Abstract
SLX4 assembles a toolkit of endonucleases SLX1, MUS81 and XPF, which is recruited to telomeres via direct interaction of SLX4 with TRF2. Telomeres present an inherent obstacle for DNA replication and repair due to their high propensity to form branched DNA intermediates. Here we provide novel insight into the mechanism and regulation of the SLX4 complex in telomere preservation. SLX4 associates with telomeres throughout the cell cycle, peaking in late S phase and under genotoxic stress. Disruption of SLX4's interaction with TRF2 or SLX1 and SLX1's nuclease activity independently causes telomere fragility, suggesting a requirement of the SLX4 complex for nucleolytic resolution of branched intermediates during telomere replication. Indeed, the SLX1–SLX4 complex processes a variety of telomeric joint molecules in vitro. The nucleolytic activity of SLX1-SLX4 is negatively regulated by telomeric DNA-binding proteins TRF1 and TRF2 and is suppressed by the RecQ helicase BLM in vitro. In vivo, in the presence of functional BLM, telomeric circle formation and telomere sister chromatid exchange, both arising out of nucleolytic processing of telomeric homologous recombination intermediates, are suppressed. We propose that the SLX4-toolkit is a telomere accessory complex that, in conjunction with other telomere maintenance proteins, ensures unhindered, but regulated telomere maintenance.
INTRODUCTION
Mammalian chromosome ends are protected by nucleoprotein structures called telomeres, and preservation of telomere integrity is essential for genome stability. Telomeres are composed of the shelterin protein complex and a double-stranded tract of short tandem repeats ending in a single-stranded 3′-overhang of the G-strand (1). It is believed that telomere DNA forms a lariat-like structure called the telomeric loop (T-loop), by invasion of the 3′-overhang into the duplex region of telomeric DNA, forming a displacement loop (D-loop) at the site of invasion (2,3). Due to their repetitive DNA sequence, unique architecture and bound shelterin proteins, telomeres pose an inherent challenge to the progression of DNA replication, repair and recombination apparatus. The major obstacle is encountering unusual alternative DNA forms such as t-loops, D-loops and Holliday Junctions (HJs), all of which may stall/delay these telomere maintenance processes. Additionally, many telomere maintenance processes are thought to involve homologous recombination (HR)-mediated resolution of telomeric DNA intermediates. For instance, HR-dependent synthesis of new telomeric DNA is believed to be responsible for telomere length maintenance in telomerase negative cancer cells through a mechanism called alternative lengthening of telomeres (ALT) (4). Furthermore, in ALT cancer cells and some telomerase positive cells, e.g. hTERT-induced cells, germline and activated T-cells, telomeres are very long and heterogeneous; and HR-dependent mechanisms of telomere shortening, called ‘telomere trimming’, ensure continuance of telomere length homeostasis in these cells (5). Hence, given the inherent tendency of branched DNA intermediates to form even in normal and undamaged telomeres, the cell must devise special attention toward the processing and removal of these telomeric intermediates to ensure timely progression of telomere replication and other maintenance processes.
The key shelterin proteins TRF1 and TRF2 directly bind to double-stranded telomeric DNA and have been proposed to recruit several different non-shelterin accessory proteins to telomeres, which enable processing of DNA joint molecule intermediates and thereby facilitate unhindered progression of telomere maintenance processes. These mainly include the helicases and the nucleases (1). Several human DNA helicases such as BLM, WRN, RecQL4, RecQL1 and RTEL1 have been implicated in telomere maintenance (6–8). Human nucleases that have roles in telomere maintenance are the exonuclease Apollo (9,10) and structure-specific endonucleases (SSEs) GEN1 and the Fanconi anemia protein SLX4-associated nucleases (11–13). Human SLX4 acts as a scaffold to assemble a DNA repair complex consisting of the SSEs SLX1, MUS81-EME1 and XPF-ERCC1. Interestingly, the SLX4 complex isolated from human cells also contained TRF2 (14). Previously, we and others have demonstrated that the SLX4-nuclease complex in human cells primarily associates with relatively long telomeres and telomeric localization of this complex is dependent on direct interaction between SLX4 and TRF2 and on a TRF2-counting mechanism (12,13).
Here, we investigate the mechanistic basis and regulation of the SLX4-assembled complex in maintaining telomere integrity, including replication. We demonstrate that SLX4 maximally associates with telomeres in late S phase and also under genotoxic stress. Disruption of the functional SLX1–SLX4 nuclease module and the SLX4–TRF2 interaction causes phenotypes associated with aberrations in telomere replication or in other joint molecule resolution-requiring processes. We also show that the SLX1–SLX4 complex-sponsored nucleolytic resolution of telomeric DNA intermediates is negatively regulated by other telomere-associated proteins. Thus, we propose that regulated SLX4-nuclease function may constitute an important layer of telomere maintenance, thereby ensuring genome stability.
MATERIALS AND METHODS
Telomere detection assays
Telomere fluorescence in situ hybridization (FISH) and Chromosome Orientation FISH (CO-FISH) were used to detect fragile telomeres and telomere sister chromatid exchanges (T-SCEs), respectively, and performed as described in (12). For indirect immunofluorescence coupled with FISH (IF-FISH), cells were stained with primary and subsequently with Alexa Fluor-labeled secondary antibodies, followed by fixation and telomere-FISH as described in (12). Telomere circle amplification (TCA) assay (15) that was used to detect telomeric circles (TCs) was performed on genomic DNA extracted from U2OS cells transiently expressing control, anti-SLX4 and/or anti-BLM siRNA for 72 h.
In vitro telomeric substrate processing assays
SLX1–SLX4-dependent nuclease reactions were performed as described in (12). SLX1–SLX4/BLM reactions contained pre-mixed enzymes and were initiated by radiolabeled substrates. For TRF1 and TRF2 protection experiments, radiolabeled substrates were pre-incubated with purified TRF1 or TRF2 on ice for 5 min, followed by addition of SLX1–SLX4 complex.
RESULTS
SLX4 differentially associates with human telomeres during cell cycle progression
Previously, we have shown that SLX4 along with its associated nucleases primarily localizes to telomeres in human cells possessing a high frequency of long telomeres, such as HeLa 1.2.11 (telomerase positive) and U2OS (telomerase negative, ALT) (12). To investigate the requirement of SLX4 in different processes of telomere maintenance and during different stages of the cell cycle, we synchronized HeLa 1.2.11 cells by a double thymidine block (Figure 1A). Indirect immunofluorescence coupled with telomere FISH (IF-FISH) detected a significant increase, albeit to varying degrees, in SLX4 foci formation in all phases of the cell cycle, compared to the asynchronized cell population (Figure 1B). It is noteworthy that a significant fraction of these SLX4 foci colocalized with telomeres in late S phase (4 h) (Figure 1B). The chromatin immunoprecipitation (ChIP) analysis of SLX4 further confirmed this trend, showing maximal significant SLX4–telomere association in late S phase (4 h), in addition to lesser, but significant association in G1/S (0 h) phase (Figure 1C). Thus, the significant association of SLX4 with telomeres throughout the cell cycle accentuates an important role for SLX4 in various processes of telomere maintenance, including during and after telomere replication.
Figure 1.
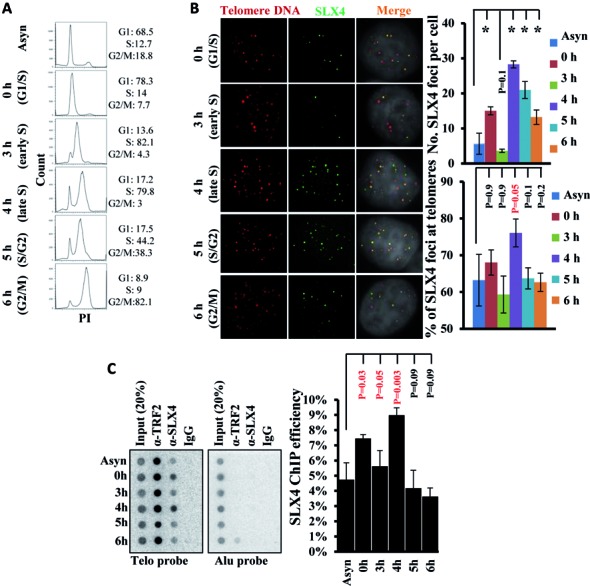
SLX4 foci formation and association with telomeres during cell cycle progression in HeLa 1.2.11 cells. (A) FACS analyses of cell cycle synchronization profile. PI indicates DNA content. Percentage of cells in G1, S and G2/M phases is shown. (B) Representative images (left) and quantification (right) showing the number of SLX4 foci (green) per cell and the percentage of foci colocalizing with telomeric DNA (red). IF-FISH was performed using anti-SLX4 rabbit antibody and a telomere-specific PNA (CCCTAA)3 probe. (C) ChIP analysis of SLX4 at telomeric DNA (Telo probe) or at a control locus (Alu probe) during cell cycle progression. SLX4 ChIP efficiency was calculated as the fraction of telomeric DNA immunoprecipitated with anti-SLX4 rabbit antibody in total DNA. Error bars represent standard deviation from three independent experiments. Phenotypes were compared between two groups using student's t test. Significant P values are either indicated in red or by an asterisk (*P < 0.0001).
Genotoxic stress induces SLX4 foci formation and their telomeric association
Because significant SLX4–telomere affiliation in S phase alluded to its importance in telomere replication, we further probed into the pattern of SLX4 foci formation and their association with telomeres in HeLa 1.2.11 cells treated with a broad spectrum of genotoxic agents, including those causing replication barriers and delays. These included replication inhibitors aphidicolin and hydroxyurea (HU), DNA interstrand cross-linkers such as mitomycin C (MMC) and DNA alkylating reagents such as methyl methanesulfonate (MMS). The number of SLX4 foci per cell and their colocalization with telomeres significantly increased after exposure to all these genotoxins, albeit to varying degrees (Figure 2A). The most substantial increase for not only the number of SLX4 foci per cell but also the fraction of SLX4 foci overlapping with telomeres was observed in aphidicolin-treated cells (Figure 2A), re-iterating a role for SLX4 in telomere replication. Furthermore, fluorescence-activated cell sorting (FACS) revealed a relative cell cycle progression block in S phase or its boundaries in response to these treatments (Figure 2B and C), which correlated with the significant SLX4–telomere association in S phase (Figure 1) or induced by these treatments (Figure 2A). Thus, SLX4 may be involved in counteracting DNA replication challenges and DNA damage at telomeres.
Figure 2.
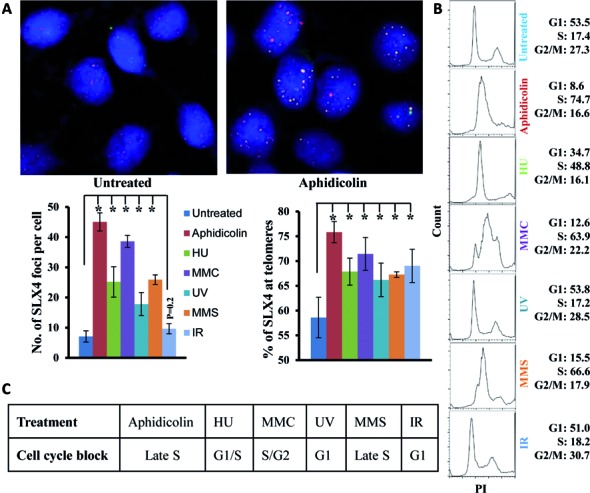
Genotoxic stress induces SLX4 foci formation and colocalization with telomeric DNA in HeLa 1.2.11 cells. (A) Representative IF-FISH image and quantification of SLX4 foci colocalizing with telomeric DNAand (B, C) FACS evaluation of cell cycle block after treatment with DNA damage reagents (0.5-μM aphidicolin, 2-mM HU, 0.3-mM MMC or 0.25-mM MMS for 16 h, 20 J/m2 UV or 10 Gray IR). Percentage of cells in G1, S and G2/M phases is indicated. Error bars represent standard error from at least three independent experiments, and PI indicates DNA content. Significant P values are indicated by an asterisk (*P < 0.0001).
SLX4 recruitment to telomeres is essential to prevent telomere fragility
Telomeres resemble genomic common fragile sites (CFS) (16) and impose an inherent challenge to the DNA replication machinery. In fact aphidicolin-induced replication stress leads to discontinuous telomere signals on metaphase spreads, which have been interpreted as a sign of fragile telomeres (17). In HT1080 supertelomerase cells, a decrease in SLX4 expression enhances the number of multi-telomeric signals at chromatid ends (18). Our observations here (Figures 1 and 2) suggest an important role for SLX4 during and after telomere replication in human cells with long telomeres. Replication-dependent telomere defects, manifested as fragile telomeres, may conceivably be compounded at long telomeres. Hence, we examined the frequency of fragile telomeres by telomere-FISH in long telomere-containing cells (U2OS, telomerase negative; HeLa 1.2.11, telomerase positive), depleted of SLX4. Transient siRNA depletion of SLX4 (Supplementary Figure S1A) in these human cells significantly increased the percentage of fragile telomeres (Figure 3A and Supplementary Figure S1B), which was further exacerbated by replication blocking agent aphidicolin (Supplementary Figure S1B, right), highlighting the necessity of SLX4 to avert telomere fragility, both in the absence and presence of exogenous replication stress.
Figure 3.
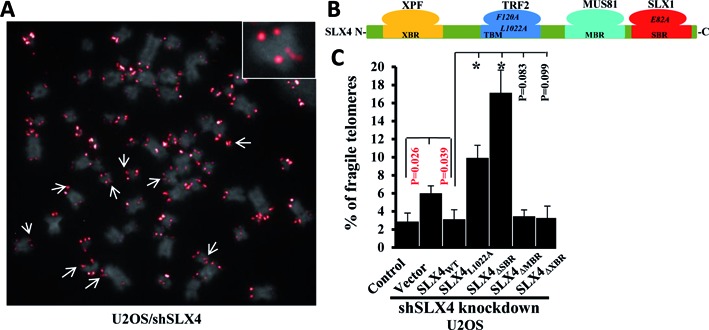
SLX4 deficiency increases telomere fragility. (A) Representative image of telomere-FISH analysis in SLX4-depleted U2OS metaphase spread showing DAPI staining (gray) and telomere fluorescence signals (red). Fragile telomeres are indicated by arrows, and also shown as an enlarged image in the inset. (B) Schematic showing interactions within the SLX4 complex. (C) Frequency of fragile telomeres in U2OS cells stably expressing shSLX4 RNA, transiently transfected with GFP (vector) or GFP-fusion SLX4 wild-type or mutant proteins. Abbreviations: XBR (XPF-binding region), MBR (MUS81-binding region), SBR (SLX1-binding region) and TBM (TRF2-binding motif). TRF2F120A and SLX4L1022A mutants independently disrupt the SLX4–TRF2 interaction. SLX1E82A is a nuclease-dead mutant of SLX1. SLX4ΔXBR, SLX4ΔMBR and SLX4ΔSBR are truncation mutants of SLX4 that specifically abolish interaction of SLX4 with XPF, MUS81 and SLX1, respectively. Error bars represent standard deviation from three independent experiments of each genotype (30 metaphases/genotype/experiment). Frequency of fragile telomeres was compared between two groups using a student's t test. Significant P values are either indicated in red or by an asterisk (*P < 0.0001).
As SLX4 assembles a multi-protein toolkit at telomeres (Figure 3B) (12), we probed further to delineate the specificities regarding the SLX4-complex requirement in suppressing telomere fragility. U2OS cells stably expressing SLX4 shRNA were transiently transfected with either GFP-empty vector or GFP-vector carrying the wild-type or respective mutant SLX4 gene. SLX4ΔSBR, SLX4ΔMBR and SLX4ΔXBR are truncation mutants of full-length SLX4 that specifically abolish interaction with the nucleases SLX1, MUS81 and XPF, respectively. The SLX4L1022A and TRF2F120A mutants each contain a point mutation that specifically and independently abrogates SLX4–TRF2 interaction and recruitment of SLX4 to telomeric DNA (Figure 3B) (12). Also, as shown in our previous report, transfection of these SLX4 mutants in SLX4-depleted U2OS cells leads to expression of the respective mutant proteins to similar levels as wild-type SLX4 (12). Whereas telomere fragility was increased in SLX4-depleted cells (vector), expression of wild-type SLX4 or SLX4ΔMBR and SLX4ΔXBR deletion mutants rescued telomere fragility to levels of control (scramble shRNA-transfected cells) (Figure 3C). In contrast, expression of the SLX4L1022A or SLX4ΔSBR mutants not only failed to rescue telomere fragility but also further elevated the fragile phenotype (Figure 3A and Figure 3C), highlighting the requirement of the interactions of SLX4 with TRF2 and SLX1 for preventing telomere fragility. Consistent with this observation, transient transfection of the TRF2F120A mutant in U2OS cells greatly aggravated telomere fragility compared to cells transiently expressing the wild-type TRF2 or the empty vector (Supplementary Figure S1C). Importantly, we have previously shown that in U2OS cells transiently expressing the TRF2F120A mutant, recruitment of SLX4 to telomeres is indeed abolished (12). Furthermore, transient transfection of U2OS cells with the nuclease-dead fusion complex SLX1E82A − SLX4WT failed to rescue telomere fragility unlike the wild-type fusion complex SLX1WT − SLX4WT (Supplementary Figure S1D), confirming the necessity of nuclease activity of SLX1 in averting telomere fragility. Taken together, these observations strongly suggest that SLX4 is required at telomeres to avert telomere fragility. To perform this function, SLX4 is critically dependent on its interaction with TRF2 and with SLX1; and on the nuclease activity of SLX1.
The SLX1–SLX4 nuclease module resolves telomeric branched DNA intermediates in vitro
Our data suggest a role for SLX4 and its associated nuclease SLX1 in counteracting DNA replication challenges and DNA damage at telomeres. Telomeres are hotspots for the formation of DNA joint molecule intermediates not only during DNA repair but also during normal DNA processing, including replication. Timely progression of telomere repair or normal DNA processing likely necessitates special attention to resolve such structural barriers. Because the SLX1–SLX4 module emerged as a player in telomere replication (Figures 1–3) and length homeostasis (12), we next probed into the mechanistic basis of SLX1-SLX4 in maintaining telomere integrity. We first investigated the efficiency and substrate specificity of SLX1-SLX4 in processing telomeric branched intermediates in vitro. We performed nuclease cleavage assays utilizing the C-terminal SLX1-binding region (SBR) of SLX4 purified in complex with the wild-type or nuclease-dead E82A mutant of full-length SLX1 (SLX1WT − SLX4SBR and SLX1E82A − SLX4SBR, respectively) (Figure 3B) (12) and various model telomeric DNA substrates constructed from oligonucleotides (Supplementary Figure S2A and Supplementary Table S1). The SLX1WT − SLX4SBR complex cleaved a wide variety of telomeric DNA intermediates, including D-loop [described in (19)], splayed arm, 3′- and 5′-flap, replication fork and static HJ, albeit with varying efficiencies (Supplementary Figure S2). In contrast, the nuclease-dead SLX1E82A − SLX4SBR mutant complex showed no detectable cleavage activity (Supplementary Figure S2A), supporting that the cleavage by the wild-type complex is indeed due to nuclease activity of SLX1. Steady state kinetic studies revealed that the SLX1WT − SLX4SBR complex preferentially cleaved the telomeric D-loop and HJ with an initial observed rate constant (kobs) of 32.6 ± 1.6 min−1 and 24.4 ± 0.9 min−1, respectively, about 3-fold higher than the kobs for the lesser preferred telomeric substrates such as the replication fork, splayed arm or 3′-flap (Supplementary Figure S2B). These results support that in vitro, the SLX1–SLX4 module is catalytically competent to nucleolytically process a miscellaneous array of telomeric joint molecule intermediates, preferentially HJs and D-loops, both of which are frequently occurring intermediates in telomere maintenance processes.
D-loops are not only regularly occurring intermediates in telomere repair or other DNA processes, but also an inherent part of the T-loop structure. Previously we had reported that SLX1-SLX4 provides the nuclease activity for processing telomeric D-loops by cleaving the invading strand (DL-IS, Figure S3, left panel) in vitro (12). However, the full mechanism of D-loop resolution, particularly with regard to nicking of the D-loop, was unknown. To address this, here we dissected the pattern of D-loop processing by the SLX1–SLX4 nuclease module. We labeled any one of the three strands of a model telomeric D-loop substrate DL-IS (67 mer), DL-B (93 mer) or DL-T (93 mer) (Supplementary Figure S3). Denaturing gel electrophoresis enabled us to determine nucleolytic cleavage events at each individual strand of the D-loop. Purified SLX1WT − SLX4SBR cleaved DL-IS (Supplementary Figure S3, left panel, lanes 2–4) and both DL-B (Supplementary Figure S3, middle panel, lanes 6–8) and DL-T (Supplementary Figure S3, right panel, lanes 10–12) on opposite sides of the melted bubble at the respective junction with the duplex arm (arrows). This in vitro pattern of D-loop cleavage favors the involvement of the SLX1–SLX4 complex in nucleolytic resolution of T-loops (Supplementary Figure S4A and B) (20,21) and provides mechanistic insight into how this process may proceed in vivo.
Telomeric DNA-binding proteins negatively regulate nucleolytic processing of telomeric joint molecules by the SLX1–SLX4 complex
While on the one hand telomeric joint molecule intermediates such as D-loops/T-loops and HJs must be processed and removed for progression of telomere maintenance processes, on the other hand the formation of T-loops is believed to sequester telomere ends from being recognized as DNA damage (1). Thus, unregulated nucleolytic activity on such telomeric intermediates constitutes a formidable threat to telomere and hence genome integrity. Moreover, because telomeres must be distinguished from double-strand breaks, repair proteins, including nucleases, must be differently regulated at telomeres (22). We thus questioned if inherent telomeric DNA-binding proteins TRF1 and TRF2, which are known to play a key role in telomere maintenance (1), could contribute toward regulation of the SLX1–SLX4 nuclease activity at telomeres.
We first confirmed that in vitro, purified TRF1 and TRF2 bound to our model telomeric D-loop and HJ substrates under SLX1–SLX4 nuclease cleavage reaction conditions. Incubation of both telomeric D-loop and HJ substrates with either TRF2 or TRF1 led to shift in substrate mobility on a native gel in electrophoretic mobility shift assays (EMSAs) (Supplementary Figure S5A). As observed in this study and by others previously (19,23–25), it is technically challenging to investigate DNA-binding by TRF2, because of retention of most of the DNA–TRF2 complex in the wells and due to lack of detectable product at low TRF2 concentrations (Supplementary Figure S5A). Nonetheless, analyses of our EMSA results showed comparable binding constants (Kd, app) of TRF2 and TRF1 with each of the telomeric substrates (D-loop and HJ) (Supplementary Figure S5A).
We then pre-bound the telomeric substrates (D-loop or HJ) with increasing concentrations (0–60 nM) of either TRF1 or TRF2, followed by nuclease cleavage reactions with catalytic amounts (0.5 nM) of purified SLX1WT − SLX4SBR. We analyzed the extent of TRF2 or TRF1 protection by separately quantifying amount of all product bands formed and also the amount of uncleaved substrate on a native gel (Figure 4A and E). The fraction of cleaved products or uncleaved substrate did not show significant changes at low concentrations of TRF2 or TRF1 [1:1 or 1:2 molar ratios of (SLX1WT − SLX4SBR:TRF2 or TRF1)], both for the telomeric D-loop (Figure 4A–D) and the HJ (Figure 4E–H). However, at higher concentrations of TRF2 or TRF1 [1:10 or higher molar ratios of (SLX1WT − SLX4SBR:TRF2 or TRF1)], protection of telomeric substrates by TRF2 or TRF1 (both in terms of cleaved products and uncleaved substrate) reached as high as 60–90% (Figure 4A–D for D-loop and Figure 4E–H for HJ). Thus, at higher concentrations, both TRF2 and TRF1 limit the extent of SLX1WT − SLX4SBR nuclease activity on telomeric substrates in vitro, implying that the extent of substrate protection offered by TRF2/TRF1 may depend on the amount of these proteins bound to telomeres at any given time. Because the purified SLX1WT − SLX4SBR complex lacked the TRF2-binding domain (TBM) of SLX4 (Figure 3B) (12), we wondered if the SLX4–TRF2 interaction would impact the observed trend of TRF2/TRF1 protection on nucleolytic processing of telomeric substrates. To address this possibility, we exogenously expressed both wild-type and full-length SLX1 and SLX4, co-immunoprecipitated the complex (SLX1WT + SLX4WT) and used it to perform similar TRF2/TRF1 protection experiments as described above. Higher concentrations of pre-bound TRF2 (or TRF1, data not shown) resulted in substrate protection and inhibition of cleavage of both telomeric HJ and D-loop by the immunoprecipitated (SLX1WT + SLX4WT) complex (Supplementary Figure S5B, HJ, lanes 1–6; D-loop, lanes 7–12), consistent with results from experiments with purified SLX1WT − SLX4SBR. Thus, telomeric substrate protection rendered by pre-bound TRF2/TRF1 is unlikely to be affected by the SLX4–TRF2 interaction.
Figure 4.
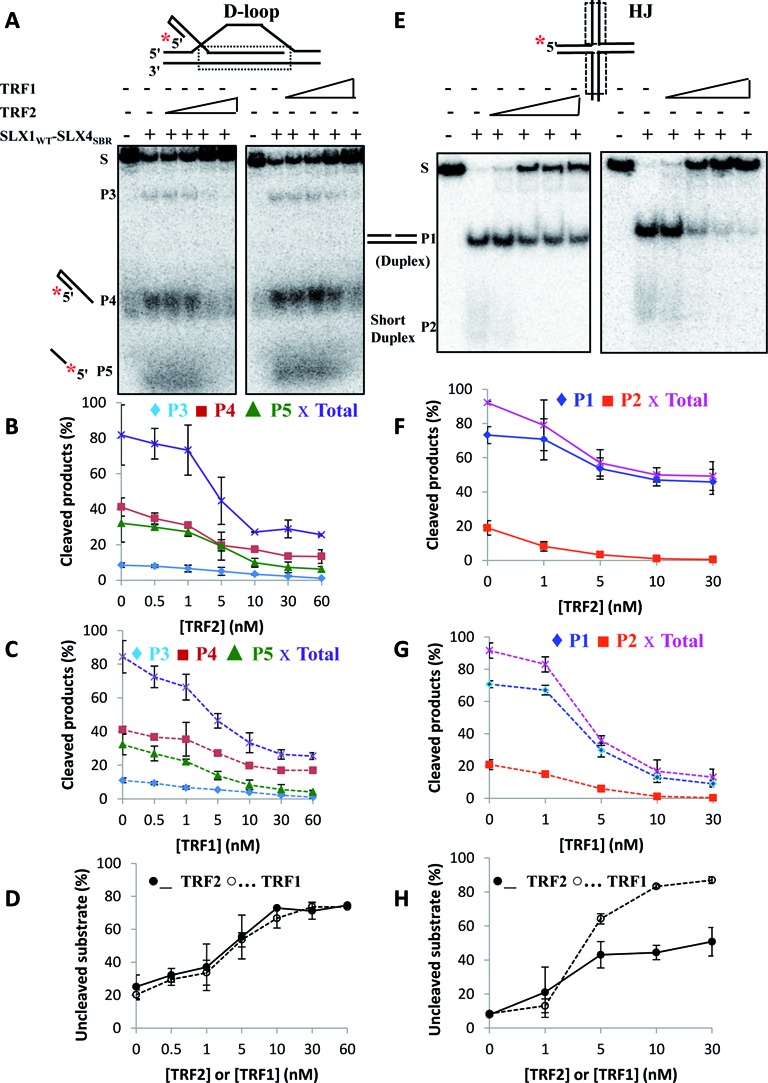
Telomeric DNA-binding proteins limit SLX1–SLX4-sponsored processing of telomeric branched DNA forms in vitro. (A–D) Telomeric D-loop and (E–H) telomeric HJ are protected from nucleolytic cleavage by the purified SLX1WT − SLX4SBR complex in vitro. Representative native gel images (A, E) and quantification of cleaved products (B, C and F, G) and uncleaved substrate (D, H) in the presence of increasing concentrations (0–60 nM) of TRF2 or TRF1. The substrate (0.5 nM) was pre-bound to TRF2 or TRF1 on ice for 5 min, followed by addition of 0.5-nM purified SLX1WT − SLX4SBR complex. S, substrate (D-loop or HJ); P1 and P2, products of HJ cleavage; P3, P4 and P5, products of D-loop cleavage; Total, total cleaved products formed due to substrate (D-loop or HJ) cleavage.
Since inhibition of telomeric substrate processing was observed in the presence of TRF2 or TRF1, we sought to further validate if this negative regulation of nuclease activity was dependent on telomeric DNA substrate binding by TRF2/TRF1. TRF1 and TRF2 share a similar architecture, characterized by a TRF homology (TRFH) and a C-terminal Myb DNA-binding domain (1). The TRFH domains of TRF2 and TRF1 have almost identical three-dimensional structures (26). Since both the N-terminal basic domain and the C-terminal Myb domain of TRF2 have been implicated in DNA-binding (27,28), we used the TRF2TRFH domain to assess the requirement of substrate interaction. When we pre-incubated purified TRF2TRFH domain (unable to bind telomeric DNA) with the telomeric substrates, it did not affect nucleolytic digestion of the substrates by the SLX1–SLX4 complex (Supplementary Figure S5C, compare lanes 4 and 5 with lane 2), implying that in the absence of physical telomeric DNA-binding by TRF2, the active site of the SLX1–SLX4 module continues to have access to the substrates. Furthermore, neither TRF2 (Supplementary Figure S5C, compare lanes 7 and 8) nor TRF1 (Supplementary Figure S5C, compare lanes 10 and 11) affected nucleolytic processing of a non-telomeric substrate (HJ) by purified SLX1WT − SLX4SBR, signifying that the limitation on nuclease activity of SLX1-SLX4 by TRF1 and TRF2 is specific for telomeric substrates. Together, these data indicate that in vitro, the telomeric DNA-binding shelterin proteins TRF1 and TRF2 impose a rein on SLX1–SLX4-sponsored nucleolytic resolution of telomeric joint molecules by physical blocking of telomeric DNA, thereby plausibly contributing to regulation of telomere recombination. This TRF2-dependent restraint on SLX1–SLX4 nuclease activity in vitro may also include contribution from the basic N-terminal domain of TRF2 (18,29).
TRF2 is instrumental in recruiting SLX4 to telomeres via its TRFH domain. Mutation in a key residue (SLX4L1022A) disrupts the SLX4–TRF2 interaction and the telomeric localization of SLX4 (12). We questioned if the SLX4–TRF2 interaction is also important for the nuclease activity of the SLX1–SLX4 complex. Exogenously expressed wild-type SLX1 together with either wild-type SLX4 or SLX4L1022A mutant (incapable of interacting with TRF2) (Figure 3B) was purified from human cells via co-immunoprecipitation. While both complexes contained comparable amounts of SLX1 and SLX4, significantly less amount of TRF2 was pulled down in the mutant complex (SLX1WT + SLX4L1022A) as compared to the wild-type complex (SLX1WT + SLX4WT) (Supplementary Figure S6A). Because the SLX1–SLX4 complex (both purified and immunoprecipitated) cleaves telomeric and non-telomeric HJ with similar efficiencies (data not shown), we used a non-telomeric HJ to compare the activities of the wild-type and mutant SLX4 complexes. This substrate would exclude the substrate-binding effect of any TRF2 pulled down in these complexes, and hence would enable us to exclusively assess the effect of SLX4–TRF2 interaction on the nuclease activity of the SLX1–SLX4 complex. We observed similar nuclease activities of these two SLX4 complexes on this substrate (Supplementary Figure S6B), suggesting that although the SLX4–TRF2 interaction is critical for the recruitment of SLX4 to telomeres, it may not be important for the nuclease activity of the SLX1–SLX4 complex. Thus, these data converge toward a dual role for TRF2 in regulating HR at telomeres: on the one hand, the direct interaction of SLX4 with TRF2 is critical for recruiting the SLX4-nuclease complex to telomeres; on the other hand, telomeric DNA-bound TRF2 (and TRF1) may restrain rampant SLX4-nuclease activity on telomeric HR intermediates, thereby potentially avoiding critical shortening or undesirable loss of telomeres. A balance between these different functions of TRF2 and TRF1 may ensure regulated nucleolytic processing of telomere joint molecule intermediates in vivo.
Helicase activity of BLM suppresses nuclease activity of SLX1-SLX4 in processing telomeric HR intermediates in vitro
T-loops and intermediates of T-SCE bear resemblance to HR intermediates and are therefore in danger of being inappropriately processed by the SLX4-nuclease complex. In fact T-loop HR, plausibly involving nucleolytic resolution of the HJ and the D-loop, could potentially cause T-loop sized telomere loss and thus formation of extrachromosomal TCs (20,21). Hence, careful regulation of T-loop HR is imperative for telomere maintenance. Among the RecQ helicases, BLM has been shown to be critical in telomere maintenance, and loss of function of BLM helicase in Bloom's syndrome patient cells causes several telomere defects (30–32). Since BLM dissolves telomeric HR intermediates (33), we questioned if this activity could constitute a potential regulatory mechanism to keep SLX4-nuclease complex-sponsored telomeric HR in check. To address this, we first assayed processing of the model telomeric D-loop substrate, labeled on DL-IS (Figure 5A) in vitro. Native gel analysis revealed that wild-type BLM alone unwound and displaced the full-length invading strand (DL-IS) from the duplex (Figure 5A, lane 2). In the presence of purified SLX1WT − SLX4SBR complex alone (Figure 5A, lane 3), or coimmunoprecipitated (SLX1WT + SLX4WT) complex alone (Figure 5A, lane 8), we observed telomeric D-loop nucleolytic cleavage products. Interestingly, in the presence of both wild-type BLM and purified SLX1WT − SLX4SBR (Figure 5A, lanes 4–6) or coimmunoprecipitated (SLX1WT + SLX4WT) (Figure 5A, lane 7), only the intact invading strand (DL-IS) was observed. Similar results were obtained for a four-way static HJ substrate. In the presence of wild-type BLM (Supplementary Figure S7, lane 2), helicase-dependent products (the two-way junction and single-stranded species) were detected. In the presence of purified SLX1WT − SLX4SBR complex alone, the expected nicked duplex cleavage product was detected (Supplementary Figure S7, lane 5). However, in the presence of both purified SLX1WT − SLX4SBR complex and wild-type BLM, only helicase-dependent products were detected (Supplementary Figure S7, lane 3). Thus, these results indicate that in vitro, the helicase activity of BLM enables suppression of nuclease activity of SLX1-SLX4 in processing telomeric D-loops and HJs. This alludes to a BLM helicase-dependent negative regulatory mechanism for SLX1–SLX4 nuclease activity during telomeric HR.
Figure 5.
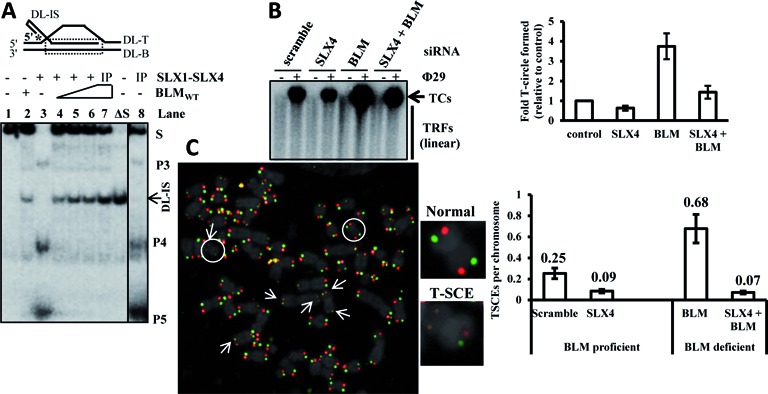
SLX4-nuclease complex-sponsored processing of telomeric HR intermediates is negatively regulated by the helicase BLM. (A) Representative native gel image showing that BLM suppresses SLX1–SLX4 complex-catalyzed nucleolytic resolution of telomeric D-loops in vitro. Reactions contained 0.5-nM radiolabeled substrate, along with 10-nM purified wild-type BLM alone (lane 2), or 2-nM purified SLX1WT − SLX4SBR complex alone (lane 3), or 0–10-nM purified BLM together with 2-nM purified SLX1WT − SLX4SBR complex (lanes 4–6). Lanes 8 and 7 contained co-immunoprecipitated (Myc-SLX1WT + HA-SLX4WT) complex alone or together with 10-nM purified BLM. The heat-denatured substrate is indicated as ΔS. The intact invading strand is indicated as DL-IS. S, substrate (D-loop); P3, P4 and P5, products due to nucleolytic cleavage of D-loop. (B) Representative gel electrophoresis image and quantification of a Ф29 polymerase-dependent TCA assay in U2OS cells transiently depleted of SLX4 alone, or BLM alone, or both. TC levels were normalized to the control of U2OS/scramble siRNA cells. Error bars were calculated from triplicate data. (C) Representative image and frequency of T-SCEs in U2OS cells transiently depleted of SLX4 alone, or BLM alone, or both, assayed by CO-FISH. Chromosome ends exhibiting T-SCE have been indicated by arrows in the image. A representative normal and T-SCE positive chromosome (encircled) have been enlarged. Error bars represent standard deviation from three independent experiments of each genotype (30 metaphases/genotype for CO-FISH). TC, telomere circle; TRF, telomere restriction fragment; TCA, telomere circle amplification; T-SCE, telomere sister chromatid exchange.
BLM helicase-depleted cells exhibit enhanced SLX4-dependent TC formation and T-SCE
To test the functional significance of the interplay between BLM and SLX4 in controlling telomere recombination, we analyzed two relevant telomere recombination events in vivo—formation of TCs and T-SCEs. We analyzed TC formation under conditions of depletion of SLX4 or BLM alone, and both in U2OS cells (Supplementary Figure S7B). For this purpose, we used a Ф29 DNA polymerase-dependent T-circle amplification (TCA) assay (15). Depletion of SLX4 alone led to dwindling levels of TCs in U2OS cells as compared to control (scramble si-RNA-treated cells). In contrast, transient depletion of BLM alone led to a significant increase in TC formation. However, the TC level was restored to that of control when SLX4 and BLM were co-depleted, signifying that enhanced TC formation in BLM-deficient cells is dependent on the SLX4-nuclease complex (Figure 5B). These data suggest that T-loop HR and TC formation in U2OS cells is suppressed in the presence of BLM. This is possibly attributable to the predominant unwinding of telomeric D-loop by the helicase activity of BLM. But, deficiency of BLM induces SLX4-nuclease complex sponsored nucleolytic resolution of telomeric D-loops, leading to augmented TC formation.
T-SCE is also dependent on the SLX4-associated nucleases in human cells (12,34). To confirm our in vitro observation that proficiency of BLM helps suppress nucleolytic processing of telomeric HR intermediates by SLX4-SLX1, SLX4 was depleted in U2OS cells in both BLM-proficient and deficient backgrounds (Supplementary Figure S7B) and analyzed for T-SCE events via chromosome orientation FISH (CO-FISH) (Figure 5C). Depletion of SLX4 in the BLM-proficient background moderately diminished T-SCEs (0.25 and 0.09 events/chromosome in scramble and SLX4-depleted cells, respectively). Depletion of BLM alone led to greatly increased T-SCEs (0.25 and 0.68 events/chromosome in scramble and BLM-depleted cells, respectively). This increase was dramatically suppressed by further exhaustion of SLX4 (0.68 and 0.07 events/chromosome in BLM alone and SLX4/BLM-depleted cells, respectively) (Figure 5C). This provides in vivo evidence that enhanced T-SCEs in BLM-deficient cells are indeed dependent on SLX4, and reinforces that when BLM is functional, SLX4-catalyzed telomeric recombination is subdued.
DISCUSSION
Telomeres are an indispensable element of genome stability, and defects or delays in telomere maintenance processes have been associated with development of several diseases including bone marrow failure and cancer (35). However, telomeres are inherently challenging regions of the genome. Owing to their highly repetitive sequence and unique architecture, telomeric DNA is a hotspot for the formation of unusual DNA intermediates during normal DNA processes, including length homeostasis, recombination and replication. It is conceivable that the longer the telomeres are, the greater is the severity of replication and other challenges. In fact very long telomeres may be prone to replication slippage and stalled replication forks, which have been proposed to be deleterious for the cell (36,37). The shelterin complex together with several accessory proteins (1), including the SLX4-assembled nuclease complex, protects and maintains mammalian telomeres (12,13).
Because SLX4 mostly locates to telomeres in human cells harboring long telomeres (12), we propose that the SLX4-nuclease toolkit performs essential functions beyond DNA repair in these cells. SLX4 is a genuine telomere accessory complex, particularly at long telomeres that require greater attention of joint intermediate-resolving enzymes for their replication and maintenance. As the occurrence of long telomeres varies among human cell types, we employed human cells possessing a high frequency of long telomeres as a model to test the hypothesis. Consistent with our hypothesis, we observed that in these human cells, SLX4 association with telomeres varies in a cell cycle phase-dependent manner, peaking in late S phase (Figure 1). A pronounced S-phase cell cycle arrest was observed after subjecting cells to replication stress, which interestingly correlated with significant increase in SLX4 foci formation and telomere association, as compared to untreated cells (Figure 2). Dysregulation of the SLX4-nuclease complex leads to an increase in multiple broken telomeric signals on metaphase spreads assayed by Telomere-FISH (Figure 3). Such structural aberrations in telomeres are believed to arise out of defects in replication (represented as fragile telomeres) and/or alterations in packaging of telomeric chromatin (17). Notably, SLX4–telomere association exhibited significant increase in late S phase. It has been reported that mammalian telomere replication may occur in two phases (one in S phase and another later) (38), raising the possibility that completion of telomere replication does not occur until late S or S/G2 phase (39). In addition, end processing of mammalian telomeres is also believed to last several hours after replication (40). Thus, it is possible that SLX4 functions in telomere replication and/or other telomere processes, e.g. telomere maturation and trimming. Furthermore, SLX4 also appears to be important for regulating telomere recombination, evident from SLX4-sponsored TC formation or T-SCE events in BLM-deficient cells (Figure 5).
Because of the high frequency of irreparable DNA lesions (41) and formation of unusual DNA intermediates including D-loops and HJs at telomeres, processing of these alternative forms of DNA is a common necessity for unhindered progression of telomere replication and other telomere maintenance processes. The shelterin protein TRF2 plays a significant role in telomere protection. One of the ways it does so is by recruiting and coordinating several non-shelterin enzymatic activities capable of processing unusual joint intermediates at telomeres (1). In a previous report, we found that the SLX4-nuclease toolkit is recruited to telomeres via direct interaction between the TRF2-binding motif (TBM) of SLX4 (SLX4TBM) and the TRFH domain of TRF2 (TRF2TRFH) (12). And indeed, in this study we find that the SLX1–SLX4 nuclease module is catalytically competent in processing a wide array of joint telomeric DNA intermediates in vitro, with kinetic preference for D-loops and HJs. In vivo, we find that disruption of the SLX4–TRF2 and SLX4–SLX1 interactions and nuclease activity of SLX1 lead to significant exacerbation of telomere fragility, suggesting that in order to alleviate telomere replication difficulties, SLX4 depends on TRF2 to localize the nuclease complex to telomeres and particularly requires the nuclease activity of SLX1. We speculate that nucleolytic activity of the SLX1–SLX4 complex may be required for processing joint molecule intermediates at telomeres that have high propensity to form such intermediates in normal maintenance processes during and after replication. It is noteworthy that TRF2 also recruits the 5′-3′ exonuclease Apollo to telomeres in S phase, when it is believed to repress aberrant telomeric intermediates (and hence DNA damage signals) during telomere replication (9,10). Interestingly, the TBM of Apollo (ApolloTBM) is conformationally identical to SLX4TBM, and both bind to TRF2TRFH in a similar fashion (12,42). While the exonuclease activity of Apollo has been proposed to contribute to strand resection of telomeric DNA during replication (10), the endonuclease complex assembled on SLX4 may be important for resolving joint telomeric DNA intermediates such as D-loops and HJs formed during telomere maintenance processes.
While the nucleases are indispensable for unperturbed telomere maintenance, their unregulated activity is a formidable threat to telomere integrity and genome stability. Hence, in our quest to investigate possible regulatory mechanisms to keep SLX4-nuclease kit activity at human telomeres in check, we first focused on the protectors of telomeres, TRF1 and TRF2. On the one hand, we find that in vitro, both TRF1 and TRF2 limit SLX1-SLX4-sponsored nucleolytic processing of telomeric joint molecule intermediates (Figure 4). This effect is dependent on telomeric DNA-binding ability of TRF2 (Figure 4 and Supplementary Figure S5), suggesting that such a telomeric substrate protection effect could potentially serve as a rein on rampant nuclease activity by the SLX4 complex. On the other hand, we know that TRF2 recruits the SLX4 complex to telomeres via its TRFH domain (12), although this protein–protein interaction between SLX4 and TRF2 may not be as important for the nuclease activity of the SLX1–SLX4 complex in vitro (Supplementary Figure S6B). How to consolidate these apparently different effects of TRF2? The explanation may lie in the multiple ways in which TRF2 not only caps telomeres but also regulates telomere processing by recruiting and orchestrating several enzymatic activities at telomeres including the SLX4 complex. TRF2 uses its TRFH domain as a docking site for several telomere accessory factors, including nucleases such as the SLX4-nuclease complex (12) and Apollo (9,10); and the helicases such as RTEL1 (8), and the RecQ helicases WRN and BLM (1). Most of these accessory proteins recruited by TRF2 are known for their roles in processing joint molecule DNA intermediates, and could be potential threats to telomere integrity as well, if left unchecked. To counteract this threat, TRF2 may negatively regulate the activities of these recruited factors. For example, as shown in Figure 4, using its telomeric DNA-binding ability, TRF2 limits endonuclease activity of the SLX4-nuclease complex on telomeric substrates in vitro. TRF2 has also been reported to use its DNA-binding ability to limit exonuclease activity of the WRN helicase (19), and its N-terminal basic domain to repress the endonucleases GEN1 (29), MUS81 and SLX1 (18) on telomeric substrates. Interestingly, it has been reported that in vivo, binding of TRF2 and its residence time at telomeres is highly dynamic (43). Whereas one fraction of TRF2 binds to telomeres transiently, another fraction binds to telomeres more stably. The transiently binding fraction of TRF2 is believed to be involved in telomere maintenance processes that require fast adaptation in recruiting and regulating nucleases and other DNA repair proteins to telomeres. The more stably residing fraction of TRF2 at telomeres is believed to be important for chromosome end protection. Thus, it is plausible that in vivo, a balance between these different effects of TRF2 ensures optimal but regulated nucleolytic processing of telomeric joint molecule intermediates (Supplementary Figure S4C). We speculate that factors contributing to this fine balance may involve cell cycle phase-dependent events such as variable association of these non-shelterin accessory factors with telomeres; phosphorylation-based regulation of enzymatic activity of these proteins (44); and control of telomeric nucleosome spacing and chromatin organization by TRF2 (45).
The second negative control mechanism we propose for the SLX4-nuclease complex activity at telomeres involves the RecQ family helicase BLM. It is important to note that BLM is believed to act on late telomere replication intermediates such as D-loops and HJs, which are also substrates for the SLX4-nuclease complex. Human cells lacking BLM have been shown to exhibit telomere defects (32). We considered if the telomeric instability observed in BLM deficient cells could be attributed, in part, to unregulated nucleolytic cleavage activity by the SLX4-nuclease complex. Our in vitro and in vivo evidence indicate that in the presence of both BLM and SLX1-SLX4, telomeric HR intermediates are preferentially processed by BLM. In addition, BLM-deficient human cells exhibit a marked increase in T-SCE and TC formation, both of which are suppressed by loss of SLX4 function (Figure 5). This suggests that under normal circumstances, human cells may employ BLM to dissolve telomeric HR intermediates. However, in the absence of functional BLM, the SLX4-nuclease complex compensates to nucleolytically resolve the remaining/persistent telomeric HR intermediates, resulting in increased T-SCEs and TC formation (Supplementary Figure S4C). It is noteworthy that telomeric localization of BLM (32) and the SLX4 complex (Figure 1) peak in late S phase, alluding to the importance of the helicase-dependent unwinding of T-loops and other telomeric joint molecules to serve as a negative regulatory mechanism to check SLX4-nuclease complex-dependent processing of telomeric intermediates. Besides BLM, the helicase activity of RTEL1 protein has been suggested to unwind G-quadruplexes and T-loops (8,46). Interestingly, however, RTEL1 has also been reported to impact telomere integrity in a telomerase-dependent manner in human cells (47). In addition, it is significant that non-telomeric roles of RTEL1 have been linked to diseased states. For example, the RING domain of RTEL1 was shown to be important for ribonucleotide protein trafficking between the nucleus and cytoplasm, defects in which have been suggested to contribute to the pathology of Hoyeraal–Hreidarsson syndrome, a severe form of dyskeratosis congenita (48,49).
In conclusion, in this study we provide novel insight into the specific elements of the SLX4 complex that are required for telomere replication and maintenance, and the mechanism and regulation of the same. We envision long telomeres facing greater telomere replication and processing challenges in terms of unusual intermediates. We thus propose that the SLX4 complex is a bona fide telomere-associated maintenance toolkit at long telomeres that in conjunction with other shelterin and non-shelterin accessory factors protects telomere integrity. We speculate that whenever long telomeres are generated/detected in a cell, the requirement for SLX4 increases at these longer telomeres. Hence we anticipate that in cells with a high frequency of long telomeres (e.g. hTERT-induced cells, germline, activated T-cells or ALT cells), the SLX4-nuclease complex may experience a functional and localization shift, at least partially, from being primarily a genomic repair toolkit to a telomere accessory maintenance complex. Furthermore, we predict that similar mechanisms of SLX4 function may be applicable to other inherently challenging genomic regions that frequently require resolution of joint DNA intermediates, such as CFS, for unhindered progression of normal DNA processing and DNA damage repair dynamics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr J. Wade Harper for the templates of SLX4 and its interacting nucleases; and Drs Robert Brosh and David Wilson for critical comments on the manuscript.
FUNDING
The Intramural Research Program of the National Institutes of Health, National Institute on Aging; the Ministry of Science and Technology of China [2013CB910402]; the National Natural Science Foundation of China [31330040]; the Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010201]. Funding for open access charge: the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Conflict of interest statement. None declared.
REFERENCES
- 1.Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.Doksani Y., Wu J.Y., de Lange T., Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesare A.J., Reddel R.R. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 5.Pickett H.A., Reddel R.R. The role of telomere trimming in normal telomere length dynamics. Cell Cycle. 2012;11:1309–1315. doi: 10.4161/cc.19632. [DOI] [PubMed] [Google Scholar]
- 6.Croteau D.L., Popuri V., Opresko P.L., Bohr V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vannier J.B., Sarek G., Boulton S.J. RTEL1: functions of a disease-associated helicase. Trends Cell Biol. 2014;24:416–425. doi: 10.1016/j.tcb.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Sarek G., Vannier J.B., Panier S., Petrini J.H., Boulton S.J. TRF2 recruits RTEL1 to telomeres in S phase to promote T-Loop unwinding. Mol. Cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Overbeek M., de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Lenain C., Bauwens S., Amiard S., Brunori M., Giraud-Panis M.J., Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Leon-Ortiz A.M., Svendsen J., Boulton S.J. Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair (Amst) 2014;19:152–162. doi: 10.1016/j.dnarep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Wan B., Yin J., Horvath K., Sarkar J., Chen Y., Wu J., Wan K., Lu J., Gu P., Yu E.Y., et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson J.S., Tejera A.M., Castor D., Toth R., Blasco M.A., Rouse J. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Rep. 2013;4:853–860. doi: 10.1016/j.celrep.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svendsen J.M., Smogorzewska A., Sowa M.E., O'Connell B.C., Gygi S.P., Elledge S.J., Harper J.W. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 16.Durkin S.G., Glover T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 17.Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saint-Leger A., Koelblen M., Civitelli L., Bah A., Djerbi N., Giraud-Panis M.J., Londono-Vallejo A., Ascenzioni F., Gilson E. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle. 2014;13:2469–2474. doi: 10.4161/cc.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opresko P.L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M.M., Bohr V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 20.Wang R.C., Smogorzewska A., de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Bucholc M., Park Y., Lustig A.J. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:6559–6573. doi: 10.1128/MCB.21.19.6559-6573.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webb C.J., Wu Y., Zakian V.A. DNA repair at telomeres: keeping the ends intact. Cold Spring Harb. Perspect. Biol. 2013;5:a012666. doi: 10.1101/cshperspect.a012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 24.Machwe A., Xiao L., Orren D.K. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 25.Popuri V., Hsu J., Khadka P., Horvath K., Liu Y., Croteau D.L., Bohr V.A. Human RECQL1 participates in telomere maintenance. Nucleic Acids Res. 2014;42:5671–5688. doi: 10.1093/nar/gku200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fairall L., Chapman L., Moss H., de Lange T., Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 27.Court R., Chapman L., Fairall L., Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouche N., Cesare A.J., Willcox S., Ozgur S., Compton S.A., Griffith J.D. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 29.Poulet A., Buisson R., Faivre-Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta-Lopez S., Bornet O., Guerlesquin F., Godet T., et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 31.Stavropoulos D.J., Bradshaw P.S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M.S. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 32.Barefield C., Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohaghegh P., Karow J.K., Brosh R.M. Jr, Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng S., Xiang T., Pandita T.K., Gonzalez-Suarez I., Gonzalo S., Harris C.C., Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calado R.T., Young N.S. Telomere diseases. N. Engl. J. Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taboski M.A., Sealey D.C., Dorrens J., Tayade C., Betts D.H., Harrington L. Long telomeres bypass the requirement for telomere maintenance in human tumorigenesis. Cell Rep. 2012;1:91–98. doi: 10.1016/j.celrep.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Y.L., Zhang F., Sun B., Du J., Sun C., Yuan J., Wang Y., Tao L., Kota K., Liu X., et al. Telomerase enzymatic component hTERT shortens long telomeres in human cells. Cell Cycle. 2014;13:1765–1776. doi: 10.4161/cc.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdun R.E., Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 39.Gilson E., Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 40.Chow T.T., Zhao Y., Mak S.S., Shay J.W., Wright W.E. Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 2012;26:1167–1178. doi: 10.1101/gad.187211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossiello F., Herbig U., Longhese M.P., Fumagalli M., d'Adda di Fagagna F. Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 2014;26:89–95. doi: 10.1016/j.gde.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Yang Y., van Overbeek M., Donigian J.R., Baciu P., de Lange T., Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 43.Mattern K.A., Swiggers S.J., Nigg A.L., Lowenberg B., Houtsmuller A.B., Zijlmans J.M. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell. Biol. 2004;24:5587–5594. doi: 10.1128/MCB.24.12.5587-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt H.D., Sarbajna S., Matos J., West S.C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Galati A., Magdinier F., Colasanti V., Bauwens S., Pinte S., Ricordy R., Giraud-Panis M.J., Pusch M.C., Savino M., Cacchione S., et al. TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS One. 2012;7:e34386. doi: 10.1371/journal.pone.0034386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vannier J.B., Pavicic-Kaltenbrunner V., Petalcorin M.I., Ding H., Boulton S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 47.Deng Z., Glousker G., Molczan A., Fox A.J., Lamm N., Dheekollu J., Weizman O.E., Schertzer M., Wang Z., Vladimirova O., et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3408–E3416. doi: 10.1073/pnas.1300600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballew B.J., Yeager M., Jacobs K., Giri N., Boland J., Burdett L., Alter B.P., Savage S.A. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum. Genet. 2013;132:473–480. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schertzer M., Jouravleva K., Perderiset M., Dingli F., Loew D., Le Guen T., Bardoni B., de Villartay J., Revy P., Londono-Vallejo A. Human regulator of telomere elongation helicase 1 (RTEL1) is required for nuclear and cytoplasmic trafficking of pre-U2 RNA. Nucleic Acids Res. 2015;18:1834–1847. doi: 10.1093/nar/gku1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


