogen NuPage MES SDS Running Buffer (20X) (Life Technologies, cat. no. NP0002-02)
Full range molecular weight Marker (Amersham, cat. no. GERPN800E)
SuperBlock T20 (TBS) Blocking Buffer (Thermo Scientific cat. no. 37536)
50% glutaraldehyde solution (Electron Microscopy Sciences cat. no. 16316)
Supersignal West Dura Chemiluminescence solution (Thermo Scientific cat. no. 34075AB)
Supersignal West Femto Chemiluminescence solution (Thermo Scientific, cat. no. 34095AB)
N-Lauroylsarcosine (Sigma-Aldrich, cat. no. SKU L5777)
NaVO3 (Sigma-Aldrich, cat. no. SKU 72060)
NaF (Sigma-Aldrich, cat. no. 201154)
Protease inhibitor mixture (Roche Life Science, cat. no. 04693159001)
Phosphatase inhibitor cocktail (Thermo Scientific, cat no. 88667)
1,10-o-phenanthroline (PNT, EMD Millipore, cat. no. M1072250010)
Phenylmethylsulfonyl fluoride (PMSF, Sigma-Aldrich, cat. no. P7626)
Formic acid (Sigma-Aldrich, cat. no. 399388)
Immun-Blot® PVDF Membrane (Bio-Rad, cat. no. 162-0177)
Nitrocellulose Membranes (0.20 Micron, 7.9 cm x 10.5 cm) (Thermo Scientific, cat. no. PE1846871)
Bio-Dot SF Filter Paper (Bio-rad, cat. no. 1620161)
A summary of the used antibodies was shown in table 2
Table 2.
Antibodies used for Western Blot analysis.
| Useful markers for | Antibody | Host | Supplier | Cat. no. | Dilution |
|---|---|---|---|---|---|
| p-tau | AT-8 | Mouse | Thermo Scientific | MN1020 | 1:40 |
| p-tau | PHF-1 | Mouse | A gift from Dr. Davis | N/A | 1:1000 |
| Total tau | anti-Tau | Rabbit | DAKO | A0024 | 1:2000 |
| Abeta | 6E10 | Mouse | Covance | SIG-39300 | 1:400 |
| Abeta | MOAB-2 | Mouse | Biosensis | M-1586-100 | 1:100 |
| Neuron (dendritic) | anti-MAP2 | Rabbit | Cell Signaling Technology | 8717 | 1:200 |
| Neuron (dendritic) | anti-MAP2 | Rabbit | EMD Millipore | AB5622 | 1:500 |
| presenilin | anti-Presenilin 1 | Rabbit | Cell Signaling Technology | 5643 | 1:2000 |
| BACE1 | anti-BACE1 | Rabbit | Cell Signaling Technology | 5606 | 1:1000 |
| Amyloid Precursor Protein | anti-C66 APP C-terminal | Rabbit | A gift from Dr. Kovacs | N/A | 1:2000 |
| Neuron | anti-NCAM | Mouse | Cell Signaling Technology | 3576 | 1:1000 |
| Neuron (synaptic) | anti-synapsin I | Rabbit | Cell Signaling Technology | 5297 | 1:500 |
| ER | anti-Calnexin | Rabbit | Cell Signaling Technology | 2679 | 1:1000 |
| Heat shock protein/control | anti-HSP70 | Rabbit | Enzo Life Sciences | ADI-SPA-812 | 1:1000 |
| human Mitochondria | anti-human mitochondrial antigen | Mouse | EMD Millipore | MAB1273 | 1:500 |
Transmission Electron Microscopy (TEM)
Mouse-anti-tau46 (Cell Signaling Technology, cat. no. 4019)
Antibody Diluent (DAKO, cat. no. S3022)
Goat-anti-mouse IgG 10 nm gold (Ted Pella, cat. no. 15751)
Phosphotungstic acid (Electron Microscopy Sciences, cat. no. 19500)
ELISA
Human/Rat Aβ40 ELISA Kit (Wako, cat. no. 294-62501)
Human/Rat Aβ42 ELISA Kit (Wako, cat. no. 290-62601)
Lactate dehydrogenase (LDH) assay
CytoTox-ONE Homogeneous Membrane Integrity Assay Kit (Promega, cat. no. G7890)
96-well white, opaque microwell assay plate (Nunc, cat. no. 236105)
EQUIPMENT
Production of FAD cells
We use a BD SORP FACSAria2 cell sorter (BD Bioscience) in a Bioprotect IV biosafety cabinet (Baker Company): equipped with 5 lasers (350, 405, 488, 561 and 640), located at the Imaging Core Facility of the Ragon Institute of MGH/MIT/Harvard (http://ragoninstitute.org/research/services/flow-cytometry/)
Cell strainer filter (70 μm Nylon, Corning/Fisher Scientific, cat. no. 352350)
3D Cell Culture
Greiner Bio-One 96-well PS uClear black TC plate, flat bottom (Greiner Bio-One, 5665-5090)
MatTek glass bottomed 96-well plates (MatTek, cat. no. P96GC-0-5-F)
Corning® Black/Clear bottom 96-well plates (Corning, cat. no. 3340)
Lab-Tek II 8-well chambered cover glass slides (Thermo Scientific, cat. no. 155409)
Lab-Tek 16-well glass chamber slides (Thermo Scientific, cat. no. 178599)
MatTek glass bottomed 35 mm plates (MatTek, cat. no. P35G-0-7-C or P35G-1.5-10-C)
T25 cell culture flasks (BD Biosciences, cat. no. 353082)
6-well cell culture plates (Fisher Scientific/Corning life sciences, cat. no. 353046)
Cell culture inserts, 0.4 μm pore size (Fisher Scientific/Falcon, cat. no. 353095)
24-well Falcon companion plates for cell culture inserts (Fisher Scientific/Falcon, cat. no. 353504)
Cell culture inserts, 0.4 μm pore size (Greiner Bio-One, cat. no. 662654) ▲CRITICAL Both cell culture inserts (Falcon and Greiner Bio-One) work well for thick-layer culture for biochemical analysis. Greiner Bio-One has a smaller internal diameter as compared to the ones from Falcon, and therefore works better with a small amount of cells to make a proper thickness suitable for thick-layer 3D culture.
0.4 μm disposable cell culture medium filter (Life Technologies, cat. no. 353095)
Sterile 15 and 50 ml conical tube (Fisher Scientific/Falcon, cat. no. 352099 and 352070)
Automated cell counter (Bio-rad, cat. no. TC10)
Cell counter slide (Bio-rad, cat. no. 1450011)
Water Bath (VWR Scientific, cat. no. 1202)
Micro-centrifuge, 4 °C (Beckman Coulter, cat. no. 367160)
Tissue culture Light microscope (Zeiss)
BL-2 biosafety culture hoods
Vortex
Sorvall T6000B Centrifuge (Brochu)
RNA extraction and RT-PCR
Sorvall Legend T+ centrifuge (Thermo Scientific, cat. no. 75004367)
RNaseZap®-rinsed, sterile forceps ▲CRITICAL Forceps should be dedicated for RNA work only and stored in a separate RNA working area
RNaseZap®-rinsed, disposable razor blades
RNase-free filter pipette tips (1, 20, 100, 1,000 μl; USA Scientific, cat. nos. 1121-3810, 1120-1810, 1120-8810 and 1126-7810, respectively)
RNase-free micro-centrifuge tubes (1.5 ml size; USA Scientific, cat. no. 1615-5500)
PTC-200 Thermal Cycler (MJ Research, cat. no. ALD-1244)
iCycler iQ™ (Bio-rad, cat. no. 170-8740)
Bio-rad Tetrad® 2 Thermal Cycler (Bio-rad, cat. no. PTC-0240G with ALS-1296GC)
NanoDrop 1000 Spectrophotometer (Thermo Scientific, cat. no. ND-1000)
Bio-rad ChemiDoc XRS System (Bio-rad, cat. no. 170-8070)
Immunofluorescence and Immunohistochemistry
Olympus IX7 fluorescence microscope equipped with spinning disk confocal unit (Olympus)
LSM Pascal 5 Carl Zeiss confocal microscope (Zeiss)
Biochemical Extraction and Western Blot
Hoefer Semi-Dry Transfer Unit (Hoefer, TE77)
Molecular imager (Bio-rad, VersaDoc MP4000)
Battery-operated spinning homogenizer (MIDSCI, cat. no. A0001)
Ultracentrifuge tubes (Beckman Coulter, cat. no. 343778)
Ultracentrifuge (Beckman, Optima TL Ultracentrifuge)
SpeedVac (Thermo Scientific, cat. no. SPD111V)
Savant RVT4104 refrigerated vapor trap with Savant SC110 SpeedVac (Savant)
TEM
JEOL JEM 1011 transmission electron microscope (JEOL)
AMT digital imaging system (Advanced Microscopy Techniques)
ELISA
ELISA plate reader (Bio-Tek Synergy 2 microplate reader)
LDH
ELISA plate reader (Bio-Tek Synergy 2 microplate reader)
REAGENT SETUP
Lentivirus
We obtained the lentiviral polycistronic vectors (CSCW-IRES-GFP and CSCW-IRES-mCherry) from Massachusetts General Hospital (MGH) viral core42, which are available through the MGH viral core (https://vectorcore.mgh.harvard.edu). CSCW-APP-GFP, CSCW-PSEN1(ΔE9)-mCherry or CSCW-APP-IRES-PSEN1(ΔE9)-IRES-mCherry vectors (Fig. 1) were constructed by inserting full length APP with FAD mutations APP (K670N/M671L (Swedish) and V717I (London)), PSEN1 with a FAD mutation, or both APP and PSEN1 with the FAD mutations 43. We have our lentiviral vectors packaged by our viral core, the by the MGH viral core 42,43. To package lentiviral vectors, first generate lentiviral particles by co-transfecting 293T cells with plasmids, the lentivirus packaging genome CMVRΔ8.91 and envelope coding plasmid (pVSV-G). 48 to 72 h post-transfection, harvest the lentiviral particles, concentrate by ultracentrifugation and titer [transducing units (t.u.)/ml] by real-time PCR using specific primers against WPRE elements as previously described42. ▲CRITICAL The aliquots of packaged viral particles can be stored at −80 °C up to one year without a significant loss of titer.
Human recombinant EGF stock solution (20 μg/ml)
Under a biosafety culture hood, add 2 ml of 0.2 μm-filtered 10 mM acetic acid to 2 mg lyophilized EGF (Sigma) (1 mg/ml), mix, make further dilution with 0.2 μm-filtered 0.1% bovine serum albumin solution to 20 μg/ml final concentration, and store 1 ml aliquots at −80 °C.
Human recombinant bFGF stock solution (25 μg/ml)
Under a biosafety culture hood, add 2 ml of 0.2 μm-filtered 10 mM Tris (pH 7.6) to 50 μg lyophilized bFGF (Stemgent), mix, make 0.2 ml aliquots and store them at −80 °C. ▲CRITICAL To avoid the activity loss by multiple freezing/thawing cycle, bFGF or EGF stocks should be kept at 4 °C once they are thawed. 4 °C stocks should be used within 2–3 weeks. The bFGF and EGF stock aliquots can be stored up to one year at −80 °C.
Cell sorting medium
Add 2% (v/v) of B27 supplement and 2% (v/v) of KnockOut serum replacement to 50 ml of D-PBS. The cell sorting media can be stored at 4 °C for 6 weeks.
Matrigel
Aliquot in 1 ml stocks on ice and keep the stocks at −80 °C. ▲CRITICAL Thaw Matrigel stock overnight at 4 °C before use. Matrigel tends to solidify above 10 °C. The frozen Matrigel stock can be stored at −80 °C up to one year.
Matrigel working solution
Mix 0.5 ml of Matrigel and 50 ml cold DMEM/F12 medium (1:100 dilution) on ice under a biosafety culture hood. ▲CRITICAL Take out a Matrigel stock (1 ml) from −80 °C freezer and place it in 4 °C refrigerator one day before the experiments. Prepare fresh Matrigel working solution.
Matrigel-coated T25 culture flask
Add 3 ml of a cold Matrigel working solution into each T25 flasks, shake gently to cover all the surface area, incubate under 37 °C CO2 incubator at least for 1 h, and remove the remaining solution. Matrigel-coated culture dishes can be stored at 4 °C for up to 2 months.
ReN differentiation medium
To prepare 500 ml of medium, combine 484.5 ml of DMEM/F12 (Gibco/Life Technologies) with 0.5 ml of Heparin (2 mg/ml stock, StemCell Technology), 10 ml of B27 (Life Technologies) and 5 ml of 100x Penicillin/Streptomycin/Amphotericin (Lonza). The cell sorting media can be stored at 4 °C for 2–3 months.
ReN proliferation medium
To prepare 100 ml of medium, combine 100 ml of ReN differentiation medium with 80 μL bFGF stock and 100 μL EGF stock. Filter the medium after adding all the reagents. ▲CRITICAL Medium with growth factors can be kept at 4 °C for several weeks, but use a fresh one if there is any noticeable decrease in cell growth rate.
10x Tris-Borate-EDTA (TBE) electrophoresis Buffer
Add 108 g Tris, 55 g Boric Acid and 7.425 g EDTA into a beaker, fill up to 1 L with distilled-deionized H2O (ddH2O) and mix to dissolve. The 10x TBE buffer is stable at least for 6 months at room temperature.
10x Tris-buffered saline (TBS) buffer
To prepare 100 ml buffer, dissolve 6.05 g Tris and 8.76 g NaCl in 80 ml of H2O. Adjust pH to 7.5 with 1 M HCl and make volume up to 100 ml with H2O. The 10x TBS buffer is stable at least for 6 months at room temperature.
1x TBS/Tween20 (TBST) buffer
To prepare 1 L buffer, add 100 ml 10x TBS buffer and 2 ml of Tween 20 and make volume up to 1 L with H2O. The 1x TBST buffer is stable at least for 3 months at room temperature.
Blocking/dilution solution for immune staining
To prepare 250 ml blocking/dilution solution, add 2.5 g bovine serum albumin (Sigma-Aldrich), 5.63g glycine, 0.25 g gelatin in 200 ml TBST, heat at 55 °C for ~10 min to dissolve gelatin, add 10 ml of donkey serum (Sigma-Aldrich) and add TBST to make the final volume to 250 ml. Filter the blocking/dilution solution with 0.4 μm filter unit (Gibco) and store the solution at 4 °C. The Blocking/dilution solution can be stored at 4 °C for 2–3 months.
0.02x Amylo-Glo working solution
Add 2 μl Amylo-Glo (100x) to 10 ml 0.9% (w/v) NaCl. Prepare fresh every time.
0.1x Amylo-Glo working solution
Add 10 μl Amylo-Glo (100x) to 10 ml 0.9% (w/v) NaCl. Prepare fresh every time.
10xTris/EDTA buffer
To prepare 100 ml buffer, dissolve 6.05 g Tris and 8.76 g NaCl in 80 ml of H2O. Add 4 ml of 0.4 M EDTA (pH 7.4) and adjust pH to 7.6 with 1 M HCl and make volume up to 100 ml with H2O. The 10x Tris/EDTA buffer is stable at least for 6 months at room temperature.
1xTBS extraction buffer
To prepare 10 ml buffer, mix 8.65 ml of distilled water (HPLC grade), 1 ml of 10xTBS buffer (pH 7.4), 1 tablet of protease inhibitor cocktail, 0.1 ml of NaVO3 (1 M), 0.1 ml of NaF (1 M), 0.05 ml of PNT (200 mM), 0.05 ml of PMSF (200 mM) and 0.05 ml of phosphatase inhibitor. ▲CRITICAL Prepare the fresh working solution before use. 1 ml aliquots of the extraction buffer without phosphatase inhibitors can be stored at −80 °C for several months.
2xTBS extraction buffer
To prepare 10 ml buffer, mix 7.5 ml of distilled water (HPLC grade), 2 ml of 10xTBS buffer (pH 7.4), 2 tablets of protease inhibitor cocktail, 0.1 ml of NaVO3 (1 M), 0.1 ml of NaF (1 M), 0.1 ml of PNT (200 mM), 0.1 ml of PMSF (200 mM) and 0.1 ml of phosphatase inhibitor. ▲CRITICAL Prepare the fresh working solution before use. 1 ml aliquots of the extraction buffer without phosphatase inhibitors can be stored at −80 °C for several months.
1xSDS extraction buffer
To prepare 10 ml buffer, mix 1 ml of SDS solution (20% stock), 1 ml of 1% Triton X-100 (10% stock), 4.5 ml of distilled water (HPLC grade), 1 ml of 10xTBS/EDTA buffer (pH 7.4), 2 tablets of protease inhibitor cocktail, 0.1 ml of NaVO3 (1 M), 0.1 ml of NaF (1 M), 0.1 ml of PNT (200 mM), 0.1 ml of PMSF (200 mM) and 0.1 ml of phosphatase inhibitor (100x). ▲CRITICAL Prepare the fresh working solution before use. 1 ml aliquots of the extraction buffer without phosphatase inhibitors can be stored at −80 °C for several months.
2xSDS extraction buffer
To prepare 10 ml buffer, mix 2 ml of SDS solution (20% stock), 2 ml of 1% Triton X-100 (10% stock), 4.5 ml of distilled water (HPLC grade), 1 ml of 10xTBS/EDTA buffer (pH 7.4), 2 tablets of protease inhibitor cocktail, 0.1 ml of NaVO3 (1 M), 0.1 ml of NaF (1 M), 0.1 ml of PNT (200 mM), 0.1 ml of PMSF (200 mM) and 0.1 ml of phosphatase inhibitor (100x). ▲CRITICAL Prepare the fresh working solution before use. 1 ml aliquots of the extraction buffer without phosphatase inhibitors can be stored at −80 °C for several months.
20 % Sarkosyl solution
Add 0.1 g N-laurylsarcosine with total 0.5 ml of distilled water (HPLC grade). The 20 % Sarkosyl solution should be made fresh before the experiments.
2xRIPA extraction buffer
To prepare 10 ml buffer, mix 0.5 ml of sodium deoxylcholate solution (10% stock), 0.2 ml of 2% NP-40 (ICABAL substitute), 7.8 ml of distilled water (HPLC grade), 1 ml of 10xTBS/EDTA buffer (pH 7.4), 02 tablets of protease inhibitor cocktail, 0.1 ml of NaVO3 (1 M), 0.1 ml of NaF (1 M), 0.1 ml of PNT (200 mM), 0.1 ml of PMSF (200 mM) and 0.1 ml of phosphatase inhibitor (100x). ▲CRITICAL Prepare fresh working solution before use. 1 ml aliquots of the extraction buffer without phosphatase inhibitors can be stored at −20 °C for several months.
High salt extraction buffer
To prepare 10 ml buffer, mix 1 ml of 10xTBS, 0.47g NaCl, 0.01 ml EGTA (1 M), 1 g sucrose, 1 tablet of protease inhibitor cocktail, 0.1 ml of NaVO3 (1 M), 0.1 ml of NaF (1 M), 0.1 ml of PNT (200 mM), 0.1 ml of PMSF (200 mM) and 0.1 ml of phosphatase inhibitor (100x). Make 10 ml with distilled water (HPLC grade). ▲CRITICAL Prepare fresh working solution before use. 1 ml aliquots of the extraction buffer without phosphatase inhibitors can be stored at −20 °C for several months.
LDH assay reagent preparation
Using a CytoTox-ONE Assay Kit, add 11 ml of Assay Buffer to each vial of Substrate Mix to prepare CytoTox-ONE Reagent. The CytoTox-One reagent is stable for 6–8 weeks at −20 °C.
EQUIPMENT SETUP
Placing tissue culture inserts on 24-well plates for 3D thick layer culture
Sterilize the forceps under the biosafety hood using ethanol and UV radiations. Place 24-well plates (Falcon), open the lids and carefully place cell culture inserts into each well with the sterilized forceps. ▲CRITICAL Always use the companion plates (Falcon) for setting tissue culture inserts (Falcon).
3D thin-layer culture with chambered coverglass slide plates or glass-bottomed 35 mm dishes
Place the coverglass slide plates (Nunc) or the glass-bottomed 35 mm dishes (MatTek) inside 150 ml culture dishes to reduce the chance of contamination.
RNA extraction from 3D differentiated cells
RNA work should be done in a dedicated area or bench. Keep a separate set of equipment only used in this area. Wipe the work surfaces with RNaseZap® or other RNase removal reagents before RNA extraction. Solutions and reagents should be stored in small aliquots to minimize contaminations with RNases. Gloves should be worn when handling RNA and should be changed frequently. The temperature in the benchtop centrifuge should be set-up to 4 °C or placed in a fridge.
PROCEDURE
ReN cell maintenance and passaging ● TIMING 30 min
-
1|
Warm the ReN proliferation medium and Accutase solution in 37 °C water bath for at least 10 min.
-
2|
Wash ~95% confluent ReN cells in T25 flask with 3 ml of D-PBS, aspirate the D-PBS, and add 0.5 ml of Accutase under a biosafety culture hood.
-
3|
Incubate the cells in 37 °C CO2 incubator for 3–5 min, add 3 ml of pre-warmed ReN proliferation medium and transfer the cell suspensions to 15 ml sterile conical tube.
-
4|
Spin down the cells at 2,000 g (Sorval T6000B) for 3 min, remove the supernatant, and resuspend the cell pellet with 12 ml of ReN proliferation medium.
-
5|
Dispense 4 ml of the ReN cell suspension into each Matrigel-coated T25 flask and incubate in 37 °C CO2 incubator.
▲CRITICAL STEP After subculture (1:3 rate), ReN cells are generally confluent again in 3–4 days. If cells are not confluent at that time, replace the medium with a fresh ReN proliferation medium and wait 1 or 2 days.
? TROUBLESHOOTING
Viral infection of ReNcell VM cells ● TIMING 96 h
-
6|
To prepare ReN cells for infection, dislodge and spin down cells from a confluent T25 flask as described in steps 1–4. Resuspend the cell pellet in 12 ml of pre-warmed ReN cell proliferation medium and dispense 2 ml of the cell suspension into each well of a Matrigel-coated 6-well plate. Gently cross-shake the dish and allow the cells to settle overnight.
-
7|
Replace the medium with 2 ml of pre-warmed ReN proliferation medium. When cells reach 70–80% confluence, add 6x106 TU (Transducing Units) of viral particles per well to achieve an approximate M.O.I. of 1. Mix gently and incubate overnight.
-
8|
Wash twice with 2 ml of pre-warmed ReN proliferation medium, then apply 2 ml of the medium. Confluence should increase over time. Expression of the transgenes can be expected 48 h after infection and should be detectable by fluorescence microscopy.
▲CRITICAL STEP Viral infection renders the cells vulnerable. Carefully monitor the culture. If abnormal cell death is observed, replace the medium immediately.
-
9|
On day 4, if the cells are confluent, passage the cells as described in steps 1–5 and recommence normal culturing. If the cells did not reach complete confluence by this point, replace the medium instead of passaging and incubate until the cells reach confluence. To generate cells expressing both APPSL/GFP and PSEN1(ΔE9)/mCherry (i.e. ReN-mGAP cells14), the transduced cells can be infected again with different lentiviral vectors after splitting, as described in steps 6–8.
-
10|
Validate the infection by microscopic detection of GFP and mCherry fluorescence and western blot analysis of overexpressed APPSL and PSEN1(ΔE9). Typically, more than 50% of infected cells visibly express the fluorescent marker.
▲CRITICAL STEP Western blot analysis of successfully transduced cell extracts will show a PSEN1(ΔE9) band (~40 kDa) and increased bands of full-length APP (695 isoform, ~110 kDa) and its C-terminal fragment (10–12 kDa). Western blot analysis can also be used to detect the elevated sAPPα levels in the supernatant as shown in Fig. 5.
■ PAUSE POINT The infected/validated cells can be frozen and stored at liquid LN2 tank at least for 1 year before FACS analyses.
? TROUBLESHOOTING
Enrich high-expressing transgenic ReN cells ● TIMING 2 h
-
11|
Culture the transduced ReN cells until confluence is reached (a 95% confluent T25 flask yields approximately 2–3 x 106 cells). While the optimal total cell count depends on the infection efficiency, at least 2–3 x 106 successfully transduced cells should be available (when assayed by FACS analysis or fluorescence microscopy).
-
12|
Detach the cells as described in steps 1–4 but resuspend the cell pellets with 4 ml of D-PBS. Determine the cell number using an automated cell counter.
-
13|
Spin down the cells, remove the supernatant and resuspend the cell pellets in ice-cold sorting medium (1 ml/107 cells) using a 1,000 μl pipette.
▲CRITICAL STEP Cells have to be resuspended in at least 200 μl of Cell sorting medium.
-
14|
To singularize the cells, aspirate the cell suspension with a 1,000 μl pipette, gently press the tip against a cell-strainer mesh at a 90° angle and empty the pipette forcefully. In case of blockage, carefully move the pipette tip across the mesh while maintaining pressure. Collect the filtered suspension in a 15 ml centrifugation tube.
■ PAUSE POINT The suspended cells can be stored on ice at least for 1 hour before FACS sorting.
-
15|
Sort cells: We perform FACS cell sorting at the Ragon Institute Imaging Core on a BD SORP FACSAria 2 cell sorter (“Special Order Research Product”) using the 100 μm nozzle and a sheath pressure of 20 psi. BD FACSDiva Software is used to acquire and analyze samples, which are viewed on a dot plot of GFP (488 nm, 200 mW excitation, 530/30 emission) vs. mCherry (561 nm, 150 mW excitation, 610/20 emission) to define sort populations for the final bulk sorting. Gates are set to select cells with high expression of APPSL/GFP alone (as shown in Fig. 5), APPSL/PSEN1(ΔE9)/mCherry alone, or APPSL/GFP and PSEN1(ΔE9)/mCherry together18.
-
16|
Immediately after sorting, plate the cells into Matrigel-coated dishes.
■ PAUSE POINT The suspended cells can be stored in a cell collection tube on ice up to 1 hour before plating.
Culturing sorted ReN cells ● TIMING 30 min
-
17|
Spin the cells down at 2,000 g for 3 min at 4 °C, then remove the supernatant and resuspend the pellet in pre-warmed ReN proliferation medium (1 ml per 2 x 106 collected cells).
▲CRITICAL STEP Determine the cell concentration after resuspension and adjust the volume accordingly. Even counts from sorting can lead to substantial overestimation.
-
18|
Seed the cells on Matrigel-coated 24-well plates at an initial density of 2 x 105 cells/cm2 and culture. Expand serially into Matrigel-coated 6-well plates, and finally into Matrigel-coated T25 flasks. Make multiple cell stocks at this stage. Passage as described in steps 1–5. When sufficient cells have been grown up, proceed to next step.
▲CRITICAL STEP High seeding density after sorting is pivotal to promote cell proliferation. At this stage, it is very important to make multiple frozen cell culture stocks and to record passage numbers. We observed that some high expressing cell lines rapidly lost APPSL and PSEN1(ΔE9) expression after 4 passages.
? TROUBLESHOOTING
3D Matrigel cultures ● TIMING 1 h
-
19|
Take out a Matrigel stock from −80 °C freezer and place it in 4 °C refrigerator 1 day before use.
-
20|
Grow ~95% confluent control and FAD ReN cells in 5 Matrigel-coated T25 flasks. An approximate number of ReN cells in confluent T25 flask is 3–4 x 106. It takes generally 2 days after passaging the cells.
-
21|
Spray down ice bucket and ice with ethanol and leave them in the hood under UV radiation for about 20 min.
-
22|
Place ReN cell differentiation medium and Matrigel stock on ice
▲CRITICAL STEP Matrigel tends to solidify above 10 °C. Thaw it at 4 °C overnight and then keep it in the ice until it is plated with the cells.
-
23|
Remove medium from the T25 flasks with aspirating pipettes. Be careful not to touch the cells with the pipettes; always use the vacuum on the non-cell side.
-
24|
Wash once with 3 ml D-PBS, then carefully remove it with the aspirating pipettes.
-
25|
Add 0.5 ml Accutase into each flask and incubate at 37 °C for 3–5 min.
-
26|
Hit the side of the container 3–5 times to help cells to detach from the coated plates; do not shake since shaking can significantly lower cell count.
-
27|
Resuspend the cells with 3 ml ReN differentiation medium and pipette up and down inside the flask at least 3 times.
-
28|
Transfer the cell suspensions from all 5 T25 flasks into a 15 ml tube.
-
29|
Centrifuge for 2 min at 2,000 g (Sorvall T6000B).
-
30|
Remove the medium with aspirating pipettes.
-
31|
Resuspend the cell pellet in 2 ml cold ReN differentiation medium and vortex for 10 sec. Set the 15 ml tubes on ice.
-
32|
Take a small aliquot of suspended cells and dilute 1:10 for cell count (10 μl suspension: 90 μl differentiation medium).
-
33|
Count the cells using a cell counter slide; dilute cells if needed (optimum concentration is about 2 x 107 cells/ml before adding Matrigel at 1:1 ratio in the next steps).
▲CRITICAL STEP It is desirable to achieve high cell concentration at this stage. We have found that 1 x 107 cells/ml cells (after adding Matrigel at 1:1 ratio; see the step 34) shows robust Aβ and p-tau accumulation based on biochemical analysis.
3D Culture cell seeding
-
34|
Follow option A to make thick-layer 3D cultures or option B for thin-layer 3D cultures (See supplementary Fig. 1 for the pictures of each 3D culture steps).
▲CRITICAL STEP Plating a high number of cells is important in order to achieve Aβ aggregates early. The desired number of cells is 1 x 107 cells/ml in 3D Matrigel mixture. Therefore, enough cells need to be grown at this stage. The passage number and the condition of the cells are also important.
(A) Thick-layer 3D culture (3–4 mm thickness) ● TIMING 2 d
Add cold Matrigel to cell suspension on ice (1:1 (vol/vol) dilution). Make sure to chill the pipette tips first by pipetting cold differentiation medium back and forth before transferring Matrigel.
Vortex for 30 sec.
Dispense 300 μl of the Matrigel/cell suspension mixture into each tissue culture insert in 24-well plates using pre-chilled pipettes.
Incubate the plates at 37 °C in CO2 incubator overnight.
Add pre-warmed (in 37 °C water bath) differentiation medium to the plates (1 ml: 500 μl to the top of the inserts and 500 μl to the bottom) and place them back in the incubator.
-
Change half volume of the medium every 3–4 days; at the beginning, medium changing may be needed every 2 days depending on the medium color. Never leave differentiated cells drying with no medium.
? TROUBLESHOOTING
-
Differentiate 3D-plated cells for 4–17 weeks, depending on the experiments. Drug treatments should be done in the last two weeks before endpoint analyses.
▲CRITICAL STEP Since it is not easy to monitor the thick-layer culture under optical microscope, it is important to perform a routine lactate dehydrogenase (LDH) release assay to check the status of the cultures (see step 36 option N). It is also strongly recommended to set up thin-layer 3D cultures from the same Matrigel/cell mixture to monitor the culture quality.
(B) Thin-layer 3D cultures (100–300 μm thickness) ● TIMING 1 h
-
Add cold Matrigel to cell suspension on ice (1:1 (vol/vol) dilution). Make sure to chill the pipette tips first by pipetting cold differentiation medium back and forth before transferring Matrigel. For 96-well plate thin-layer cultures, further dilute 1:1 Matrigel/cell mixture by adding 5 volume of a cold ReN differentiation medium (1:11 dilution final) and vortex for 30 sec. The same dilution rate can be used for thin-layer 3D cultures in 8-well/16-well chambered cover glass slides or MatTek glass bottomed dishes. We also found that a small volume of 1:1 Matrigel/cell mixture can be directly loaded onto these glass bottomed dishes and these quasi thin-layer cultures can be used for confocal immunofluorescence analysis.
▲CRITICAL STEP Each lot of Matrigel has a different protein concentration and therefore needs to be pre-tested. We generally test 1:10, 1:15, and 1:20 dilutions and pick the best dilution rate for each lot.
? TROUBLESHOOTING
Plate 100 μl of the Matrigel/cell suspension mixture per each well of 96-well plates using pre-chilled pipettes. If a thicker 3D culture is desired, use 2 drops per each well (160 μl using a multichannel pipette). A volume of 200 μl is recommended for 8-well chambered cover glass slides and 300 μl for glass-bottomed dishes.
Incubate the plates at 37 °C in CO2 incubator overnight.
Next day, add 2 drops of pre-warmed ReN differentiation medium to each well of the 96-well plates (160 μl using a multichannel pipette), 200 μl for 8-well chambered cover glass slides and 300 μl for glass-bottomed dishes.
-
Change half volume of the medium every 3–4 days; at the beginning, medium changing may be needed every 2 days depending on the medium color. Never leave differentiated cells drying with no medium. Drug treatments should be done in the last two weeks before endpoint analyses.
▲CRITICAL STEP ReN cell differentiation in thin-layer 3D cultures can be closely monitored by optical and fluorescence microscopy. Some of the cultures can be fixed with 4% paraformaldehyde at 2–4 weeks and tested for neural marker expressions by immunofluorescence staining or immunohistochemistry.
Endpoint analysis
-
35|
The following options describe processing of the 3D culture to produce DNA, RNA, protein samples or fixed cultures, as required for further analysis. To extract RNA from a thick layer culture, follow option A. To extract RNA from a thin layer culture, follow option B. To fix thin layer 3D cultures for immunofluorescence/histochemical/Amylo-Glo staining, follow option C. To quantitate Aβ and p-tau levels using biochemical analyses, initially follow option D to prepare a matrigel cell pellet. Alternatively, proceed directly to step 36 for further analyses of the collected 3D culture media with Aβ ELISA and LDH release assay..
(A) RNA Extraction from thick layer 3D cultures ● TIMING 1–2 h
▲CRITICAL Extraction from one thick layer culture insert usually yields enough RNA for quantitative RT-PCR analyses.
Place the cell culture dish on ice. Using forceps, carefully take out the inserts and place them on top of a RNase-free micro-centrifuge tube.
-
Release the 3D Matrigel culture from the insert by cutting a circle around the outermost edge of the bottom of the insert. Place the insert back on top of the micro-centrifuge tube and centrifuge at 3,200 g (Beckman Coulter cat. no. 367160) for 30 sec at room temperature (~24 °C).
▲CRITICAL STEP Do not cut a full circle around bottom. Leave an overhang of ~1 mm, thereby keeping the membrane of the insert attached to the insert. Be sure that the resulting opening is large enough to release the Matrigel into the micro-centrifuge tube. Be aware that the insert is not placed loosely as it might fall down during centrifugation.
Gently remove the media supernatant by pipetting carefully as to not disrupt or remove parts of the Matrigel.
-
Resuspend the Matrigel with one half volume of cold RNase-free D-PBS.
▲CRITICAL STEP Cut a 1,000 μL RNase-free pipette tip with RNaseZap®-rinsed scissors to obtain an opening of 2mm in diameter.
-
Centrifuge the resuspended 3D culture at 12,900 g (Beckman Coulter cat. no. 367160) for 5 min at 4 °C. Remove the supernatant.
■ PAUSE POINT The resulting pellet can be directly processed or stored at −80 °C for several months.
-
Add 350 μL Buffer RLT to the pellet and mix thoroughly by repeatedly pipetting up and down. Transfer the lysate into a QIAshredder spin column and centrifuge at 12,900 g (Beckman Coulter cat. no. 367160) for 1 min at room temperature.
▲CRITICAL STEP Add 10 μl 2-Mercaptoethanol per ml Buffer RLT before use. Make sure that the Matrigel/cell mixture is thoroughly disrupted and homogenized. The viscosity should be greatly reduced to achieve a homogenous lysate as failure in doing so may result in the clogging of the QIAshredder spin column and reduced RNA concentration and purity. The steps using the RNeasy Mini spin column protocol should be done at room temperature.
-
Perform RNA extraction according to the manufacture’s protocol using the RNeasy Mini Kit. Measuring the RNA concentration using the NanoDrop 1,000 Spectrophotometer.
■ PAUSE POINT The isolated RNA can be directly processed or stored at −80 °C for 6 months.
(B) RNA extraction from thin layer 3D cultures
▲CRITICAL While extraction from one thick layer culture insert usually yields enough RNA for quantitative RT-PCR analyses. If needed, pool together 2–3 wells from thin layer cultures (96-well format).
Place the cell culture dish on ice. Do not remove the media but resuspend the thin layer 3D culture with the cell culture media. Transfer the mixture into an RNase-free micro-centrifuge tube and place on ice.
-
Clean each well by adding 100–400 μl cold RNase-free D-PBS. Repeatedly pipette up and down and transfer the resulting lysate into the same micro-centrifuge tube.
▲CRITICAL STEP Observe the cell culture plate under the tissue culture microscope for any remaining sample and repeat the D-PBS step if necessary. Centrifuge at 12,900 g (Beckman Coulter cat. no. 367160) for 5 min at 4 °C.
-
Gently remove the supernatant by pipetting. Resuspend the pellet in one half volume cold RNase-free D-PBS of the resulting pellet. Centrifuge at 12,900 g (Beckman Coulter cat. no. 367160) for 5 min at 4 °C. Remove the supernatant.
■ PAUSE POINT The resulting pellet can be directly processed or stored at −80 °C for more than several months.
Add 350 μL Buffer RLT to the pellet and mix thoroughly by repeatedly pipetting up and down. Transfer the lysate into a QIAshredder Spin Column and centrifuge at 12,900 g (Beckman Coulter cat. no. 367160) for 1 min at room temperature.
Perform RNA extraction according to the manufacture’s protocol using the RNeasy
-
Mini Kit. Measure the RNA concentration using the NanoDrop 1000 Spectrophotometer.
■ PAUSE POINT The resulting pellet can be directly processed or stored at −80 °C for 6 months.
(C) Fixation of thin-layer 3D cultures
Wash cell layer once with D-PBS.
Add 100 μl of 4% paraformaldehyde solution per each well and incubate at room temperature overnight.
-
Remove the paraformaldehyde solution and wash three times with D-PBS.
■ PAUSE POINT The fixed 3D culture plate can be stored at 4 °C for 6 months. Make sure to tightly wrap the plates to avoid evaporation of the D-PBS in each well.
(D) Preparation of Matrigel/cell pellet from thick layer 3D cultures ● TIMING 1 h
Place the cell culture dish on ice. Using forceps, carefully take out the inserts and place them on top of a micro-centrifuge tube.
-
Release the 3D Matrigel culture from the insert by cutting a circle around at the outermost edge of the bottom of the insert. Place the insert back on top of the micro-centrifuge tube and centrifuge at 3,200 g (Beckman Coulter cat. no. 367160) for 30 sec at room temperature (~24 °C).
▲CRITICAL STEP Do not cut a full circle around bottom. Leave an overhang of ~1 mm, thereby keeping the membrane of the insert attached to the insert. Be sure that the resulting opening is large enough to release the Matrigel into the micro-centrifuge tube. Be aware that the insert is not placed loosely as it might fall down during centrifugation.
Remove the insert and centrifuge again at 8,000 g for 5 min at 4 °C.
-
Retain the pellet (Matrigel/cell pellet).
■ PAUSE POINT The Matrigel/cell pellet can be frozen and stored at −80 °C for several months
-
36|
If you have extracted RNA, proceed if desired to RT-PCR (option A), qRT-PCR (option B) or 3R/4R Tau analysis (option C). If you have fixed sections, proceed to immunofluorescence (option D), immunohistochemistry (option E) or Amylo-Glo staining (option F). For biochemical analyses of Aβ peptide and p-tau, follow options G-K. For EM analysis of Sarkosyl insoluble tau fibrils, first perform Sarkosyl extraction (option I) and proceed to immuno EM (option L). The protocols for Aβ ELISA and LDH release assay from 3D culture media are described in option M and N.
? TROUBLESHOOTING
(A) RT-PCR ● TIMING 8 h
-
Perform cDNA synthesis according to the manufacture’s protocol using the SuperScript®III first-strand synthesis system and the Oligo(dT)20 primer. Use the same total amount of RNA for each sample.
■ PAUSE POINT Synthesized cDNA can be directly processed in downstream application or stored at −20 °C for later use.
-
Perform a PCR amplification step for the relative 3R/4R tau expression analysis in a final volume of 20 μL per reaction. To prepare a Master Mix (1 Reaction), place the following components into an RNase-free micro-centrifuge tube and mix by pipetting. Scale-up the volume of the reaction mixture according to the number of reaction to be performed.
Component Amount Final concentration 3R4R tau forward primer (10 μM) 1 μL 500 nM 3R4R tau reverse primer (10 μM) 1 μL 500 nM SYBR® Select Master Mix (2X) 10 μL 1X ddH2O 7 μL ▲CRITICAL STEP Mix and briefly spin the reaction tubes to ensure components are at the bottom of the tubes without bubbles.
-
Add 1 μL cDNA per reaction. Quickly centrifuge the samples and perform the PCR amplification step under the following conditions using the Bio-rad Tetrad® 2 thermal cycler:
Cycle number Denature Anneal Extend Final 1 95 °C, 15 min 2–31 94 °C, 30 s 60 °C, 30s 74 °C, 90 s 32 74 °C, 10 min 33 4 °C Analyze RT-PCR products by agarose-gel electrophoresis using a 2% w/v agarose gel in 1x TBE buffer. To prepare samples, mix 15 μL of the RT-PCR product with 3 μL DNA loading dye (6x). Load 18 μL into the gel pockets. For size estimation of the products, load 5 μL Quick-Load® 2-Log DNA ladder.
Using a 150 mL agarose gel, run the electrophoresis at 150 V for 50 min to separate the RT-PCR products.
Image RT-PCR product using Biorad ChemiDoc XRS System.
(B) qRT-PCR for neuronal markers ● TIMING 8 h
-
To prepare a Master Mix (1 Reaction), place the following components into an RNase-free micro-centrifuge tube and mix by pipetting. Scale-up the volume of the Master Mix according to the number of reactions to be performed.
Component Amount Final concentration Neural marker primers F+R (5 μM) 0.4 μL 0.1 μM SYBR® Select Master Mix (2x) 10 μL 1X ddH2O 8.96 μL Aliquot 19 μL of Master Mix into each well of a 96-well plate and then add 1 μL of cDNA.
Use β-actin or GADPH as the housekeeping gene (same Master Mix as neural marker primer).
Seal the plate with a film and roll it.
-
Centrifuge down the plate at 2, 000 g (Sorvall Legend T+, Thermo Scientific, cat. no. 75004367) for 2 min.
▲CRITICAL STEP Check if there are bubbles. If present, try to remove them by flicking the tubes and centrifuge again.
-
Perform the PCR amplification step under the following conditions using the Bio-rad®
Table 7.
Cycle number Denature Anneal Extend Final 1 95 °C, 10 min 2–62 95 °C, 10 s 58 °C, 45s 72 °C, 30 s Melting curve 95 °C, 60s 55 °C, 60s followed by 55 + 0.5 °C, 10s each Hold 4 °C Determine the threshold cycles of the each gene using the iCycler IQ software (Bio-rad). Normalize gene expression levels against β-actin levels in each sample and calculate the percentage changes using the 2−ΔΔCT method as previously described44.
(C) qRT-PCR for 3R/4R Tau ● TIMING 8 h
-
To prepare a Master Mix (1 Reaction) for 3R or 4R Tau, add the components in the table into an RNase-free microcentrifuge tube and mix by pipetting. Scale-up the volume of the Master Mixes according to the number of reactions to be performed.
Component Amount Final concentration Tau (3R or 4R) forward primer (10 μM) 1 μL 500 nM Tau (3R or 4R) reverse primer (10 μM) 1 μL 500 nM SYBR® Select Master Mix (2x) 10 μL 1X ddH2O 7 μL Aliquot 19 μL of Master Mix in each well of a 96 well plate and then add 1 μL of cDNA.
Use β-actin or GADPH as the housekeeping gene (same Master Mix as neural marker primer).
Seal the plate with a film and roll it.
-
Centrifuge the plate at 2,000 g (Sorvall Legend T+, Thermo Scientific, cat. no. 75004367) for 2 min.
▲CRITICAL STEP Check if there are bubbles. If present, try to remove them by flicking the tubes and centrifuge again.
-
Perform the PCR amplification step under the following conditions using the Bio-rad® iCycler:
Cycle number Denature Anneal Extend Final 1 95 °C, 10 min 2–62 95 °C, 10 s 58 °C, 45s 72 °C, 30 s Melting curve 95 °C, 60s 55 °C, 60s followed by 55 + 0.5 °C, 10s each 64 65 Hold 4 °C Determine the threshold cycles of the each gene using the iCycler IQ software (Bio-rad). Normalize gene expression levels against β-actin levels in each sample and calculate the percentage changes using the 2−ΔΔCT method as previously described44.
(D) Immunofluorescence staining of thin-layer 3D culture ● TIMING 2 d
? TROUBLESHOOTING
Permeabilize the fixed 3D cultures by incubating with 200 μl of TBST containing 0.5% Triton X-100 and 4% goat IgG at room temperature for 1 h.
Block the culture by incubating with 200 μl of blocking/dilution solution supplemented with 4% goat IgG at 4 °C overnight with gentle rocking.
Wash with TBST once for 10 min.
Incubate with primary antibodies (see Table 1) in blocking/dilution solution with 4% goat IgG at 4 °C overnight with gentle rocking.
-
Wash with TBST 5 times for 10 min per washing.
■ PAUSE POINT This step can be extended by performing one of the 5 washes as an overnight washing at 4 °C.
-
Incubate with AlexaFluor secondary antibodies for 5 h at room temperature with gentle rocking (see Table 1).
▲ CRITICAL STEP The signals from AlexaFluor350 antibodies are relatively weak, especially in 96-well plates due to the UV absorption by plastic-based materials in these plates. Glass-bottomed dishes will perform better for AlexaFluor350 antibody staining. Use of Cy5-labeled antibodies may be another option.
■ PAUSE POINT This step can be extended to overnight at 4 °C.
Add a drop of anti-fade gold on top of the fixed/stained thin-layer 3D culture sections to avoid fluorescence quenching before imaging.
Capture the fluorescence images using an Olympus DSU confocal microscope; perform image analysis and 3D reconstitution. We used ImageJ (NIH), IPlabs (IP Labs) or MetaMorph (Olympus).
(E) Immunohistochemical staining of thin-layer 3D culture ● TIMING 2 d
? TROUBLESHOOTING
Permeabilize the fixed 3D cultures by incubating with 200 μl of TBST containing 0.5% Triton X-100 and 4% goat IgG at room temperature for 1 h.
Block the culture by incubating with 200 μl of blocking/dilution solution supplemented with 4% goat IgG at 4 °C overnight with gentle rocking.
Incubate the cultures with 0.3% (vol/vol) H2O2 solution in TBS for 5 min at room temperature to block endogenous peroxidase activities.
Wash with TBST for 10 min 3 times.
-
Incubate with primary antibodies (see Table 1) in blocking/dilution solution with 4% goat IgG at 4 °C for 1 day with gentle rocking.
▲CRITICAL STEP We found that BA27 (Aβ40) and BC05 (Aβ42) antibodies work well to detect Aβ deposits in 3D culture with minimal background. For BA27, we recommend overnight incubation at 4 °C, while for BC05, a 2 h room temperature incubation, to avoid over-staining.
Wash with TBST for 10 min 5 times.
Incubate with ImmPRESS anti-mouse or -rabbit Ig HRP polymer conjugates (1:1 diluted with blocking/dilution solution supplemented with 4% goat IgG) at room temperature for 30 min.
Wash with TBST for 10 min 5 times.
Develop the antibody signal using DAB peroxidase substrate kits.
(Optional) Counterstain the DAB-stained cultures with Hematoxylin.
(F) Amylo-Glo staining of thin-layer 3D culture ● TIMING 2 d
? TROUBLESHOOTING
Wash the culture wells 3 times with 0.9% (w/v) NaCl solution.
-
Add 100 μl of 0.02x Amylo-Glo working solution and incubate for 5 min at room temperature.
▲ CRITICAL STEP Amylo-Glo over-stains easily. Therefore, we recommend using 0.02x Amylo-Glo working solution to avoid the background. If the signal is too weak, try the 0.1x solution instead.
Remove the staining solution.
Add 200 μl of 0.9% saline and incubate for 5 min.
Wash the wells 3 times with ddH2O.
(Optional) Add 200 μl of propidium iodide nuclear counter staining solution and incubate for 5 min.
Wash 3 times with 0.9% (w/v) NaCl solution.
Add 100 μl of Anti-fade gold on top of the wells.
Image the wells under 4x, 10x, and 40x magnification.
(G) TBS/SDS/Formic acid extraction of thick-layer 3D culture ● TIMING 2 d
Thaw the Matrigel/cell pellet (from step 35, option D) on ice for 10 min.
Add same volume of 2x TBS extraction buffer (~100–200 μl in general).
-
Homogenize using disposable-tip rotor-driven homogenizer 10 times up and down. Gels should become clear/mildly cloudy.
▲ CRITICAL STEP Do not lift tip too far up the tube at risk of losing sample.
Sonicate for 10 min at 4 °C using a sonic cleaning water bath.
Spin down at 100,000 g (Beckman, Optima TL Ultracentrifuge) for 1 h at 4 °C.
Retain the supernatant as TBS-soluble fractions and measure protein levels. Freeze the TBS-soluble fractions at −20 °C.
Homogenize the pellet with 50 μl of 1x SDS extraction buffer using rotor-driven homogenizer and then sonicate for 10 min at 4 °C.
Spin down at 100,000 g (Beckman, Optima TL Ultracentrifuge) for 1 h at 4 °C.
Save the supernatant as TBS-insoluble/2% SDS-soluble fractions and freeze at −20 °C.
Wash the pellet briefly with 100 μl 1x SDS extraction buffer.
Extract the pellet with 10 μl of 70% formic acid on ice.
Centrifuge for 1 h at 100,000 g (Beckman, Optima TL Ultracentrifuge) and collect the supernatant (TBS-insoluble/2% SDS-insoluble/formic-acid-soluble fractions).
(Optional) Evaporate formic acid in SpeedVac to concentrate the formic acid fractions.
Neutralize the formic acid samples by adding 10 volumes of 2M Tris. Mix with a quarter volume of 4x LDS buffer with 8% β-mercaptoethanol.
Run SDS-PAGE of desired fractions as described in option H.
(H) SDS-PAGE and Western Blot Analysis of Aβ ● TIMING 2 d
? TROUBLESHOOTING
Run the extracted samples from option G on 12% NuPAGE gel.
-
Transfer proteins from the SDS-PAGE gels to PVDF membranes.
▲CRITICAL STEP For Aβ detection, incubate the membrane with 0.5% glutaraldehyde solution for 10 min before blocking.
Block the membrane with SuperBlock blocking solution for 1 h at room temperature.
Add primary antibody solution (in 4% BSA) and incubate at 4 °C overnight with rocking (for primary antibody concentration, please see table 2).
Wash with TBST 3 times.
Add secondary antibody solution and incubate for 1 h at room temperature with gentle rocking.
Wash with TBST 3 times.
Develop the blot with SuperSignal Dura or Femto ECL solutions.
(I) Sarkosyl Extraction of thick-layer 3D culture ● TIMING 2 d
Thaw the Matrigel/cell pellet (from step 35, option D) on ice for 10 min.
Add same volume of 2x TBS extraction buffer (~100 μl in general).
Homogenize using disposable-tip rotor-driven homogenizer 10 times up and down.
Sonicate for 10 min at 4 °C using a sonic cleaning water bath.
Mix the TBS-extracted samples with equal volume of 2x RIPA buffer.
Homogenize using rotor-driven homogenizer on ice.
Incubate on ice for 15 min.
Sonicate 2 times for 5 min each.
Centrifuge at 10,000 g with a tabletop centrifuge for 5 min at 4 °C.
Transfer the supernatant into a new tube.
Add 80 μl of 1x RIPA to the pellets in the tubes, resuspend by pipetting and sonication.
Centrifuge at 10,000 g with a tabletop centrifuge for 5 min at 4 °C.
Mix the two supernatant samples from x and xii.
(optional) The RIPA-insoluble pellets can be further extracted with a high-salt extraction buffer. Follow the same protocols (xi–xii) and proceed to xv.
Add 1/20th volume of 20% Sarkosyl solution to the supernatant and incubate at room temperature for 60 min in rotary mixer.
Centrifuge at 150,000 g (Beckman, Optima TL Ultracentrifuge) for 1 h at 4 °C.
Transfer the supernatant into new tubes and measure protein levels (Sarkosyl-soluble fractions).
Wash the pellet briefly with 100 μl 2x RIPA buffer once.
-
Wash the pellet with 0.5 ml PBS 3 times (Sarkosyl-insoluble fractions).
▲ CRITICAL STEP Avoid disrupting the pellets during the PBS washing.
Add 1x LDS sample buffer with 10 M Urea and 2% β-mercaptoethanol.
Heat at 95 °C for 5 min.
Run SDS-PAGE as described in option J, or dot blot as described in option K on a small sample prior to TEM as described in option L.
(J) SDS-PAGE and Western Blot Analysis of p-tau ● TIMING 2 d
? TROUBLESHOOTING
Run on 4–12% gel for p-tau analysis.
Transfer proteins from the SDS-PAGE gels to PVDF membranes.
-
Block the membrane with either 4% BSA or SuperBlock blocking solution for 1 h at room temperature.
▲CRITICAL STEP Do not use skim milk for blocking p-tau blot since it contains phosphatase.
Add primary antibody solution and incubate at 4 °C overnight with rocking (for primary antibody concentration, please see table 2).
Wash with TBST for 10 min 3 times.
Add secondary antibody solution and incubate for 1 h at room temperature with gentle rocking.
Wash with TBST for 10 min 3 times.
Develop the blot with SuperSignal Dura or Femto ECL solutions.
(K) Dot Blot Analysis ● TIMING 2 d
▲CRITICAL Dot blot analysis can be a useful for quickly testing p-tau levels before EM analysis.
▲CRITICAL Users of vacuum-based systems will need to modify these steps.
Wet the nitrocellulose membrane in 1X TBS buffer for 5 min.
Locate the membrane on top of a pre wetted Bio-rad blotting paper.
Load 0.5–1 μl of protein samples.
(optional) A dot blot kit can be also used for loading the samples.
Incubate the membrane for 20 min until the protein samples are completely absolved into the membrane.
Block the membrane with either 4% BSA or SuperBlock blocking solution for 1 h at room temperature.
Add the primary antibody solution and incubate at 4 °C overnight with rocking (for primary antibody concentration, please see table 2).
Wash with TBST for 10 min 3 times.
Add the secondary antibody solution and incubate for 1 h at room temperature with gentle rocking.
Wash with TBST for 10 min 3 times.
Develop the blot with SuperSignal Dura or Femto ECL solutions.
(L) Immunogold staining of Sarkosyl-insoluble tau for Transmission Electron Microscopy (TEM) analysis ● TIMING 3 h
? TROUBLESHOOTING
Resuspend the Sarkosyl-insoluble fractions in 20–40 μl of PBS (from option I). The volume will depend on the concentration of the fibrils in the fraction obtained).
Pipet 7–10 μl of the resuspended samples onto formvar-carbon coated 200 mesh Ni grids held in forceps in a dry and dust-free container.
Allow the sample to adsorb onto grid for 10–20 min. Do not allow grids to dry. Adjust adsorption time for smaller sample volumes.
Blot or wick away sample droplets and place grids directly on top of 15 μl drops of primary antibody (mouse-anti-tau46) diluted 1:25 or 1:50 in antibody diluents. Incubate in a humidified chamber for 1 h.
Rinse grids on drops of 1x PBS for 5 min 3 times.
Blot and place grids on top of 15 μl drops of secondary antibody (goat-anti-mouse IgG 10 nm gold) diluted 1:15 in antibody diluents. Incubate in the humidified chamber for 1 h.
Blot and rinse grids on drops of distilled water for 5 min 3 times.
Blot and place grids on drops of 2% aqueous phosphotungstic acid for 1 min.
Blot and transfer grids to drops of distilled water briefly to rinse.
Blot and allow to air-dry before proceeding with TEM analysis.
We examine grids in a JEOL JEM 1011 transmission electron microscope at 80 kV. We collect digital images using an AMT digital imaging system.
(M) Media harvesting protocol for Aβ peptide quantification
-
Collect culture media either from the inserts of the thick layer cultures or from the wells of the thin layer cultures.
▲CRITICAL STEP If the cell differentiation is on-going, be careful to collect the media in sterile conditions, using sterile tips and pipettes under the hood. If the differentiation time is at the end and the cell cultures are ready to be used for different analyses, place the plates on ice during the collection of the media.
Collect half volume of media for each insert/well in a micro centrifuge tube.
Spin down the media at 4 °C for 5 min at 6,000 g (Beckman Coulter, cat. no. 367160).
Collect supernatant in a new tube.
-
Run 30 μL on SDS-PAGE or load 100 μL in a WAKO Aβ 40/42 ELISA kit 96-well plate.
■ PAUSE POINT The remaining media can be stored at −80 °C for several months.
(N) Lactate dehydrogenase (LDH) assay of thick and thin-layer 3D culture media ● TIMING 2 d
▲CRITICAL In the control ReN cell 3D cultures, we found that levels of LDH release are below 3% of total LDH levels.
Collect culture media from the plastic inserts of thick-layer cultures and/or the wells of thin-layer cultures.
(Optional) Add 100 μl of lysis buffer (provided by Cytotox kit) to a couple of wells and incubate 30 min at 37 °C. This is used to determine total LDH levels.
Transfer 100 μl of each culture media sample to a 96-well white, opaque assay plate.
Add 100 μl CytoTox-ONE reagent to each sample.
Mix by gently pipetting up and down and incubate for 30 min to 1 h at room temperature.
Measure fluorescence signal (Ex 560 nm / Em 590 nm) using a Biotek Synergy 2 microplate reader powered by Gen5 analysis software.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3.
Troubleshooting.
| Steps | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | Cells do not proliferate and begin to differentiate. | Activity of EGF and/or bFGF is lost. | Avoid multiple freezing/thawing cycle for EGF/bFGF stocks. |
| 10 | The transfection rate is low. | Low titer of lentiviral vector, poor cell condition, negative impact on cell proliferation of APPSL or PSEN1(ΔE9). | Increase titer of lentiviral vector. Do multiple transfections with the same lentiviral vector. |
| 18 | FACS-sorted cells are not proliferating or apoptotic. | Expression of FAD mutations obtained with the FACS sorting is too high. | 1) Plate the FACS-sorted cells in small-scale dishes to increase the cell density (the high cell density helps to accelerate ReN cell proliferation); 2) Enrich cells with lower fluorescence signal intensities. |
| 34 (A) vi | The color of the ReN differentiation medium rapidly changes into yellow in 1–2 days. | The cell density is too high. | 1) Change the 3D cell culture medium more frequently in early period (every 2 days); After 2–3 weeks, switch to a normal medium change schedule (every 3–4 days); 2) Reduce the number of cells at step 33. |
| 34 (B) i | ReN cells sink down to the bottom and look like 2D culture. | Matrigel concentration is too low (since each lot of Matrigel has a different protein concentration, the optimal dilution rate should be determined every time). | 1) Use different dilution rate to increase the final Matrigel concentration (Even if the cell body seems to be at the bottom, we observed that the Matrigel still formed 3D gels on top of the cells, making 3D neurite outgrowth and displaying Aβ and tau pathologies. Actually, low Matrigel concentration is desirable for immunohistochemical staining to reduce background). Pre-test 1:10, 1:15 and 1:20 dilutions and pick the best dilution rate for each lot; 2) Prewarm the plates and move them to 37 °C immediately after plating. This will accelerate the gelation of Matrigel before the cells sink down. |
| 36 | Aβ aggregates/p-tau pathologies are delayed or not evident. | Number of cells is low. | The starting number of cells/ml is between 3–10 million/ml, but number of cells desired is 10 million/ml. |
| Aβ42 levels (Aβ42/40 ratio) are not high. | 1) Enrich cells further with higher fluorescence signal intensities; 2) Start 3D culture again using high quality culture of cells with earlier passage number or start again from the viral transfection; 3) We also found that additional transfection with PSEN1(ΔE9) vector could further elevate Aβ levels. | ||
| 36 (D) (E) | Strong backgrounds | Non-specific interaction between the primary/secondary antibodies and the fixed Matrigel matrix. Uneven solidification of the Matrigel in thin-layer 3D cultures. | Increase permeabilization, blocking and washing time. Decrease secondary antibody concentrations. Use lower Matrigel concentrations (1:15 or 20) during thin-layer 3D culture generation. |
| 36 (F) | Strong backgrounds (Amylo-Glo) | 1) The concentration of Amylo-Glo working solution is too high. 2) Presence of autofluorescent aggregates. | Reduce the Amylo-Glo working solution concentration and incubation time. Check the fluorescence background in other flouropore filters including FITC/Rhodamine. |
| 36 (H) (J) | Normalization of total protein levels for Western blot does not fit to control protein levels including GAPDH. | The total protein extracts are contaminated by the solubllized Matrigel components liberated during the extraction. | Use the human specific protein markers such as human-specific mitochondrial antigen (H-mito, see Table 2) to renormalize the samples. Use dot blot analysis. |
| 36 (L) | Poor fibrils imaging | Tau fibril structures are damaged during the extraction. Tau fibrils are not mature. | Reduce the Sarkosyl extraction time at room temperature. Use 3D culture samples differentiated more than 14 weeks. |
| Contamination with other non-specific structures. | Sarkosyl extraction was not enough. | Increase the Sarkosyl extraction time. Re- centrifuge the samples before Sarkosyl extraction. |
ANTICIPATED RESULTS
For reproducible output, we found that it is very important to plate a high number of cells in order to achieve early Aβ aggregation. In addition, the passage number and the condition of the cells are also important. Especially for the highly enriched cell lines expressing APPSL and PSEN1ΔE9, we strongly recommend using only cells with very early passage numbers (up to 4) to ensure that a robust Aβ and tau pathology is consistently observed. FAD ReN cell lines with high Aβ42/40 ratio will show denser amyloid deposits in 3D culture (e.g. ReN-mGAP cells that express both GFP:APPSL and mCherry:PSEN1ΔE9 as shown in Fig. 5d and 7; ReN-mAP cells that express mCherry:APPSL:PSEN1ΔE9 in Fig. 8; HReN-mGAP (enriched) cells that express both GFP and mCherry:APPSL:PSEN1ΔE9 in Fig. 9 and supplementary fig. 2) while the cell lines with mid/low Aβ42/40 ratio will develop mostly diffuse or no obvious deposits despite these cells having secreted high levels of total Aβ (e.g. ReN-GA2 cells in Fig. 5b–d). In general, we observed that the ReN cells secreting high levels of Aβ, preferentially Aβ42, develop more robust p-tau pathology.
To enhance Aβ and p-tau pathologies, we used FACS sorting to enrich ReN cells expressing high levels of transgene. As shown in Fig. 3, FACS sorting of the top 2, 10 or 50% levels of GFP signals (Fig. 3a) lead to the generation of ReN-GA cells (APPSL/GFP) expressing high, mid and low APPSL/GFP (ReN-GA2, -GA10 and -GA50; Fig. 3b). The control ReN-G cells with comparable levels of GFP expression were also prepared (ReN-G2, -G10 and -G50; Fig. 3a–b). As expected, ReN-GA2 and the control ReN-G2 cells differentiated comparably in the thin-layer 3D culture condition into neurons and glial cells (Fig. 6). Western blot analysis (Fig. 5a) and Aβ ELISA (Fig. 5b–d) showed robustly elevated Aβ levels in ReN-GA2 cells as compared to the control ReN-G2 cells. Aβ ELISA also detected an elevated level of Aβ in conditioned medium collected from 1-week differentiated thick-layer 3D cultures of ReN-GA2 cells (Fig. 5c). However, the Aβ42/40 ratio was moderately increased in these cells because they do not express PSEN1ΔE9, which alters γ-secretase structures to produce more Aβ42. As a result, we observed that ReN-mGAP (enriched) cell lines displayed much more robust Aβ deposits as compared to ReN-GA (enriched) cells (Fig. 5d). Similar results were also observed in ReN-mAP (enriched) cell lines (data not shown).
The quality of the ReN cell 3D cultures can be monitored by LDH releases assay, qRT-PCR/RT-PCR analyses of adult neural marker expression and immunofluorescence staining (as shown in Fig. 6). Based on immunofluorescence staining, we estimate that ~68% ReN cells differentiate into MAP2-positive neurons and ~30% into GFAP-positive astrocytes following 2–4 weeks of 3D differentiation. We also found a robust increase in adult neuronal markers, particularly 4-repeat adult tau isoforms in 3D culture conditions, which are essential for replicating tauopathy in human neuronal cultures14.
With the proper cell density and passage number, extracellular β-amyloid deposits start developing after 6 weeks in FAD ReN cell 3D cultures with high levels of Aβ42 (Fig. 7) and the number and size of β-amyloid aggregations increases with longer culture. Amylo-Glo45, a fluorescent amyloid dye can also be used to detect Aβ deposits, particularly after 8-weeks differentiation (Supplementary Fig. 2). Accumulation of p-tau in cell bodies and neurites can be detected by immunostaining with p-tau antibodies, including AT-8 and PHF-1, after 8 weeks (Fig. 8) while robust accumulation of p-tau aggregates are detected after 10–14 weeks14. PHF-like tau filaments can be detected after 14 weeks in Sarkosyl-insoluble fractions of FAD ReN cells with highly robust Aβ levels (preferentially Aβ42) by electron microscopy.
Supplementary Material
Acknowledgments
This work is supported by the grants from the Cure Alzheimer’s fund to D. Y. K. and R. E. T. and National Institute of Health grants 1RF1AG048080-01 (D.Y.K. and R.E.T.), 5P01AG15379 (D.Y.K. and R.E.T.), 2R01AG014713 (D.Y.K.) and 5R37MH060009 (R.E.T.). We appreciate Drs. Bradley T. Hyman, Oksana Berezovska, Dora M. Kovacs (Massachusetts General Hospital, Boston, USA), John Hardy (NIH, Bethesda, MD, USA) and Peter Davies (Albert Einstein College of Medicine, Bronx, USA) for providing cDNAs and antibodies. We also would like to acknowledge Dr. Bakhos A. Tannos at MGH Viral Vector Core (supported by NIH/NINDS P30NS04776), Mr. Michael Waring at Ragon Institute’s Imaging Core facility (part of the Harvard CFAR Immunology Core), Ms. Mary L. McKee and Ms. Diane Capen at MGH Microscopy Core of the Center for Systems Biology for providing the detailed protocols and revised the manuscript.
Footnotes
DECLARATION OF INTEREST
The authors report no competing financial interest.
AUTHOR CONTRIBUTIONS
D.Y.K. and R.E.T. were equally responsible for experimental design and supervising the whole project. Y.H.K., S.H.C., C.D. and D.Y.K., mainly contributed to writing and revising the manuscript. C.D., E.B., K.J.W., M.H. and D.Y.K. performed the experiments. M.H., O.B., J.B.K. and C.S. contribute to writing the manuscript.
References
- 1.Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Götz J, Ittner LM. Animal models of Alzheimer’s disease and frontotemporal dementia. Nat Rev Neurosci. 2008;9:532–544. doi: 10.1038/nrn2420. [DOI] [PubMed] [Google Scholar]
- 3.Chin J. Selecting a mouse model of Alzheimer’s disease. Methods Mol Biol. 2011;670:169–189. doi: 10.1007/978-1-60761-744-0_13. [DOI] [PubMed] [Google Scholar]
- 4.Oddo S, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 5.Yagi T, et al. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 6.Israel MA, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, et al. A human stem cell model of early Alzheimer’s disease pathology in Down syndrome. Sci Transl Med. 2012;4:124ra29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Sproul AA, et al. Characterization and Molecular Profiling of PSEN1 Familial Alzheimer’s Disease iPSC-Derived Neural Progenitors. PLoS ONE. 2014;9:e84547. doi: 10.1371/journal.pone.0084547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muratore CR, et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi SH, Tanzi RE. iPSCs to the rescue in Alzheimer’s research. Cell Stem Cell. 2012;10:235–236. doi: 10.1016/j.stem.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young JE, Goldstein LSB. Alzheimer’s disease in a dish: promises and challenges of human stem cell models. Hum Mol Genet. 2012;21:R82–9. doi: 10.1093/hmg/dds319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi SH, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donato R, et al. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007;8:36. doi: 10.1186/1471-2202-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffrogge R, et al. 2-DE proteome analysis of a proliferating and differentiating human neuronal stem cell line (ReNcell VM) Proteomics. 2006;6:1833–1847. doi: 10.1002/pmic.200500556. [DOI] [PubMed] [Google Scholar]
- 17.Beyer S, et al. Neuroproteomics in stem cell differentiation. Proteomics Clin Appl. 2007;1:1513–1523. doi: 10.1002/prca.200700324. [DOI] [PubMed] [Google Scholar]
- 18.Le MTN, et al. MicroRNA-125b Promotes Neuronal Differentiation in Human Cells by Repressing Multiple Targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan PJ, et al. Protection of neurons derived from human neural progenitor cells by veratridine. Neuroreport. 2009;20:1225–1229. doi: 10.1097/WNR.0b013e32832fbf49. [DOI] [PubMed] [Google Scholar]
- 20.Lange C, et al. Small molecule GSK-3 inhibitors increase neurogenesis of human neural progenitor cells. Neurosci Lett. 2011;488:36–40. doi: 10.1016/j.neulet.2010.10.076. [DOI] [PubMed] [Google Scholar]
- 21.Hernández-Benítez R, Vangipuram SD, Ramos-Mandujano G, Lyman WD, Pasantes-Morales H. Taurine enhances the growth of neural precursors derived from fetal human brain and promotes neuronal specification. Dev Neurosci. 2013;35:40–49. doi: 10.1159/000346900. [DOI] [PubMed] [Google Scholar]
- 22.Mazemondet O, et al. Quantitative and kinetic profile of Wnt/β-catenin signaling components during human neural progenitor cell differentiation. Cell Mol Biol Lett. 2011;16:515–538. doi: 10.2478/s11658-011-0021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liedmann A, Rolfs A, Frech MJ. Cultivation of human neural progenitor cells in a 3-dimensional self-assembling peptide hydrogel. Journal of visualized experiments : JoVE. 2012:e3830. doi: 10.3791/3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovakimyan M, et al. Survival of transplanted human neural stem cell line (ReNcell VM) into the rat brain with and without immunosuppression. Ann Anat. 2012;194:429–435. doi: 10.1016/j.aanat.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Hübner R, et al. Differentiation of human neural progenitor cells regulated by Wnt-3a. Biochem Biophys Res Commun. 2010;400:358–362. doi: 10.1016/j.bbrc.2010.08.066. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger A, Baake J, Weiss DG, Kriehuber R. Glycogen synthase kinase-3beta regulates differentiation-induced apoptosis of human neural progenitor cells. Int J Dev Neurosci. 2013;31:61–68. doi: 10.1016/j.ijdevneu.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhry ZL, Ahmed BY. Caspase-2 and caspase-8 trigger caspase-3 activation following 6-OHDA-induced stress in human dopaminergic neurons differentiated from ReNVM stem cells. Neurol Res. 2013;35:435–440. doi: 10.1179/1743132812Y.0000000135. [DOI] [PubMed] [Google Scholar]
- 28.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 29.Ortinau S, et al. Effect of 3D-scaffold formation on differentiation and survival in human neural progenitor cells. Biomed Eng Online. 2010;9:70. doi: 10.1186/1475-925X-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaPlaca MC, Vernekar VN, Shoemaker JT, Cullen DK. Three-Dimensional Neuronal Cultures. Methods in Bioengineering: 3D Tissue Engineering. 2010:187–204. [Google Scholar]
- 31.Suga H, et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Wijekoon A, Leipzig ND. 3D differentiation of neural stem cells in macroporous photopolymerizable hydrogel scaffolds. PLoS ONE. 2012;7:e48824. doi: 10.1371/journal.pone.0048824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liedmann A, Frech S, Morgan PJ, Rolfs A, Frech MJ. Differentiation of human neural progenitor cells in functionalized hydrogel matrices. Biores Open Access. 2012;1:16–24. doi: 10.1089/biores.2012.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang-Schomer MD, et al. Bioengineered functional brain-like cortical tissue. Proc Natl Acad Sci USA. 2014;111:13811–13816. doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falk A, et al. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS ONE. 2012;7:e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, et al. A 3D Alzheimer’s disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials. 2014;35:1420–1428. doi: 10.1016/j.biomaterials.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 37.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 38.Mussolino C, Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr Opin Biotechnol. 2012;23:644–650. doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iovino M, Patani R, Watts C, Chandran S, Spillantini MG. Human stem cell-derived neurons: a system to study human tau function and dysfunction. PLoS ONE. 2010;5:e13947. doi: 10.1371/journal.pone.0013947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingelsson M, et al. No alteration in tau exon 10 alternative splicing in tangle-bearing neurons of the Alzheimer’s disease brain. Acta Neuropathol. 2006;112:439–449. doi: 10.1007/s00401-006-0095-3. [DOI] [PubMed] [Google Scholar]
- 42.Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Choi SH, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Schmued L, et al. Introducing Amylo-Glo, a novel fluorescent amyloid specific histochemical tracer especially suited for multiple labeling and large scale quantification studies. J Neurosci Methods. 2012;209:120–126. doi: 10.1016/j.jneumeth.2012.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.