Abstract
RNA-based temperature sensing is common in bacteria that live in fluctuating environments. Most naturally-occurring RNA thermosensors are heat-inducible, have long sequences, and function by sequestering the ribosome binding site in a hairpin structure at lower temperatures. Here, we demonstrate the de novo design of short, heat-repressible RNA thermosensors. These thermosensors contain a cleavage site for RNase E, an enzyme native to Escherichia coli and many other organisms, in the 5′ untranslated region of the target gene. At low temperatures, the cleavage site is sequestered in a stem–loop, and gene expression is unobstructed. At high temperatures, the stem–loop unfolds, allowing for mRNA degradation and turning off expression. We demonstrated that these thermosensors respond specifically to temperature and provided experimental support for the central role of RNase E in the mechanism. We also demonstrated the modularity of these RNA thermosensors by constructing a three-input composite circuit that utilizes transcriptional, post-transcriptional, and post-translational regulation. A thorough analysis of the 24 thermosensors allowed for the development of design guidelines for systematic construction of similar thermosensors in future applications. These short, modular RNA thermosensors can be applied to the construction of complex genetic circuits, facilitating rational reprogramming of cellular processes for synthetic biology applications.
INTRODUCTION
The ability to sense and respond to temperature is essential for survival, and accordingly, a variety of mechanisms to achieve this task can be observed in nature. Protein-based regulation systems, such as sigma factors specific to heat-shock proteins (1,2) and chaperone proteins that aid in a variety of heat- and cold-shock responses (3–7), allow organisms to respond to temperature changes at both transcriptional and post-translational levels. RNA-based, temperature-responsive regulation systems, which function at a translational level, are also common throughout nature (8,9). They exploit the natural tendency of single-stranded RNA molecules to change their secondary structure in response to temperature shifts, resulting in altered RNA stability or translation rates.
RNA thermosensors can be described as heat-inducible or heat-repressible, meaning that they turn on or off gene expression at high temperatures, respectively. Most naturally-occurring RNA thermosensors are heat-inducible, and they function by sequestering the ribosome binding site (RBS) in a hairpin structure at low temperatures and exposing the RBS upon hairpin destabilization at high temperatures (9). One example of such an RNA thermosensor is a regulatory element known as ROSE (Repression Of heat-Shock gene Expression) (10). The predicted structure of the 5′ UTR sequesters not only the Shine-Dalgarno (SD) sequence but also the start codon at low temperatures. Research has shown that the thermosensor hairpin does not unfold completely at high temperatures, but rather that structural perturbations at high temperatures are sufficient for translation initiation to occur (9,11). A computational model has recently been developed to predict temperature-dependent perturbations in RNA secondary structure and might provide insight into the mechanisms of such RNA thermosensors (12).
Heat-repressible RNA thermosensors function by a variety of mechanisms. For example, expression of the RpoS sigma factor in Escherichia coli is regulated by a trans-acting asRNA called DsrA (13). DsrA can take two structural conformations, one of which (the F form) will bind to the target mRNA and expose the RBS. The F form has increased stability at low temperatures, allowing for heat-repressible expression of the target gene. However, the mechanism for the stability difference between the two forms is unknown (13). Another well-studied heat-repressible RNA thermosensor regulates translation of the cspA mRNA in E. coli (14). At low temperatures, the mRNA takes a stable conformation that is more efficiently translated. The entire length of the mRNA (428 nt), not just the 5′ UTR, participates in this structural rearrangement, making the coding region an integral part of the mechanism (14,15).
Naturally-occurring RNA thermosensors, though abundant in nature, can be difficult to implement in engineered systems. For example, the RpoS mechanism is complex and poorly understood, and the cspA thermosensor is very large and requires the participation of the cspA coding sequence. These characteristics can prevent naturally-occurring thermosensors from being effectively implemented in synthetic biology applications (i.e. limited reusability or modularity). On the other hand, a de novo design strategy offers the potential to develop minimal size thermosensors with a simple, well-understood mechanism. RNA regulators lend themselves to de novo design because they form predictable secondary structures (16,17) and have a well-understood structure–function relationship (18,19), which are characteristics that contribute to their scalability (20). Furthermore, synthetic RNA thermosensors can be designed to respond to a pre-determined temperature. However, very few studies have attempted to design synthetic RNA thermosensors thus far. Two studies in 2008 used computational tools and experimental screening to design heat-inducible RNA thermosensors de novo that unfolded a stem–loop to expose the SD sequence at high temperatures (21,22). To our knowledge, the only example of a designed heat-repressible RNA thermosensor was published more recently, which did not use a de novo design strategy, but simply fused naturally-existing RNA sequences (23). In this work, we demonstrate the first, heat-repressible RNA thermosensors designed de novo (Figure 1 and Table 1).
Figure 1.
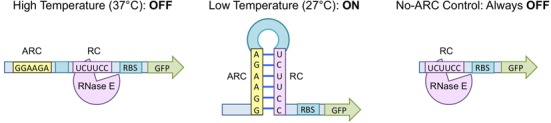
Mechanism of RNA thermosensors. At high temperatures (37°C), the RNase E cleavage site (RC – purple) is exposed, mRNA is cleaved by RNase E, and expression is ‘off’. At low temperatures (27°C), the RC binds to the anti-RNase E cleavage site (ARC – yellow) and forms a hairpin. This structure sequesters the RC, and expression is turned ‘on’. The No-ARC control lacks an ARC, and thus it is unable to form a hairpin structure. This control is expected to be in the ‘off’ state at all temperatures.
Table 1. Sequences of the 24 thermosensors used in this work (see Supplementary Table S1 for their design parameters and Supplementary Figure S1 for their predicted structures).
| Thermosensor | Sequence |
|---|---|
| No-ARC control | AAAAAAUUAAAAUCUUCCGCUCUUCC |
| A1 | AGAAAGACGAGAAAGAAAAUAAUCUUCCGCUCUUCC |
| A2 | AGAAAGACGAGAAAGAAAAAAUAUAAAUCUUCCGCUCUUCC |
| A3 | AGAAAGACGAGAAAGAAAAAAUAUAUAUAAAAUCUUCCGCUCUUCC |
| B1 | GUUGAAUGAGUUCGGUUAAGAAAUAAUCUUCCGCUCUUCC |
| B2 | GUUGAAUGAGUUCGGUUAAGAAAAAUAUAAAUCUUCCGCUCUUCC |
| B3 | GUUGAAUGAGUUCGGUUAAGAAAAAUAUAUAUAAAAUCUUCCGCUCUUCC |
| C1 | AGAAAGACGAGAAAGACUCAAUAAGAAACUCUUCCGCUCUUCC |
| C2 | AGAAAGACGAGAAAGACUCAAAAUAUAAAGAAACUCUUCCGCUCUUCC |
| C3 | AGAAAGACGAGAAAGACUCAAAAUAUAUAUAAAAGAAACUCUUCCGCUCUUCC |
| D1 | GGAAGAAAAUAAUUCUUCC |
| D2 | GGAAGAAAAAAUAUAAAUUCUUCC |
| D3 | GGAAGAAAAAAUAUAUAUAAAAUUCUUCC |
| E1 | GGAAGAAAUAAUCUUCC |
| E2 | GGAAGAAAAAUAUAAAUCUUCC |
| E3 | GGAAGAAAAAUAUAUAUAAAAUCUUCC |
| F1 | AGGAAGAAAUAAUCUUCCU |
| F2 | AGGAAGAAAAAUAUAAAUCUUCCU |
| F3 | AGGAAGAAAAAUAUAUAUAAAAUCUUCCU |
| G1 | AAGGAAAGAAAGCAAGGAAAAGAAAUAAUCUUCCGCUCUUCC |
| H1 | AGGUUAAGAGCCCGGUUAAGAAAUAAUCUUCCGCUCUUCC |
| I1 | AGGAAUGAUUGCUUGGUUAAGAGACUCAAUAAGAAACUCUUCCGCUCUUCC |
| J1 | AGAGAACGAUGCCGGCCAAGAAAUAAUCUUCCGCUCUUCC |
| K1 | AAGAGAACGAUGCCGGCAAAGAAAUAAUCUUCCGCUCUUCC |
| L1 | AGAGAACGAUGCCGGUUAAGAAAUAAUCUUCCGCUCUUCC |
| Common UTR | CAAAGUCGGUGACAGAUAACAGGAGUAAGUAAUG |
The RNase E cleavage site (RC) is underlined and the Anti-RNase E cleavage site (ARC) is shown in bold. Each thermosensor is directly followed by a common UTR containing the ribosome binding site (RBS), shown in the last row. The start codon of gfp is also shown in bold.
The RNA thermosensors developed here have a small size and a simple mechanism, allowing for construction of temperature-responsive, complex genetic circuits and potential implementation in synthetic biology applications. At low temperatures, a hairpin sequesters a cleavage site for the native ribonuclease, RNase E in the 5′ untranslated region (UTR) of the target mRNA (Figure 1). At high temperatures, the hairpin is destabilized, allowing RNase E to degrade the transcript and turn off expression. RNase E was chosen for several reasons among a variety of ribonucleases, including RNase III, PNPase and RNase P. First, RNase E is an endoribonuclease, with a preference for regions of single-stranded RNA (24). This allows for targeted degradation of RNA in its unfolded form, which occurs at higher temperatures. Second, RNase E is native to E. coli, alleviating the need for the expression of a heterologous protein. Finally, both RNase E and its homologue RNase G are common in β- and γ-proteobacteria (25) as well as cyanobacteria (26), and about half of all eubacteria outside of these groups have at least one of these enzymes on its chromosome (25). This provides reason to believe that implementation of these thermosensors in other organisms is possible with host-specific optimization. Our data show that gene expression can be regulated through the use of small, cis-acting, heat-repressible RNA thermosensors designed de novo. In addition, by analyzing thermosensor behaviors, we provide insights into their design principles.
MATERIALS AND METHODS
De novo design and construction of RNA thermosensors
Thermosensors were initially under the control of a strong constitutive Anderson promoter (http://parts.igem.org/Part:BBa_J23104) and were constructed using blunt end ligation. Each thermosensor sequence was inserted downstream of the transcription start site and upstream of the RBS, as shown in Table 1. A template plasmid containing constitutive gfp was amplified with primers containing the thermosensor sequence. After digestion with DpnI (New England Biolabs), the amplified fragment was phosphorylated with T4 polynucleotide kinase (New England Biolabs), ligated using T4 DNA ligase (New England Biolabs), and electroporated into E. coli DH10B (27). The subsequent replacement of the constitutive promoter with pTet was accomplished using Golden Gate assembly (28). All plasmids, strains, and key DNA sequences (i.e. genes, promoters and UTRs) used in this work are summarized in Supplementary Tables S2, S3 and S4, respectively.
Thermosensor structures (Supplementary Figure S1) and parameters (Supplementary Table S1) were estimated using the Mfold Web Server (17). The RNA folding form provided the predicted secondary structures and ΔG values as shown in Supplementary Table S1. Melting temperatures (Tm = 25.6–37.8°C) were estimated using the ‘Two-state melting (hybridization)’ application on the DINAMelt Web Server (http://mfold.rna.albany.edu/?q=DINAMelt/Two-state-melting) (29). This application predicts the melting temperature of two separate strands of RNA. In this case, the RC and the ARC were considered to be the two separate strands, as shown in Table 1 and Supplementary Figure S1. The effects of loop size were neglected in this approximation. Supplementary Table S1 and Supplementary Figure S1 show that the design parameters are varied within the 24 thermosensors. It is worth noting that the number of RCs and the total number of bulges are linked parameters. Bulges on either side of the stem were introduced in order to tune melting temperatures to the desired range (Tm = 25.6–37.8°C). With two RCs, it is necessary to include bulges in the stem because a perfectly complementary stem of that length has an estimated melting temperature of 76°C, which exceeds an appropriate growth temperature for E. coli. On the other hand, including bulges in thermosensor stems with only one RC reduces the estimated melting temperature to less than ∼15°C, which is below our testing temperatures. For this reason, all of the thermosensors with two RCs contain stem bulges, and all of the thermosensors with one RC contain no stem bulges.
Experimental characterization of RNA thermosensors
Cells were grown overnight in LB media (Miller) and diluted 100X with M9 minimal media with 4 g/l glucose (for cultures of the cells that contain the three-input composite circuit, see below; given catabolite repression and its effect on the pBAD promoter, glycerol, instead of glucose, was used as a carbon source for characterization of the three-input composite circuit). After 2 h of growth at 37°C and 250 rpm, cells were centrifuged and resuspended in M9 minimal media with 4 g/l glucose. These cultures were grown with supplementation of anhydrotetracycline (aTc) at the appropriate testing temperature and 250 rpm until stationary phase. Stationary phase occurred 20, 22 and 25 h after induction for cells growing at 37, 32 and 27°C, respectively. The same time points were used in all experiments unless otherwise indicated. To determine the optimum level of transcription, cells were induced with a gradient of aTc concentrations, ranging from 3.2 pg/ml to 250 ng/ml (Supplementary Figure S2). An aTc concentration of 1 ng/ml was used unless otherwise indicated. Kanamycin (20 μg/ml), ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) were added as appropriate.
Measurements were taken with a Tecan Infinite M200 plate reader. Absorbance (Abs) was measured at 600 nm to monitor cell growth. GFP was measured at excitation = 483 nm and emission = 530 nm. In order to normalize the data, a series of controls were included in each experiment. Escherichia coli DH10B was grown to provide a background fluorescence level. First, fluorescence was divided by absorbance (abs) to provide an approximate ‘per cell’ fluorescence measure. Any GFP/Abs value within one standard deviation of the value of DH10B was indicated with an asterisk in all figures. The background GFP/Abs value, determined by measuring fluorescence and absorbance from DH10B, was subtracted from thermosensors’ values. To account for differences in promoter activity due to temperature, as well as differences in protein folding and degradation rates at different temperatures, the GFP/Abs value for each culture was divided by that of the positive control. The positive control contained the same promoter as the thermosensor testing construct, but lacked the thermosensor entirely. For the three-input composite circuit, the positive control was psicA-GFP (lacking a thermosensor), with the same input plasmids (pBAD-sicA* and pTet-invF) (Figure 6D). For all other experiments, the positive control was pTet-GFP with no thermosensor. The final normalized fluorescence was calculated as follows, where TS = thermosensor, + = positive control, and 0 = E. coli DH10B or BL21 Star (DE3): normalized GFP = [(GFP/Abs)TS − (GFP/Abs)0]/[(GFP/Abs)+ − (GFP/Abs)0].
Figure 6.

Implementation of RNA thermosensors in genetic circuits. (A) Diagram for two-input composite circuit shows that gfp is under the control of the pTet promoter and an RNA thermosensor. (B) Truth table for two-input composite circuit. For temperature, ‘0’ = 37°C and ‘1’ = 27°C. aTc was used at a concentration of 1 ng/ml. (C) Results of two-input composite circuit with the B1 thermosensor. Data is the average of six biological replicates, over two separate days. The asterisk (*) indicates that the GFP/Abs value was within one standard deviation of the background DH10B GFP/Abs value (Materials and Methods). Error bars represent standard error of the mean (SEM). (D) Circuit diagram for three-input composite circuit. pBAD and pTet control the expression of SicA* (chaperone) and InvF (transcription factor), respectively. These two proteins form a complex that is necessary to activate the psicA promoter, which controls the transcription of gfp with an RNA thermosensor. (E) Truth table for three-input composite circuit. For temperature, ‘0’ = 37°C and ‘1’ = 27°C. aTc was used at a concentration of 2 ng/ml and arabinose (Ara) was used at a concentration of 0.32 μM. (F) Results of three-input composite circuit with the E3 thermosensor. Data is the average of three biological replicates, over two separate days. The asterisk (*) indicates that the GFP/Abs value was within one standard deviation of the background DH10B GFP/Abs value (Materials and Methods section). Error bars represent SEM.
For magnesium and pH testing, cells were grown overnight in LB media and diluted 100× with M9 minimal media with 4 g/l glucose, appropriate antibiotics, and 1 ng/ml aTc. Test conditions for the magnesium experiments were 2 mM Mg2+ or 2 μM Mg2+. Magnesium was added in the form of MgSO4, and missing SO42− in the 2 μM Mg2+ condition was supplemented with Na2SO4 to 2 mM. Test conditions for pH experiments were pH5 or pH 7. Media was acidified with HCl. Cultures were grown at 27°C until stationary phase, and measurements were taken as described above.
To construct the RNase E rescue strain, the coding sequence for RNase E (rne) along with its native promoters and 5′ UTR was PCR-amplified from the E. coli MG1655 genome (2550362–2554197; http://blast.ncbi.nlm.nih.gov) and cloned on a plasmid (Supplementary Tables S2 and S4). An alternative version of the plasmid containing no rne was used as a control. Escherichia coli BL21 Star (DE3) [F− ompT hsdSB (rB−mB−) gal dcm rne131 (DE3)] was co-transformed with one thermosensor plasmid and either the plasmid containing rne or the alternative control plasmid with no rne (Supplementary Table S3). Transcriptional scanning was repeated as described in Supplementary Figure S2, and 2 ng/ml aTc was identified to give the optimum transcription level for thermosensor function in these strains. Fluorescence measurements were taken at stationary phase as described above.
RT-qPCR
RT-qPCR was performed with four thermosensors (D2, E3, F2 and F3), as well as the No-ARC control and the positive control (pTet-GFP). These four thermosensors were chosen because they had demonstrated significant increases in fold change upon introduction of the RNase E rescue plasmid based on fluorescence data (Supplementary Figure S6). Each thermosensor and each control was tested in both the BL21 Star (DE3) strain and the RNase E rescue strain, and at 27°C and 37°C, for a total of 24 samples. To prepare samples for RT-qPCR, temperature induction was performed as described above. Samples were treated with rifampicin (300 μg/ml) as described previously (30) at stationary phase. RNA was immediately isolated from two biological replicates of each sample with a total culture volume of 1.5 ml per replicate (48 samples total). RNA isolation was performed using TRIzol Reagent (Life Technologies), followed by DNase treatment using the DNA-free Kit (Life Technologies). Finally, cDNA libraries were generated using the AffinityScript QPCR cDNA synthesis kit (Agilent Technologies). Details of each of these steps are included in the Supplementary Methods.
Reference genes and their primers for RT-qPCR were chosen based on literature. The cysG, hcaT and idnT genes were found to be stably expressed in the BL21 (DE3) strain, specifically at different temperatures, and their primer sequences were taken from literature (31). Primers for gfp were generated using the NCBI Primer-BLAST tool to ensure specificity. All amplicons are 100–150 nt in length. All primers were ordered from Integrated DNA technologies and sequences are shown in Supplementary Table S5. Details of primer optimization and efficiency can be found in the Supplementary Methods.
RT-qPCR was performed using the Power SYBR Green PCR Master Mix (Life Technologies) according to manufacturer's instructions, using a 50 μl reaction and 50 nM primers. The CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) was used with the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and then fluorescent detection. This was immediately followed by a melting curve (65–95°C, incrementing 0.5°C for 5 s, plate reading). The melting curve analysis confirmed the absence of non-specific products. For each sample, data are representative of two biological and two technical replicates (qPCR stage).
Quantification cycles (Cq) were determined using The CFX96 Touch™ Real-Time PCR Detection System software (Bio-Rad Laboratories, Inc.). Cq of the target gene (gfp) was normalized to the geometric mean of that of the reference genes (cysG, hcaT and idnT) (31). The relative expression level of each sample was normalized to that of the positive control (pTet-gfp) in that strain and at that temperature. Corrections were applied (log transformation, mean centering, and autoscaling) to account for variation associated with biological replicates, in accordance with MIQE guidelines (32,33). For each sample, biological and technical replicates were averaged and the standard error of the mean was calculated.
Construction and characterization of three-input composite circuits
To construct the three-input composite circuit, seven different thermosensors (B1, C1, D1, E1, E3, F1, F3) along with the common UTR (Table 1) were inserted upstream of gfp and downstream of the transcription start site of the psicA promoter (Supplementary Table S4) using blunt end ligation as described above. The plasmids containing pBAD-sicA* and pTet-invF had been constructed previously (34). Cells were grown overnight in LB media and diluted 100× with M9 minimal media with 0.4% glycerol, 2 g/l casamino acids, and 0.3 g/l thiamine hydrochloride. After 2 h of growth at 37°C and 250 rpm, cells were centrifuged and resuspended in M9 minimal media with 0.4% glycerol, 2 g/l casamino acids, and 0.3 g/l thiamine hydrochloride. These cultures were grown, with supplementation of 2 ng/ml aTc and 0.32 μM arabinose when necessary, at the appropriate testing temperature and 250 rpm until stationary phase. Measurements were taken as described above.
RESULTS
Design of heat-repressible RNA thermosensors
The RNA thermosensors described here consist of a fluorescence reporter (i.e. GFP), a common RBS, and an RNase E cleavage site (RC) sequestered by an anti-RNase E cleavage site (ARC) in a stem–loop at low temperatures and exposed at high temperatures (Figure 1 and Table 1). At low temperatures, the mRNA will be protected from degradation by the stem–loop formation, and translation will occur unhindered, resulting in an ‘on’ state. As the temperature increases, the stem–loop will unfold, exposing the RNase E cleavage site and allowing the transcript to be degraded. Thus, at high temperatures, expression will be ‘off’. The No-ARC control does not contain an ARC and is not expected to form a stem–loop at any temperature. Thus, at all temperatures, this control is expected to be in the ‘off’ state.
RNase E was chosen for several reasons. First, it was necessary to choose an endoribonuclease so that an internal location on the transcript could be cleaved. It was also essential to choose an enzyme that cleaves single-stranded RNA, instead of double stranded RNA, so that the transcript is degraded at high temperatures when the stem–loop is unfolded. Additionally, an enzyme that has some sequence specificity allows for rational design of the thermosensor. Finally, an enzyme that is conserved throughout many bacterial species provides the potential for this thermosensor to be applied in other organisms in the future. RNase E met all these requirements, it is well studied, and an RNase E mutant strain is available (24,25,35,36).
The mechanism shown in Figure 1 is a simplified version of the true behavior of RNA within the cell. While RNA hairpin folding is sometimes treated as a two-state process (folded and unfolded) (37,38), a wealth of kinetic data indicates that it is actually a multi-state process (39–42). Furthermore, the structures shown in Supplementary Figure S1 are one of several potential folded states, and different secondary structures may dynamically coexist within the cell (17). As a simple shorthand for this complex process, we will consider the folded and unfolded states shown in Figure 1 to be two ‘model-predicted’ states, recognizing that there are various dynamic structures that may occur at any given temperature.
De novo design of RNA thermosensors began with the RNase E cleavage site (RC) (UCUUCC), identified in literature (36). This sequence does not appear elsewhere within the gfp transcript (Supplementary Table S4). Thermosensor sequences contain either one RC, or two RCs separated by a GC spacer (Table 1). The anti-RC (ARC) was constructed by taking the reverse complement of the RC, and then modifying it to achieve a predicted melting temperature within the 25 - 38°C temperature range. Thermosensors were named such that those that share a letter in their name (e.g. A1, A2 and A3) share a stem structure, and due to our estimation method, also share an estimated melting temperature. The loop region separating the ARC and RC consists of A's and U's and has lengths ranging from 5 to 16 nucleotides. Thermosensors that share a number in their name (e.g. A1, B1 and C1) share an approximate loop size, where X1 = 5–6 nt loop, X2 = 10–11 nt loop and X3 = 15–16 nt loop. The 24 thermosensors also vary in their estimated ΔG and the number of bulges that they contain in each side of the stem. These parameters are summarized for each thermosensor in Supplementary Table S1, and all predicted thermosensor structures are shown in Supplementary Figure S1. To ensure that there was no potential downstream interaction that would prevent sequestration of the RC, secondary structures were also predicted with the common UTR included (Table 1). It was found that there was no major deviation from the structure shown in Supplementary Figure S1.
Optimization of thermosensor function
The thermosensors were initially tested with a strong constitutive promoter, Bba_J23104 (http://parts.igem.org/Part:BBa_J23104). Although they demonstrated increased fluorescence at low temperatures, the fold change was very small and in some cases, only observable when the temperature was reduced to 17°C. It was hypothesized that at such a high level of transcription, the large number of thermosensor-containing mRNAs overwhelmed the capacity of the native RNase E. Furthermore, it was reasoned that there would be an optimum transcription level that would allow for the maximum fold-change of the temperature response. In order to find the optimum transcription level, the constitutive promoters were replaced with the inducible pTet promoter. The No-ARC control and the positive control, which contain no thermosensor, were induced at 37°C at a variety of aTc concentrations. Because the No-ARC control does not form a hairpin at any temperature, it was expected to mimic the ideal ‘off’ behavior. Conversely, the positive control does not contain an RC and is expected to mimic the ideal ‘on’ behavior. By scanning expression levels (i.e. measuring reporter fluorescence) for maximum fold change between the two controls, we could identify the optimum transcription level for thermosensor function (Supplementary Figure S2). There was a strong peak in the fold change (positive control/No-ARC control) between 0.08 and 2 ng/ml aTc. After narrowing down an appropriate range of expression levels, follow-up experiments were performed. Further experimentation identified 1.0 ng/ml as the optimum aTc concentration.
Further optimization was necessary in the measurement and normalization procedures. Because the rates of growth, transcription, translation, and RNA and protein degradation can vary with temperature, measurements were taken at early stationary phase, when fluorescence and absorbance (at 600 nm) values were stable. Furthermore, all data was normalized to the positive control (pTet-GFP) to account for differences in transcription, translation, and degradation rates (see Materials and Methods).
Thermosensor response to temperatures
Once the transcription level and induction protocols had been optimized, thermosensor function was measured at 27 and 37°C (Figure 2). Asterisks are shown for expression levels that are ‘completely off,’ meaning the fluorescence is within one standard deviation of the background (E. coli DH10B). Thermosensors functioned as expected, with a tightly-regulated ‘off’ state at 37°C and a clear ‘on’ state at 27°C. Some thermosensors (D2, E2, E3, F1, F2, H1, I1 and L1) showed leaky expression at 37°C. The No-ARC control confirms the importance of the stem–loop structure for temperature sensing. Because this control is unable to sequester the RC in a stem–loop at low temperatures, expression is ‘off’ at both temperatures.
Figure 2.
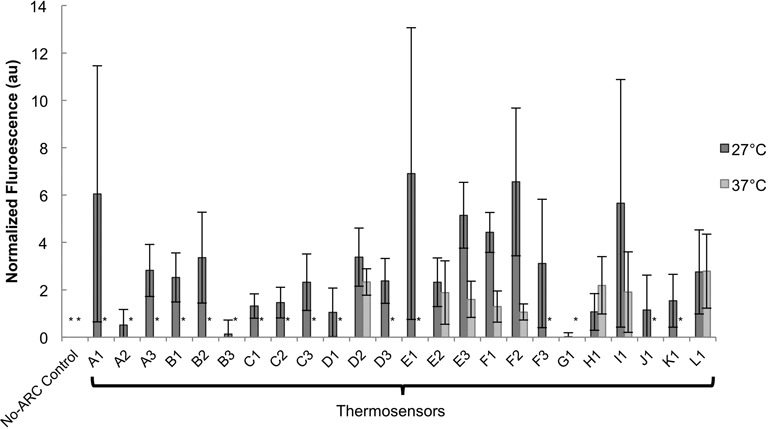
Thermosensor response in E. coli DH10B. Normalized fluorescence of thermosensors at 27 and 37°C is shown (see Supplementary Figure S3 for raw data, without normalization applied, including data for an intermediate temperature at 32°C). Fluorescence was normalized to pTet-GFP output at each temperature (1 ng/ml aTc). The asterisk (*) indicates that the GFP/Abs value was within one standard deviation of the DH10B GFP/Abs value (Materials and Methods section). This means that expression was completely ‘off’ and that these thermosensors are not leaky at 37°C. As expected, the No-ARC control is completely ‘off’ at both temperatures. Data is the average of six biological replicates, over two different days. Error bars represent standard error of the mean (SEM). A one-tailed, unpaired, Student's t-test was performed to see if expression was significantly higher at 27°C than at 37°C. The increase in fluorescence was significant for the A3, B1, C1, E3 and F1 thermosensors (P < 0.05). If the criterion is relaxed, the increase in expression at 27°C from the B2, D3 and F2 thermosensors (P < 0.07) as well as the K1 thermosensor (P < 0.09) can also be considered significant. Thermosensors A1, A2, C2, C3, D1, D2, E1, F3, G1 and H1 had P-values <0.25, and thermosensors B3, E2, I1, J1 and L1 had P-values >0.25.
The behaviors exhibited in Figure 2 were analyzed with respect to the design characteristics of each thermosensor, shown in Supplementary Table S1. Statistical analysis showed that certain design parameters could be correlated with thermosensor behavior in vivo (Figure 3). A total of 24 thermosensors were constructed and tested at 37 and 27°C, and all 24 thermosensors were included in the statistical analysis. Most importantly, a tradeoff was observed between reduced leakiness and a high maximum expression level in the ‘on’ state. In other words, a thermosensor that had a very tight ‘off’ state would have a lower expression level in the ‘on’ state, which suggests the importance of a delicate balance when selecting design parameters. To provide guidelines for thermosensor design, detailed statistical analysis was performed as discussed below.
Figure 3.
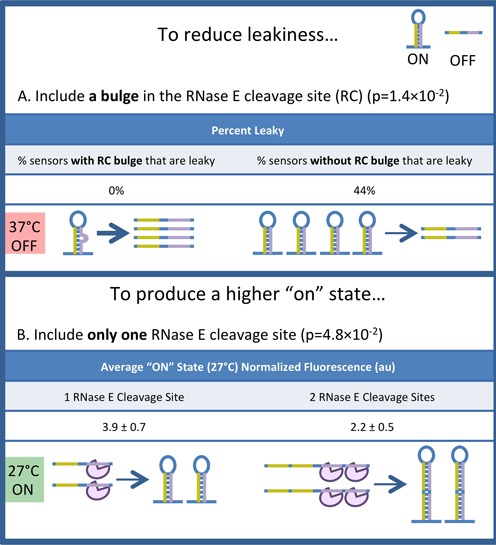
Analysis of thermosensor parameters. The parameters describing all 24 thermosensors were analyzed with a two-tailed, unpaired Student's t-test to determine which parameters had a significant impact (P < 0.05) on thermosensor function. The parameters described here are correlated with reduced leakiness (A), or a higher ‘on’ state expression level (B). (A) Reduced leakiness is correlated with the presence of a bulge in the RC. Stem bulges contribute to instability of the secondary structure, which will cause equilibrium to shift more transcripts to the model-predicted unfolded ‘off’ state at high temperatures, reducing leakiness. The weights of the arrows indicate the relative abundance of the two structures at equilibrium. The P-value (1.4 × 10−2) indicates that there is a significant difference in the average number of RC bulges between ‘leaky’ and ‘not leaky’ thermosensors. (B) A higher ‘on’ state is correlated with a single RC. The purple object represents RNase E. Increasing the number of RCs will increase the probability that an unfolded transcript will be degraded and turned off. The P-value (4.8 × 10−2) indicates that there is a significant difference in average ‘on’ state normalized fluorescence between thermosensors with one RC and thermosensors with two RCs.
Reduced leakiness was analyzed as a potentially desirable thermosensor characteristic. A thermosensor was considered ‘not leaky’ if the fluorescence of the ‘off’ state at 37°C was within one standard deviation of the white cells (DH10B). A thermosensor was considered ‘leaky’ if the fluorescence of the ‘off’ state exceeded one standard deviation of the white cells. Thermosensors that were not leaky were likely to contain a bulge in their RC (P = 1.4×10−2; unpaired, two-tailed Student's t-test). In fact, none of the thermosensors that contained a bulge in their RC were leaky, while 44% of the thermosensors lacking a bulge in their RC were leaky (Figure 3A and Supplementary Figure S4). A possible explanation for this trend relates to stability. Thermosensors containing a bulge are less stable, meaning that the equilibrium between the two model-predicted structures would be driven toward the model-predicted unfolded ‘off’ state, especially at high temperatures. It is interesting to note that bulges in the ARC had no effect on leakiness; only mismatches that would cause bulges on the RC side of the stem were correlated with reduced leakiness. This suggests that the bulge may improve RNase E access to the RC, providing an alternative explanation for this correlation.
Depending on the ultimate application, thermosensors with high expression in the ‘on’ state may be more useful than thermosensors with reduced leakiness. The inclusion of a single RC instead of two was correlated with a higher ‘on’ state (P = 4.8×10−2) (Figure 3B and Supplementary Figure S5). Increasing the number of RCs will increase the probability of RNase E-mediated cleavage, regardless of the proportion of transcripts in the model-predicted unfolded state. Thus, as the number of RCs is reduced, the chance of cleavage decreases, more transcripts are left intact, and expression increases. Because all of the thermosensors with one RC also have perfectly complementary stems, it is difficult to say whether the higher ‘on’ state is due to the lower number of RCs or the lack of bulges in the stems. Thermosensor variants with these parameters decoupled (e.g. with one RC and stem bulges, or two RCs and no bulges) were not tested because the estimated melting temperatures of such designs were outside the range of temperatures that can be tested in vivo.
Some parallels can be drawn between the synthetic heat-repressible RNA thermosensors developed here and the naturally-occurring heat-inducible ROSE thermosensors. It has been observed that ROSE thermosensors contain a conserved G-bulge across from the SD sequence when it is sequestered in a stem–loop at low temperatures. When this bulge was eliminated, two changes in thermosensor function were observed (43). First, the thermosensor is not de-repressed at high temperatures. Second, there is increased repression at low temperatures. Both of these behaviors can be explained by an increased proportion of transcripts in the model-predicted folded state at each temperature. Similarly, we observed that thermosensors containing no RC bulge had leaky expression and a higher ‘on’ state. Again, this behavior would suggest that an increased proportion of transcripts are in the model-predicted folded state, consistent with observations of the ROSE thermosensors. Thus, conserved features observed in nature can provide insights into the function of synthetic RNA devices.
Specificity of temperature response
While temperature is known to cause changes in RNA secondary structure, there are other environmental conditions that can also affect RNA stability. In order to confirm that the RNA thermosensors respond specifically to temperature, and are not inadvertently activated by other variations in RNA stability, the effects of pH and magnesium starvation were assessed. It has been observed that some RNA thermosensors, such as the Salmonella fourU-type RNA thermometer, behave differently as magnesium concentration is altered (44). However, other RNA thermosensors, such as ROSE thermosensors, are unaffected by magnesium concentration (45). RNA is a negatively charged molecule, and intramolecular repulsive forces can prevent correct RNA folding. Divalent cations, especially Mg2+, play an important role in alleviating these repulsive forces and allowing RNA structures to form (46,47). Thus, a magnesium-limited environment may prevent the hairpin structure from forming at 27°C, causing transcripts to remain in the model-predicted unfolded ‘off’ state, leading to a lower ‘on’ state. In order to test this hypothesis, responses of thermosensors were measured at 27°C at 2 mM Mg2+ or 2 μM Mg2+.
The second stability variable tested was pH. RNA shows increased stability in slightly acidic environments (48). Furthermore, although cells are known to maintain homeostasis, research has shown that E. coli can reduce its intracellular pH in an acidic environment (49–51). Thus, it is plausible to suggest that these thermosensors, though they function intracellularly, might be influenced by media pH in addition to temperature. In order to test this hypothesis, cells were grown in neutral and acidic media (pH 7 and pH 5, respectively). Based on a range of [H+]internal/[H+]external ratios (0.025–6.3) reported by literature and supported by thermodynamic modeling (52), the intracellular pH is expected to be between 6.2 and 8.6 for an external pH of 7 and between 4.2 and 6.6 for an external pH of 5. If the thermosensors were to respond to low pH, it would be expected that a higher proportion of thermosensors would be in the stable, model-predicted folded state at 27°C, resulting in a higher ‘on’ state at pH 5.
In Figure 4A, the average fold changes for the A3, B1, D3, E1, E3, F1 and F2 thermosensors are compared when exposed to three different stimuli. A fold change of one (shown by the dashed line) indicates that there is no response to the stimulus in question. The fold changes for both the pH experiment (pH 7/pH 5, 2 mM Mg2+, 27°C) and the magnesium experiment (2 mM Mg2+/2 μM Mg2+, pH 7, 27°C) were not significantly different from one (P > 0.05; one-mean, two-tailed Student's t-test). On the other hand, the fold change for the temperature response (27°C/37°C, 2 mM Mg2+, pH 7) was significantly different from one (P < 0.05; one-mean, two-tailed Student's t-test). This means that the thermosensors respond specifically to temperature and are unlikely to be activated by RNA stability variations brought on by magnesium starvation or low pH (Figure 4). However, because intracellular pH was not directly measured or manipulated in this experiment, the possibility that the intracellular pH remained constant between the two cultures cannot be disregarded.
Figure 4.
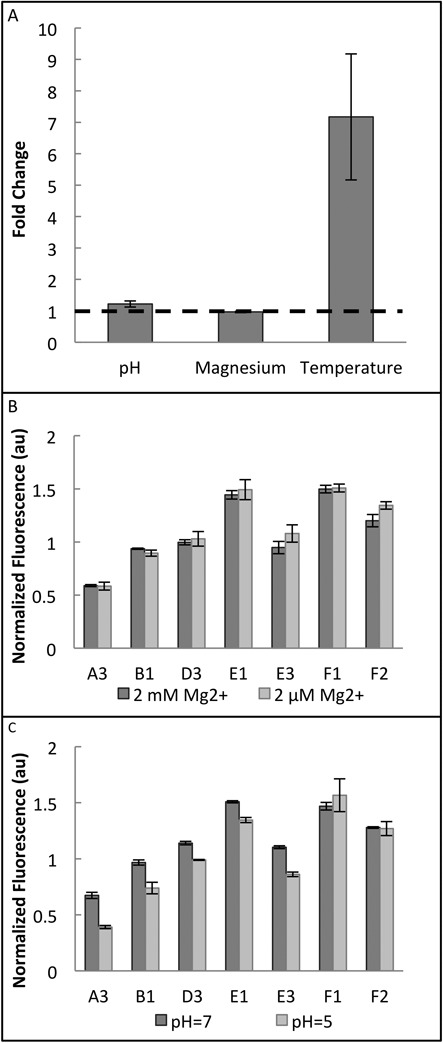
Specificity of RNA thermosensors. (A) Average fold changes are shown for the A3, B1, D3, E1, E3, F1 and F2 thermosensors. Fold changes are the ratios of normalized fluorescence at the two conditions (pH fold change = pH 7/pH 5; magnesium fold change = 2 mM Mg2+/2 μM Mg2+; and temperature fold change = 27°C/37°C). A fold change of one, shown by the dashed line, is the expected fold change if there is no response to the stimulus. A one-mean, two-tailed Student's t-test was used to determine if the average fold change was significantly different from one. The average fold change was not significantly different from one for low pH and magnesium starvation (P > 0.05), but it was significantly different from one for the temperature change (P < 0.05). (B) Thermosensor response to magnesium starvation at 27°C for individual thermosensors. There is no apparent response to magnesium starvation. (C) Thermosensor response to pH change at 27°C for individual thermosensors. There is no apparent response to pH changes. Data for pH and magnesium experiments is the average of three biological replicates; temperature data is the average of six biological replicates, over two separate days. Error bars represent standard error of the mean (SEM).
Confirmation of RNase E participation in the temperature-sensing mechanism
In order to confirm that RNase E does in fact play a central role in the temperature-sensing mechanism (Figure 1), the thermosensors were tested in the BL21 Star (DE3) strain of E. coli. This strain contains a truncated RNase E that is unable to cut RNA (53). According to the hypothesized mechanism, the thermosensors would not be able to turn off without a fully-functional RNase E. Thus, in BL21 Star (DE3), the RNA thermosensors would be expected to lose their ability to respond to temperature and instead remain in the ‘on’ state at all temperatures. Given that extensive optimization was required to see a clear temperature response in DH10B, it was important to verify that any failure to sense temperature in the BL21 Star (DE3) strain was due to the absence of a functional RNase E, not a lack of optimization. To ensure that this was the case, an RNase E rescue strain was constructed by expressing the wild-type RNase E gene (rne) on a plasmid in BL21 Star (DE3). The RNase E rescue strain would be expected to show a temperature response due to the presence of a functional version of RNase E, providing a control for thermosensor behavior in BL21 Star (DE3).
Thermosensor behavior was initially assessed with fluorescence data, as had been done for the DH10B strain. However, the results from the fluorescence data were unclear. A complete loss of temperature sensing in BL21 Star (DE3) would result in a fold change of one, which was not observed (Figure 5A). Though 27°C/37°C fold changes were consistently higher in the RNase E rescue strain than in the BL21 Star (DE3) strain, as would be expected, fold changes in BL21 Star (DE3) were greater than one for all tested thermosensors (Figure 5A). Furthermore, small changes in fluorescence were observed in the No-ARC control, which was expected to display a constant fold change close to one in both strains.
Figure 5.
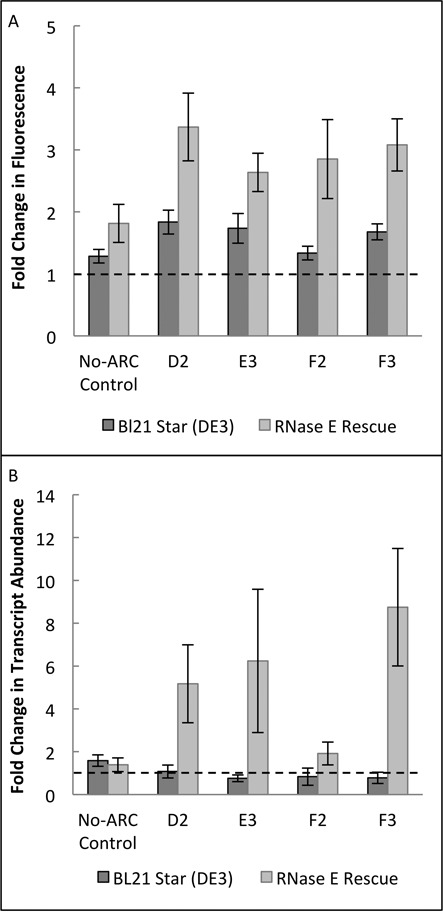
Thermosensor response in BL21 Star (DE3) and RNase E rescue strains. BL21 Star (DE3) contains a mutated version of RNase E that is unable to cut RNA. The RNase E rescue strain was constructed by expressing RNase E on a plasmid in BL21 Star (DE3). Fold changes are expected to be equal to one in the BL21 Star (DE3) strain (shown by dashed lines), indicating a loss of the ability to sense temperature, and greater than one in the RNase E rescue strain, demonstrating the recovery of the ability to sense temperature. (A) Fluorescence fold change (27°C fluorescence/37°C fluorescence) of the D2, E3, F2 and F3 thermosensors, all of which demonstrate a significant increase in the 27°C/37°C fluorescence ratio from the BL21 Star (DE3) strain to the RNase E rescue strain (P < 0.05; two-tailed, unpaired, Student's t-test). P-values are as follows: No-ARC control = 0.13, D2 = 0.02, E3 = 0.03, F2 = 0.04 and F3 = 0.01. Data is the average of 14 biological replicates, over a total of three different days. Data for the fluorescence in each strain at each temperature is provided in Supplementary Figure S6. (B) Fold change in transcript abundance (27°C transcript abundance/37°C transcript abundance) in the D2, E3, F2 and F3 thermosensors as well as the No-ARC control, based on RT-qPCR data. The thermosensors demonstrate a low 27°C/37°C fold change in BL21 Star (DE3), indicating that the temperature response has been removed. The response is recovered by introducing a functional version of RNase E, as evidenced by the increased fold change in the RNase E rescue strain. As expected, the No-ARC control has a low fold change in both strains. Data was normalized to the positive control (pTet-gfp) in that strain and at that temperature, and corrections were applied (log transformation, mean centering, and autoscaling) in accordance with MIQE guidelines (32,33). The data shown is from two biological and two technical replicates. Error bars represent standard error of the mean (SEM). P-values were calculated with a one-tailed, unpaired Student's t-test, and are as follows: No-ARC = 0.33, D2 = 0.05, E3 = 0.10, F2 = 0.08 and F3 = 0.03. Data for the relative transcript levels in each strain at each temperature is provided in Supplementary Figure S7.
To gain a clearer understanding of the effect of RNase E on the stability of thermosensor-containing transcripts, RT-qPCR was performed with the two strains. Because the hypothesized mechanism functions on the transcript stability level, directly measuring transcript levels provides better experimental support for the mechanism than does fluorescence data, which measures protein levels. Furthermore, it has been reported that there is no clear linear relationship between mRNA levels and protein levels (54,55), so by examining transcript abundance directly with RT-qPCR, we can neglect differences in translation rate or protein stability that may contribute to unexpected differences between strains in the fluorescence data. This will allow us to directly observe the impacts of RNase E on thermosensor-containing transcript abundance.
RT-qPCR analysis was performed with the D2, E3, F2 and F3 thermosensors because they each showed a significant increase in fold change between the BL21 Star (DE3) strain and the RNase E rescue strain based on fluorescence data (Figure 5A). Four other thermosensors that were tested did not show a significant change in fold change, possibly due to a lack of optimization in the new strain (Supplementary Figure S6). Transcript abundance levels based on RT-qPCR analysis are shown in Figure 5B. The 27°C/37°C fold change is expected to be one in the BL21 Star (DE3) strain, because this strain is lacking a functional RNase E and is not expected to respond to temperature. This level is shown by the dotted line. As expected, in each of the four thermosensors as well as the No-ARC control, the reported fold change is very close to one in BL21 Star (DE3). Upon introduction of a functional RNase E in the RNase E Rescue strain, all four thermosensors showed an increase in fold change, though the increase was small in the F2 thermosensor (fold changes in RNase E rescue strain: D2 = 5.2, E3 = 6.2, F2 = 1.9 and F3 = 8.7). This indicates that RNase E does in fact play an important role in the temperature sensing mechanism. Additionally, the No-ARC control showed a low fold change in the RNase E rescue strain. Although this fold change was not exactly equal to one (No-ARC fold change = 1.6 in BL21 Star (DE3) and 1.4 in RNase E rescue strain), the difference in fold change between BL21 Star DE3 and the RNase E rescue strain was not significant (P = 0.33, Figure 5B), indicating the RNase E does not have a major impact on the 27°C/37°C fold change of the No-ARC transcript abundance.
While RT-qPCR provides the most relevant data for elucidating the mechanism of these thermosensors on the level of transcript stability, fluorescence data is more relevant for assessing whether or not these thermosensors have the potential for implementation in a real system. For most synthetic biology applications, protein expression level is the central outcome, whether that protein is an enzyme in a metabolic pathway, a pathogen-killing toxin, or a transcription factor in a complex genetic circuit. This means that while transcript abundance provides important insights into the mechanism, fluorescence data would be more important for determining the potential of these thermosensors to be applied in engineered systems. Because thermosensors had been optimized to function in DH10B, behavioral differences between thermosensors in DH10B and BL21 Star (DE3) strains are not unexpected. Further optimization would be required to show that these thermosensors can function as well on the protein (fluorescence) level in BL21 Star (DE3) (the RNase E rescue strain) as they do in DH10B.
Construction and characterization of multi-input composite circuits
A benefit of regulators that act on the transcript stability level is that they can be combined with regulation at multiple levels to build complex logic operations. In order to demonstrate that the RNA thermosensors developed in this study can be used in complex circuits, the existing construct was first demonstrated to function as a two-input composite circuit. We define a composite circuit as a circuit that utilizes regulation mechanisms at more than one level (e.g. transcription and transcript stability). Because the thermosensors are under the control of pTet (Figure 6A), we can consider this construct a two-input composite circuit, with temperature and aTc as the two inputs. A response would only be expected when the two inputs are present (aTc = 1 ng/mL and temperature = 27°C) (Figure 6B). Using the B1 thermosensor, which was the best performing two-input composite circuit that was also tested in a three-input composite circuit, we can see that the two-input composite circuit functions as expected (Figure 6C). Results for two-input composite circuits using all 24 thermosensors are shown in Supplementary Figure S8.
To build on this concept and further demonstrate the modularity and composability of the RNA thermosensors, a three-input composite circuit was constructed (Figure 6D). This circuit used components from the type III secretion system in Salmonella typhimurium (56). In this system, the chaperone (SicA) and transcription factor (InvF) form a complex which is required to activate transcription from the sicA promoter. This system has been previously optimized to function in a two-input AND gate (34). The previously published AND gate was modified to form a three-input composite circuit by adding a thermosensor downstream of the psicA promoter to control the expression of the reporter, GFP (Figure 6D). Seven variants of this circuit were generated by inserting seven different thermosensors. These thermosensors were selected so as to represent a wide variety of behaviors, specifically with respect to three criteria (see Figure 2): (i) a high ‘on’ state fluorescence level (>3 normalized fluorescence units), (ii) a low P-value (<0.05) and (iii) leakiness. Two thermosensors (E3 and F1) met all three criteria, two thermosensors (E1 and F3) met criterion (i) only, two thermosensors (B1 and C1) met criterion (ii) only, and one thermosensor (D1) met none of the three criteria.
The three-input composite circuit operates on three regulatory levels: the transcriptional (promoter-mediated), the post-translational (protein–protein interaction-mediated), and the transcript stability (RNA-mediated) level. Results for the E3 thermosensor circuit, which was the best performing three-input composite circuit, are shown in Figure 6F. This circuit performed as expected, with the highest level of expression occurring when all three inputs are present (Ara = 0.32 μM; aTc = 2 ng/ml; temperature = 27°C) (Figure 6E and F). A 5.1-fold change was achieved between the leakiest ‘off’ state [1 1 0] and the ‘on’ state [1 1 1]. It is notable that this circuit demonstrates leakiness under certain conditions. This behavior is consistent with previous data, since the E3 thermosensor was slightly leaky in the two-input composite circuit (Supplementary Figure S8). However, thermosensors that did not exhibit leakiness in the two-input composite circuit exhibited levels of leakiness in the three-input composite circuit similar to that of the E3 thermosensor circuit (Supplementary Figure S8 for two-input and Supplementary Figure S9 for three-input). Still, because the thermosensors are functioning in a different genetic context (i.e. under the control of the different promoter psicA, instead of pTet), slight differences in behavior would not be unexpected. The behavior of each thermosensor was compared in the three-input and two-input circuits. A slight correlation (r2 = 0.72) was found between the fold change of the 3-input circuit and the ‘on’ state of the two-input circuit (Supplementary Figure S10).
DISCUSSION
RNA thermosensors have a wide variety of potential applications in synthetic biology and metabolic engineering. As synthetic biology transitions from lab-scale genetic circuit demonstrations to industrial, medical, and environmental applications, chemical inducers such as arabinose and aTc will have to be replaced. Not only are these inducers irrelevant in situ (e.g. inside the human body or at a potential bioremediation site), but their cost is also inhibitory when it comes to scale-up. On the other hand, temperature can be a meaningful signal in the environment, and temperature-responsive systems will not require expensive chemical inducers. Furthermore, metabolic engineers often need to consider temperature variations within large-scale bioreactors and adjust host cells’ metabolism accordingly. For example, hotspots are a common problem in solid-state fermentation (57), and photobioreactors can overheat in the afternoon (58). Heat-repressible RNA thermosensors with customized melting temperatures could be implemented to down-regulate product synthesis pathways that divert the cells’ resources away from survival during such undesirable periods.
In this study, heat-repressible RNA thermosensors were designed de novo and demonstrated to function in E. coli (Figure 2). They have a stem–loop in the 5′ UTR upstream of the RBS that unfolds at high temperatures to expose an RNase E cleavage site. The exposed RNase E cleavage site allows for degradation of the transcript, turning off expression (Figure 1). Several experiments were conducted to confirm the hypothesized mechanism outlined in Figure 1. The No-ARC control is not expected to form a stem–loop at any temperature, leaving it consistently prone to RNase E degradation. This control is off at both 27 and 37°C, demonstrating the importance of the stem–loop structure for a functional thermosensor (Figure 2). The magnesium and pH experiments summarized in Figure 4 ensure that the thermosensor responds specifically to temperature and does not respond to conditions that affect RNA stability in general. Finally, the RT-qPCR experiments with BL21 Star (DE3) support the proposed mechanism outlined in Figure 1. These experiments show that thermosensors show no temperature response in a strain with a non-functional RNase E, but regain function upon introduction of a fully functional RNase E (Figure 5). This result supports the hypothesis that RNase E plays an important role in the temperature-sensing mechanism. A thorough knowledge of the mechanism will allow for streamlined implementation of these thermosensors in other genetic systems and potentially in other organisms.
A potential complication to the mechanism presented in Figure 1 is the participation of the ribosome. Each of the 24 thermosensors was designed to share an SD sequence (Table 1) in order to maintain relatively consistent translation rates. However, because context effects are well known to affect translation rates (59), it is likely that the actual RBS strengths varied among thermosensors. One factor that may cause differences in translation rates among thermosensors is the secondary structure of the thermosensors themselves. A recent paper investigates the effect of secondary structure of long 5′ UTRs on ribosome binding (60). The authors found that the ribosome can bind to standby sites, which are structurally similar to the model-predicted folded state of the thermosensor. However, because a structured 5′ UTR introduces a binding free energy penalty, it is energetically more difficult for a ribosome to bind to a structured 5′ UTR than an unstructured 5′ UTR. Thus, higher translation rates would be expected in the unfolded ‘off’ state, which counters the prediction made by our proposed mechanism (Figure 1), and is not supported by experimental data (Figure 2). While the structure of the 5′ UTR may impact translation rates, it is likely that the structural effect is overshadowed by the effects of RNase E-mediated transcript degradation.
However, differences in translation rates due to secondary structure can provide an alternative explanation for the higher observed ‘on’ state in thermosensors containing only one RC site, instead of two. While this trend could be explained by considering that a transcript with two RCs will be less stable than a transcript with only one RC (Figure 3B), it is also possible that the longer hairpins in two-RC thermosensors reduces translation rates (60). Still, RT-qPCR data indicates that transcript stability plays a major role in the observed changes in gene expression. If differential translation rates were solely responsible for temperature-induced changes in expression, we would not expect to see the changes in mRNA abundance that are shown in Figure 5. Furthermore, the correlation between RC bulges and a reduction in leakiness indicates that RNase E access may play a more important role than secondary structure itself (Figure 3A), since no correlation was observed between leakiness and bulges in the ARC.
Another factor to consider is the impact of ribosome binding on the structure of the transcript. The ribosome footprint is approximately 30 nt (61,62). Though a distance of about 20 nt was left between the SD sequence and the thermosensor hairpin to account for ribosome binding, the possibility exists that the binding of the ribosome would cause partial unfolding of the thermosensor hairpin (60). Furthermore, in the model-predicted unfolded state at high temperatures, it is possible that either the ribosome or RNase E could bind to the transcript, though simultaneous binding is unlikely due to steric hindrance. Thus, translation can occur at high temperatures (in the ‘off’ state) until RNase E binds to the transcript. Once RNase E binds and cleaves the transcript at the RC site, the remaining transcript fragments will be quickly degraded to nucleotides by exoribonucleases and accessory factors including RNase II, RNase R, PNPase and RhlB, among others (63). Because RNase E cleavage is the limiting factor in the RNA degradation process, it is unlikely that the mRNA fragments would be stable enough for significant translation to occur after RNase E cleavage. Although these factors should be considered to gain a more complex understanding of the mechanism, the simple mechanism presented in Figure 1 can still be used to describe the general behavior of these thermosensors.
An understanding of this mechanism, in addition to the results shown in Figure 2, can be used to evaluate assumptions made during the design phase. For instance, the assumption was made that loop size could be neglected when estimating melting temperature (Methods). Current RNA secondary structure prediction programs calculate stem stability using nearest-neighbor approximations, and account for hairpin loops with an additional free energy term that reduces the stability of the structure (64,65). In general, the stability of the hairpin decreases with increasing loop size (66), which is reflected in the ΔG estimations obtained from Mfold that considered loop size (Supplementary Table S1). Had loop size been considered in melting temperature approximations, it is expected that melting temperature estimates would have decreased with increasing loop size. However, the analysis of experimental data showed no relationship between thermosensor behavior and loop size.
Besides the ‘on’ and ‘off’ states shown in Figure 2, another potentially important characteristic of these thermosensors is their response time. To determine how quickly a thermosensor would respond to a change in temperature, the response time of the F1 thermosensor was measured. The F1 thermosensor was chosen due to its high ‘on’ state (normalized fluorescence = 4.4 au) and low P-value (P = 1.5 ×10−2; Figure 2). In order to characterize the response time, Welch's t-test was used to find the time at which the fluorescence of the thermosensor at 27°C was significantly higher than that of the No-ARC control at 27°C (Supplementary Figure S11 and Supplementary Methods) (67). Based on this method, the response time for the F1 thermosensor was 1.2 h (72 min) after the temperature shift from 37 to 27°C. This is in line with a recent study on an in vivo RNA regulator controlling RFP expression, which reported response times of 41.7–72.7 min, depending on the speed of the response element (67). The lower growth temperature in our experiment may have contributed to a slow response time.
After characterizing ‘on’ and ‘off’ states (Figure 2), response specificity (Figure 4), thermosensor mechanism (Figure 5), and the response time (Supplementary Figure S11), the ability of thermosensors to participate in more complex systems can be evaluated. Construction of a three-input composite circuit demonstrates the modularity and composability of the RNA thermosensors (Figure 6). They remain functional in a different genetic context (i.e. with two different promoters pTet and psicA) and can be combined with other genetic devices to form complex logic operations. Modularity and composability are fundamental to the scalability of any genetic device (68). Considering these characteristics alongside the designability of the RNA thermosensors and their proposed generality to diverse hosts, it is reasonable to suggest that there is wide potential applicability for these RNA thermosensors in synthetic biology.
There are several recent examples of RNA-based devices being implemented in genetic circuits, including the construction of an RNA-mediated transcriptional cascade (69), cotranscriptional in vitro RNA circuits (70), logic gates consisting of RNA toehold switches (71), and a pathway diverter utilizing an RNA transducer together with a promoter to determine cell fate (72). In addition, several recent studies have used the CRISPR-Cas system, which utilizes small guide RNAs (sgRNAs) and associated proteins, in the design of genetic circuits (73–75). The composite circuit in this study uses a unique combination of genetic controls on a variety of regulatory levels. Namely, this circuit implements inducible promoters at the transcriptional level, interacting proteins at the post-translational level, and an RNA thermosensor at the transcript stability level. By diversifying the levels of circuit regulation, the potential for different circuit architectures is expanded while the metabolic burden and overall circuit size for a given logical operation are reduced. For example, an analogous three-input AND gate demonstrated previously requires three layers, five transcription units and seven protein regulators (chaperones and transcription factors) (34). The three-input composite circuit demonstrated here requires only two layers, three transcription units and four protein regulators. While the circuit complexity was reduced, the same logical operation was maintained. By extension, a much larger genetic circuit could be similarly simplified by using a combination of transcriptional and post-transcriptional controls, providing opportunities for future circuit development.
In this work, we have demonstrated that a simple stem–loop structure could be designed to act as a heat-repressible RNA thermosensor in E. coli. These thermosensors are small, have a simple mechanism, and can be designed to have a very tightly regulated ‘off’ state. Because of these characteristics, they can be more easily implemented into complex genetic circuits than can natural RNA thermosensors. To our knowledge, this is the first demonstration of heat-repressible RNA thermosensors designed de novo. Although the structure of these thermosensors is simple, design and optimization were not trivial. Insights gained from this study regarding design choices and optimization protocol will be invaluable in the implementation of these or similar thermosensors in future work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Hani Zaher for advice during this work.
FUNDING
Gates Foundation [OPP1087549 to T.M.]; National Science Foundation [MCB-1331194 to A.H. and T.M. and CBET-1350498 to T.M.]; Mr and Mrs Spencer T. Olin Fellowship for Women in Graduate Study (to A.H.). Funding for open access charge: National Science Foundation [CBET-1350498].
Conflict of interest statement. None declared.
REFERENCES
- 1.Grossman A.D., Erickson J.W., Gross C.A. The htpR gene product of E. coli is a sigma factor for heat-shock proteins. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 2.Yura T., Nagai H., Mori H. Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 3.Craig E.A., Weissman J.S., Horwich A.L. Heat shock proteins and molecular chaperones: mediators of protein conformation and turnover in the cell. Cell. 1994;78:365–372. doi: 10.1016/0092-8674(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 4.Muga A., Moro F. Thermal Adaptation of Heat Shock Proteins. Curr. Protein Peptide Sci. 2008;9:552–566. doi: 10.2174/138920308786733903. [DOI] [PubMed] [Google Scholar]
- 5.Guisbert E., Yura T., Rhodius V.A., Gross C.A. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol. Mol. Biol. Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones P.G., Inouye M. The cold-shock response – a hot topic. Mol. Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 7.Horn G., Hofweber R., Kremer W., Kalbitzer H.R. Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 2007;64:1457–1470. doi: 10.1007/s00018-007-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortmann J., Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 2012;10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 9.Narberhaus F., Waldminghaus T., Chowdhury S. RNA thermometers. FEMS Microbiol. Rev. 2006;30:3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- 10.Narberhaus F., Kaser R., Nocker A., Hennecke H. A novel DNA element that controls bacterial heat shock gene expression. Mol. Microbiol. 1998;28:315–323. doi: 10.1046/j.1365-2958.1998.00794.x. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury S., Ragaz C., Kreuger E., Narberhaus F. Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem. 2003;278:47915–47921. doi: 10.1074/jbc.M306874200. [DOI] [PubMed] [Google Scholar]
- 12.Chursov A., Kopetzky S.J., Bocharov G., Frishman D., Shneider A. RNAtips: analysis of temperature-induced changes of RNA secondary structure. Nucleic Acids Res. 2013;41:W486–W491. doi: 10.1093/nar/gkt486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Repoila F., Gottesman S. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 2001;183:4012–4023. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giuliodori A.M., Di Pietro F., Marzi S., Masquida B., Wagner R., Romby P., Gualerzi C.O., Pon C.L. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell. 2010;37:21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 15.Freischmidt A., Hiltl J., Kalbitzer H.R., Horn-Katting G. Enhanced in vitro translation at reduced temperatures using a cold-shock RNA motif. Biotechnol. Lett. 2013;35:389–395. doi: 10.1007/s10529-012-1091-4. [DOI] [PubMed] [Google Scholar]
- 16.Liu C.C., Arkin A.P. Cell biology. The case for RNA. Science. 2010;330:1185–1186. doi: 10.1126/science.1199495. [DOI] [PubMed] [Google Scholar]
- 17.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappell J., Takahashi M.K., Meyer S., Loughrey D., Watters K.E., Lucks J. The centrality of RNA for engineering gene expression. Biotechnol. J. 2013;8:1379–1395. doi: 10.1002/biot.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carothers J.M., Goler J.A., Juminaga D., Keasling J.D. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 20.Lucks J.B., Qi L., Whitaker W.R., Arkin A.P. Toward scalable parts families for predictable design of biological circuits. Curr. Opin. Microbiol. 2008;11:567–573. doi: 10.1016/j.mib.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Waldminghaus T., Kortmann J., Gesing S., Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol. Chem. 2008;389:1319–1326. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- 22.Neupert J., Karcher D., Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008;36:e124. doi: 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saragliadis A., Krajewski S.S., Rehm C., Narberhaus F., Hartig J.S. Thermozymes synthetic RNA thermometers based on ribozyme activity. RNA Biol. 2013;10:1009–1016. doi: 10.4161/rna.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouvier M., Carpousis A.J. A tale of two mRNA degradation pathways mediated by RNase E. Mol. Microbiol. 2011;82:1305–1310. doi: 10.1111/j.1365-2958.2011.07894.x. [DOI] [PubMed] [Google Scholar]
- 25.Condon C., Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron J.C., Gordon G.C., Pfleger B.F. Genetic and genomic analysis of RNases in model cyanobacteria. Photosynth. Res. 2015 doi: 10.1007/s11120-015-0076-2. doi:10.1007/s11120–11015–10076–11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durfee T., Nelson R., Baldwin S., Plunkett G. 3rd, Burland V., Mau B., Petrosino J.F., Qin X., Muzny D.M., Ayele M., et al. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoo S.M., Na D., Lee S.Y. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat. Protoc. 2013;8:1694–1707. doi: 10.1038/nprot.2013.105. [DOI] [PubMed] [Google Scholar]
- 30.Moller T., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhou K., Zhou L., Lim Q., Zou R., Stephanopoulos G., Too H.P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011;12 doi: 10.1186/1471-2199-12-18. doi:10.1186/1471–2199–1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 33.Willems E., Leyns L., Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Analyt. Biochem. 2008;379:127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 34.Moon T.S., Lou C., Tamsir A., Stanton B.C., Voigt C.A. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpousis A.J., Luisi B.F., McDowall K.J. Endonucleolytic initiation of mRNA decay in Escherichia coli. Progr. Mol. Biol. Transl. Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 36.Caron M.-P., Bastet L., Lussier A., Simoneau-Roy M., Masse E., Lafontaine D.A. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3444–E3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuznetsov S.V., Ren C.C., Woodson S.A., Ansari A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008;36:1098–1112. doi: 10.1093/nar/gkm1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ansari A., Kuznetsov S.V., Shen Y. Configurational diffusion down a folding funnel describes the dynamics of DNA hairpins. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7771–7776. doi: 10.1073/pnas.131477798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung J., Van Orden A. A three-state mechanism for DNA hairpin folding characterized by multiparameter fluorescence fluctuation spectroscopy. J. Am. Chem. Soc. 2006;128:1240–1249. doi: 10.1021/ja0560736. [DOI] [PubMed] [Google Scholar]
- 40.Bowman G.R., Huang X., Yao Y., Sun J., Carlsson G., Guibas L.J., Pande V.S. Structural insight into RNA hairpin folding intermediates. J. Am. Chem. Soc. 2008;130:9676–9678. doi: 10.1021/ja8032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W.B., Chen S.J. Exploring the complex folding kinetics of RNA hairpins. I. General folding kinetics analysis. Biophys. J. 2006;90:765–777. doi: 10.1529/biophysj.105.062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnykov A.V., Nayak R.K., Hall K.B., Van Orden A. Effect of loop composition on the stability and folding kinetics of RNA hairpins with large loops. Biochemistry. 2015;54:1886–1896. doi: 10.1021/bi5014276. [DOI] [PubMed] [Google Scholar]
- 43.Nocker A., Hausherr T., Balsiger S., Krstulovic N.P., Hennecke H., Narberhaus F. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 2001;29:4800–4807. doi: 10.1093/nar/29.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinnenthal J., Klinkert B., Narberhaus F., Schwalbe H. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 2011;39:8258–8270. doi: 10.1093/nar/gkr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhury S., Maris C., Allain F.H., Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole P.E., Yang S.K., Crothers D.M. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972;11:4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- 48.Oivanen M., Kuusela S., Lonnberg H. Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by Bronsted acids and bases. Chem. Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 49.Foster J.W. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 50.Richard H., Foster J.W. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diez-Gonzalez F., Russell J.B. The ability of Escherichia coli O157:H7 to decrease its intracellular pH and resist the toxicity of acetic acid. Microbiology. 1997;143:1175–1180. doi: 10.1099/00221287-143-4-1175. [DOI] [PubMed] [Google Scholar]
- 52.Henry C.S., Broadbelt L.J., Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys. J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez P.J., Marchand I., Joyce S.A., Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 54.Lu P., Vogel C., Wang R., Yao X., Marcotte E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 55.Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darwin K.H., Miller V.L. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 2001;20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang E., Han B., Li S. Numerical simulation of transient radial temperature distribution in rotating drum bioreactor for solid state fermentation. 2013 International Conference on Materials for Renewable Energy and Environment. 2013:291–294. [Google Scholar]
- 58.Bechet Q., Shilton A., Fringer O.B., Munoz R., Guieysse B. Mechanistic modeling of broth temperature in outdoor photobioreactors. Environ. Sci. Technol. 2010;44:2197–2203. doi: 10.1021/es903214u. [DOI] [PubMed] [Google Scholar]
- 59.Salis H.M. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42. doi: 10.1016/B978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- 60.Espah Borujeni A., Channarasappa A.S., Salis H.M. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li G.-W., Oh E., Weissman J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484 doi: 10.1038/nature10965. 538-U172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carpousis A.J., Luisi B.F., McDowall K.J. Endonucleolytic Initiation of mRNA Decay in Escherichia coli. Mol. Biol. RNA Process. Decay Prokaryotes. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 64.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 65.Giese M.R., Betschart K., Dale T., Riley C.K., Rowan C., Sprouse K.J., Serra M.J. Stability of RNA hairpins closed by wobble base pairs. Biochemistry. 1998;37:1094–1100. doi: 10.1021/bi972050v. [DOI] [PubMed] [Google Scholar]
- 66.Groebe D.R., Uhlenbeck O.C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 1988;16:11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V., Spring K.J., Al-Khabouri S., Fall C.P., Noireaux V., et al. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth. Biol. 2014 doi: 10.1021/sb400206c. doi:10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi L.S., Arkin A.P. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat. Rev. Microbiol. 2014;12:341–354. doi: 10.1038/nrmicro3244. [DOI] [PubMed] [Google Scholar]
- 69.Lucks J.B., Qi L., Mutalik V.K., Wang D., Arkin A.P. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhadra S., Ellington A.D. Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers. Nucleic Acids Res. 2014;42:e58. doi: 10.1093/nar/gku074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Green A.A., Silver P.A., Collins J.J., Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galloway K.E., Franco E., Smolke C.D. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341:1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nielsen A.A.K., Voigt C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014;10 doi: 10.15252/msb.20145735. doi:10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas Toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nature Commun. 2014;5 doi: 10.1038/ncomms6393. doi:10.1038/ncomms6393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


