Abstract
Convergent (or parallel) evolution provides strong evidence for a deterministic role of natural selection: similar phenotypes evolve when independent populations colonize similar environments. In reality, however, independent populations in similar environments always show some differences: some non-convergent evolution is present. It is therefore important to explicitly quantify the convergent and non-convergent aspects of trait variation, and to investigate the ecological and genetic explanations for each. We performed such an analysis for threespine stickleback (Gasterosteus aculeatus) populations inhabiting lake and stream habitats in independent watersheds. Morphological traits differed in the degree to which lake-stream divergence was convergent across watersheds. Some aspects of this variation were correlated with ecological variables related to diet, presumably reflecting the strength and specifics of divergent selection. Furthermore, a genetic scan revealed some markers that diverged between lakes and streams in many of the watersheds and some that diverged in only a few watersheds. Moreover, some of the lake-stream divergence in genetic markers was associated within some of the lake-stream divergence in morphological traits. Our results suggest that convergent evolution, and deviations from it, are primarily the result of natural selection, which corresponds in only some respect to the dichotomous habitat classifications frequently used in such studies.
Keywords: adaptive radiation, ecological speciation, natural selection, constraint, adaptive divergence, genomic, parallel evolution
INTRODUCTION
Independent populations that colonize similar environments and then evolve similar traits provide strong evidence for a deterministic response to natural selection (Endler 1986; Schluter 2000). The resulting pattern has been variously called “convergent” or “parallel” depending on similarity among the starting populations and on one's preferred definition (Futuyma 1986; Arendt and Reznick 2008; Losos 2011; Wake et al. 2011). We will here use the term convergent, although the key concepts apply also to parallel evolution in its various guises. Convergent evolution has been documented for a large number of taxa, with some classic examples from animals including Anolis lizard ecomorphs on different islands (Losos 2009), fishes in similar predation regimes in different rivers (Reznick et al. 1996; Langerhans and DeWitt 2004), fishes in benthic versus limnetic environments in different lakes (Schluter and McPhail 1992; McPhail 1993; Landry and Bernatchez 2010), fishes in freshwater versus marine environments (Bell and Andrews 1997), and many species living in a variety of caves (Jones et al. 1992). These and many other examples of convergent evolution have helped to build a general acceptance of natural selection as the primary driver of the evolution of biological diversity.
Although convergent evolution is commonly reported, it is not all pervasive. For example, independent “replicates” of a particular ecotype or ecomorph often show substantial differences in phenotype, as is clear in the above-cited studies and also in more recent work (Landry and Bernatchez 2010; Ożgo 2011; Romero 2011; Rosenblum and Harmon 2011). In more extreme cases, mean phenotypes of different populations in two environment types show considerable overlap: i.e., divergence is sometimes in the “wrong,” meaning unexpected, direction. These non-convergent aspects to evolution should not be surprising for a number of reasons that we here group into four broad categories: ecological, genetic, functional, and sexual. Although only the first two will be considered in the present study, we introduce them all so as to provide a general overview.
The ecological explanation for phenotypic non-convergence is that the classification of environments into discrete types (e.g., high versus low predation, benthic versus limnetic, cave versus surface) ignores selective factors that differ among replicates of a given environment “type.” This explanation can be evaluated by relating trait differences among populations to quantitative differences in important ecological variables, such as prey availability (e.g., Landry and Bernatchez 2010), diet (e.g., Schluter and McPhail 1992; Berner et al. 2008), or the type and abundance of predators (e.g., Endler 1978; Millar et al. 2006). For example, a given predation “regime” (high versus low) might differ among locations in the type and number of predators (Endler 1978; Reimchen 1994; Reznick et al. 1996), as well as in other environmental factors (Endler 1978; Grether et al. 2001).
The genetic explanation for phenotypic non-convergence is that variation among populations in genetic architecture can bias responses to selection. In some cases, this bias might make the difference between evolution toward alternative fitness peaks on a phenotypic adaptive landscape (Schluter 2000; Losos 2009). The specific genetic effects are several. First, different founding populations might have different genetic architectures that favour different evolutionary trajectories (Cohan and Hoffmann 1989; Schluter 1996; de Brito et al. 2005; Simoes et al. 2008). Second, evolution in different populations could occur via different new mutations at the same gene (e.g., Chan et al. 2010) or different genes (e.g., Steiner et al. 2009), either of which could have different phenotypic effects. Third, some populations might be influenced by genetic drift after their founding (Barton and Charlesworth 1984; de Brito et al. 2005). Fourth, replicate populations in a given environment might experience different levels of gene flow from populations in other environments (Hendry and Taylor 2004; Bolnick and Nosil 2007). Fifth, populations can differ in the extent to which phenotypes are plastic (Wund et al 2008), which could increase or decrease trait convergence beyond that expected from genotypes. These possibilities can be assessed by relating phenotypic differences to various types of genetic variation.
The main functional explanation for phenotypic non-convergence is that fitness is more closely related to integrated aspects of organismal “performance,” such as swimming speed or foraging ability, than it is to any of the traits that contribute to performance (Arnold 1983; Walker 2007; Irschick et al. 2008). Hence, when different trait combinations can generate equivalent performance, convergence in function need not entail convergence in traits (Alfaro et al. 2005; Wainwright et al. 2005). Indeed, this is one potential source of the alternative fitness peaks mentioned earlier (Schluter 2000; Losos 2009). This explanation for non-convergence can be assessed by relating performance variation to trait variation (e.g., Alfaro et al. 2004, 2005).
The sexual explanation for phenotypic non-convergence recognizes that some traits under natural selection are also influenced by sexual selection. These two aspects of selection sometimes can be closely aligned, in which case the contributions of sexual selection could simply reflect and enhance ecological differences. In other cases, however, sexual selection might diverge among populations for reasons not directly related to ecology, such as in sexual conflict (Chapman et al. 2003). The result could be divergence in traits in ways that are not easily predictable from ecological differences. This explanation for non-convergence can be assessed by relating phenotypic differences to measures of, or surrogates for, both natural and sexual selection (e.g., Svensson et al. 2006).
The above explanations for non-convergence are not mutually exclusive. Consequently, although convergent evolution is a strong hallmark of adaptation, non-convergence could reflect adaptation, neutrality, or mal-adaptation. The typical over-emphasis on convergent aspects of phenotypic evolution has thus led to missed opportunities for valuable insights into evolutionary process. Fortunately, recent studies that specifically consider non-convergence are begin to generate such insights (e.g. Berner et al. 2008; Landry and Bernatchez 2010; Rosenblum and Harmon 2011; Ożgo 2011).
We here explore both convergence and non-convergence through a study of threespine stickleback (Gasterosteus aculeatus). (It is typical in stickleback research to use the term parallel rather than convergent. We will continue to use convergent but the phenomenon is the same as that considered by previous workers to be parallel.) This species occupies a diversity of freshwater environments within many separate watersheds (reviews: Bell and Foster 1994; McKinnon and Rundle 2002), and a number of these situations represent independent radiations following colonization by similar marine ancestors (Lindsey 1962; Bell 1976; Withler and McPhail 1985; McPhail 1993; Thompson et al. 1997; Taylor and McPhail 2000; Berner et al. 2009). When colonizing the different environments, stickleback typically show dramatic adaptation in a number of phenotypic traits, such body shape, trophic morphology, and defensive armor (Bell 1974; Bell et al. 1985; Francis et al. 1986; Bell 1987; Schluter and McPhail 1992; Bell et al. 1993; Lavin and McPhail 1993; Bell and Orti 1994; Spoljaric and Reimchen 2007). Of particular relevance here, patterns of adaptation seemingly show aspects that are both convergent and non-convergent (Schluter and McPhail 1992; Hendry and Taylor 2004; Berner et al. 2008; Hendry et al. 2009).
The present study focuses on lake and stream stickleback from six watersheds on Vancouver Island, British Columbia, Canada. Taking advantage of these independent instances of divergence between lake and stream habitats, we quantify the extent and nature of divergence in ecology (diet), morphology (body shape, armor, and gill rakers), and genetics (microsatellite makers). Although all of our morphological data come from wild-caught individuals, we have reason to believe that the observed patterns have a strong genetic basis, as will be outlined below. In addition, we examine how these different aspects of variation are correlated with each other, with an eye toward informing the drivers of phenotypic divergence and something of its genetic basis.
MATERIAL AND METHODS
Populations and sampling
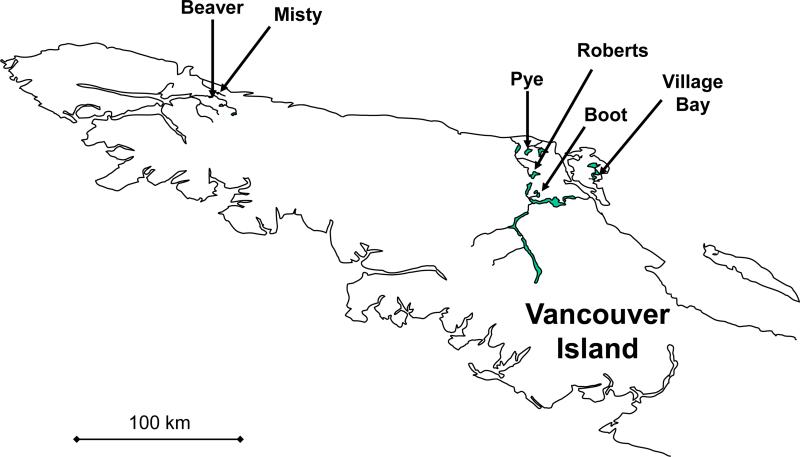
In May 2008, we collected threespine stickleback from paired lake and stream sites in each of six independent watersheds on Vancouver Island, British Columbia, Canada (12 sites in total). The specific sites (Fig. 1; Appendix S1) were chosen based on prior evidence of (1) strong lake-stream divergence in morphology, and (2) their independent origins from marine ancestors (Hendry and Taylor 2004; Berner et al. 2008, 2009). For each site, we used unbaited minnow traps to capture and retain 40 individuals (Appendix S1). These fish were euthanized with MS222, and their left side was photographed with a digital camera (Nikon D100). We then weighed each fish (±0.01 g) and preserved part of the pectoral fin in 95% ethanol. To enable isotope analyses (see below), white muscle tissue from the back of each fish was preserved on ice and later dried in an oven for 36 hours at 72 °C. Each fish was also dissected to determine sex and to remove the stomach for diet analysis. The stomach (and its contents) and the remaining carcass were preserved separately in 95% ethanol.
Figure 1.
Map of the studied watersheds on northern Vancouver Island, British Columbia, Canada. Other watersheds and tributaries are omitted for clarity.
Ecological divergence
We estimated ecological divergence between lake and stream fish based on their diet. This indirect approach to inferring divergent selection was chosen because comparisons of prey availability are compromised by the necessarily different sampling procedures in lakes versus streams. Our use of this indirect method is supported by studies showing strong associations between prey availability, diet, and morphological traits in lake stickleback (Gross and Anderson 1984; Schluter and McPhail 1992; Robinson 2000; Ingram et al. 2011). In short, diets are a good way of capturing the divergent selection pressures that ultimately drive morphological evolution in threespine stickleback (Schluter and McPhail 1992; Bolnick et al. 2008; Snowberg and Bolnick 2008; Berner et al. 2008, 2009). To consider short-term diets, food items from each stomach were categorized as either limnetic or benthic following Schluter and McPhail (1992). For each individual, we then estimated the “proportion of limnetic prey” (PLP) as the number of limnetic prey items divided by the total number of identified prey items (limnetic plus benthic) (Schluter and McPhail 1992). Following previous studies (Berner et al. 2008, 2009), we expected limnetic prey to be more important (relative to benthic prey) for lake populations that stream populations.
To consider long term diets and ecological niches (i.e., resource and habitat), we used stable isotopes (Post 2002; Newsome et al. 2007). We specifically estimated 1) the relative importance of different sources of primary production for each individual (δ13C), and 2) the trophic position of each individual (δ15N). This inferential method is commonly applied in studies of stickleback diet divergence (Bolnick et al. 2008; Reimchen et al. 2008; Snowberg and Bolnick 2008; Matthews et al. 2010). Weighed and dried muscle samples were sent to the UC Davis Stable Isotope Facility, where isotopes were measured with a PDZ Europa ANCA-GSL elemental analyzer, interfaced to a PDZ Europa 20-20 isotope ratio mass spectrometer (Sercon Ltd., Cheshire, UK). The resulting δ13C and δ15N values were expressed relative to international standards: PDB (PeeDee Belemnite) for carbon and air for nitrogen. Lipid often has a lower δ13C than do other biochemical tissues (Post et al. 2007; Logan et al. 2008), and so we lipid-corrected our δ13C values using Post et al. (2007, equation 3): δ 13Ccorreted = δ 13Cuntreated – 3.32 + 0.99 x C:N, where C:N is the ratio of the mass of carbon to nitrogen.
Using the isotope data, we estimated the trophic position of each individual and the mean trophic position for each population. We controlled for environmental sources of variation in carbon and nitrogen by analyzing snails and mussels and then applying the appropriate formula in Post (2002). Specifically, Tpos = λbase + [δ15Nfish – (αlittoral x δ15Nsnails + (1- αlittoral ) x δ15Nmussels)]/ΔN, where αlittoral is the proportion of littoral carbon for an individual ([δ13Cfish - δ13Cmussels]/[δ13Csnails - δ13Cmussels]), λbase is the trophic position of the baseline primary consumer (λbase = 2), and ΔN is the trophic fractionation (ΔN = 3.4‰, from Post 2002). In the few sites where mussels or snails were unavailable (mussels: Misty Inlet; snails: Misty Lake and Inlet and Beaver Lake), we applied a correction based on slopes and intercepts obtained from the other populations (see Appendix S2 for details). Given that trophic position is essentially a linear transformation, or “corrected” measure, of δ15N, we present the results for trophic position but not δ15N. Given the expectation of different stickleback diets in lakes and streams (see above), we expected consistent divergence in stable isotopes between lake and stream stickleback
Morphological divergence
We chose several morphological traits frequently subject to divergent selection between freshwater environments: body shape, armor traits (plates and spines), and trophic traits (gill raker number and length). Although plasticity can contribute to these traits (e.g., Sharpe et al. 2008; Wund et al. 2008), the patterns seen in wild-caught fish are expected on several grounds to have a strong genetic basis. First, three independent sets of experiments with stickleback from the Misty watershed have shown that lake-stream differences in body shape, spine length, and gill raker traits are almost perfectly maintained through multiple generations in the laboratory (Lavin and McPhail 1993; Hendry et al. 2002; Sharpe et al. 2008; Hendry et al. 2011; Berner et al. 2011). Second, a large genetic component for differences in these same traits has been documented in a number of other threespine stickleback comparisons, such as benthic versus limnetic (e.g. Hatfield 1997) and freshwater versus anadromous (e.g., Schluter et al. 2004). Third, QTL studies have found strong genetic determinants for population differences in these same traits (e.g., Peichel et al. 2001; Colosimo et al. 2004; Cresko et al. 2004; Shapiro et al. 2004; Albert et al. 2008). We will thus proceed under the assumption that among-population patterns have a strong genetic basis – while acknowledging that we cannot provide formal confirmation of this beyond the Misty system.
Body shape
Body shape often differs between lake and stream stickleback, probably because of divergent selection related to diet, foraging mode, and swimming performance. In lakes, stickleback commonly feed on limnetic prey in the open water (Berner et al. 2008, 2009), a situation where selection for sustained swimming should lead to the evolution of streamlined bodies (Webb 1984; Walker 1997). In streams, stickleback feed more commonly on benthic prey in complex environments (Berner et al. 2008, 2009), a situation where selection for high manoeuvrability should lead to the evolution of deeper bodies (Webb 1984; Walker 1997; Blake 2004; Hendry et al. 2011). These lake-stream differences in body shape have been confirmed in many studies (Reimchen et al. 1985; Lavin and McPhail 1993; Hendry et al. 2002; Hendry and Taylor 2004; Berner et al. 2008, 2009; Aguirre 2009), and so we also here expected consistent lake-stream divergence in body shape.
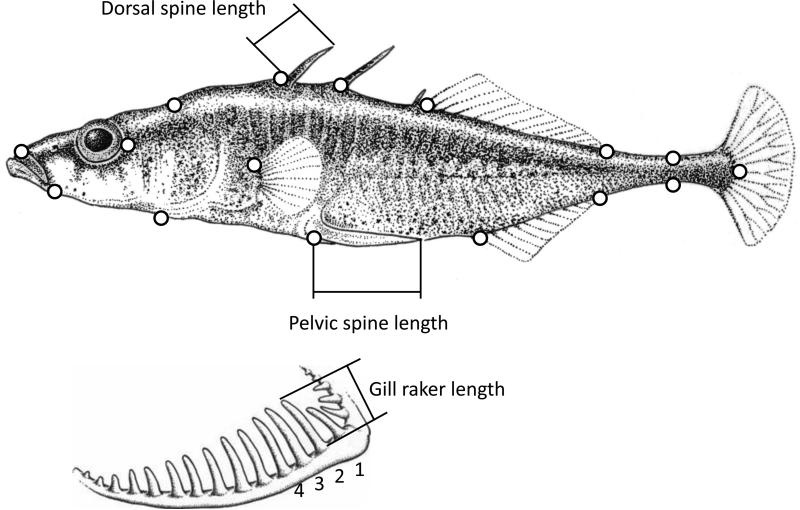
To analyze stickleback body shape, we used geometric morphometrics (e.g., Baumgartner et al. 1988; Walker 1997; Spoljaric and Reimchen 2007; Albert et al. 2008; Sharpe et al. 2008; Hendry et al. 2011). TpsDig software (Rohlf 2006) was used to place 16 landmarks onto the digital image of each fish (Fig. 2). These landmarks are the same as those used in Berner et al. (2009). TpsRelw (Rohlf 2005) was used to estimate the centroid size of each fish (a standardized measure of body size), as well as the affine (uniform) and non affine (partial warp) components of body shape (Zelditch et al. 2004). TpsRelw was also used to extract relative warps (principal components) of shape variation across all individuals in the study.
Figure 2.
Positions of landmarks used to quantify divergence in body shape between lake and stream stickleback. Also shown are the spine measurements. Drawing modified from Berner et al. (2008).
Armor traits
Spines and lateral plates are thought to differ among stickleback populations as a result of predation and a variety of other factors (Giles 1983; Reimchen and Nosil 2006; Reimchen et al. 2008). For instance, the presence of vertebrate predators is associated with longer spines and more numerous lateral plates (Moodie 1972; Hagen and Gilbertson 1972, 1973a, 1973b; Reimchen 1992, 1994). By contrast, a variety of other biotic and abiotic factors are associated with short spines and fewer later plates (Bergstrom 2002; Reimchen et al. 2008; Marchinko and Schluter 2007; Marchinko 2009; Myhre and Klepaker 2009). However, we did not expect consistent divergence between lake and stream fish, partly because predators do not differ in the same way between all lake and stream habitats (R. Kaeuffer, D. Bolnick, and A.P. Hendry, pers. obs.).
We counted lateral plates on the left side of each fish and used digital calipers to measure the lengths of the left pelvic spine and the first and second dorsal spines along the anterior side of the spine (Fig. 2). Each trait was measured three times, repeatability was high (correlation between pairs of measurements was ≥ 0.98); to further ensure accuracy, the mean of the three measurements was used for analysis.
Gill raker traits
Gill raker traits are closely related to diet: gill raker length and number are often lower for fishes foraging more often on benthic (macro-invertebrate) prey in littoral habitats than for fishes foraging more often on limnetic (zooplankton) prey in the open water of lakes (Bentzen and McPhail 1984). These differences are thought to arise because longer and more numerous gill rakers are better suited for feeding on smaller food items, whereas fewer and shorter gill rakers are better suited for feeding on larger food items (Hagen and Gilbertson 1972; Bentzen and McPhail 1984; Schluter and McPhail 1992; Bolnick 2004; Matthews et al. 2010). Given that lake and stream stickleback differ in their degree of limnetic versus benthic foraging, it is not surprising that they often show divergence in gill raker traits (Hendry and Taylor 2004; Berner et al. 2008, 2009). Based on this previous work, we expected consistent lake-stream divergence in gill raker traits, although previous work suggested it would be stronger for gill raker number than for gill raker length (Hendry and Taylor 2004; Berner et al. 2008, 2009).
Following Berner et al. (2008, 2009), we counted the gill rakers on the left gill arch and measured the length of the second to fourth gill raker from the epibranchial-ceratobranchial joint on the ceratobranchial. Each gill raker was measured three times at 45× magnification on a stereomicroscope with a micrometer (maximal precision of 0.01 mm). The mean was calculated from the three measurements per gill raker and across the three gill rakers. Each trait was measured three times, repeatability was very high (correlation between pairs of measurement was ≥ 0.98); to further ensure accuracy, the mean of the three measurements was used for analysis.
Trait standardization and divergence metrics
Some of the measured traits are correlated with body size, and so allometric size standardizations were applied. For body shape, this was done by including centroid size as a covariate in the analyses. For the other traits, this was done by calculating MSTD = M0 (63.53 / CS0)b (Reist 1986), where b is the common within-group slope from ANCOVA of log10 transformed trait size on log10 transformed centroid size, M is the measured trait size, CS is body (centroid) size, and 63.53 is the mean centroid size across all individuals in the data set. The response variables in ANCOVA were log10 transformed trait sizes, and the predictor variables were population, centroid size, and their interaction. The interactions were non-significant in all cases and so were removed to calculate b (Reist 1986).
Statistically, we first examined how the total morphological variation could be partitioned among effects of watershed, habitat type (lake or stream), body size (centroid), and all possible interactions. Habitat type and watershed were both fixed effects as this allowed more direct comparison of effect sizes and because the specific sites were chosen based on a priori information (see above.) The analysis involved two MANCOVAs, one for body shape and one for the linear measurements. For body shape, response variables were the two uniform components and the 16 partial warps. For linear measurements, response variables were gill raker size, gill raker number, pelvic spine length, and plate number. We then also used two-way ANOVA to estimate the effect of watershed and habitat on several key traits: relative warp 1 (RelW1) from the body shape data (more details below), gill raker number, gill raker length, plate number, dorsal and pelvic spine length. In all of these analyses, the traits were size-standardized as described above.
We next estimated the proportion of the variance among populations relative to the total variation. This estimate was similar to Qst (Lande 1992; Spitze 1993), but was based on phenotypic measurements and so is referred to as Pst. The calculation follows Pst = δ2GB /(δ2GB+δ2GW), where δ2GB and δ2GW are the between population and within population variance components for a given trait. Pst has particular value because it allows direct comparisons across traits and population types. First, it is directly analogous to the common genetic divergence metric Fst. Second, it is a unitless proportional measure and so is easy to compare across variables and populations with very different units, measurement scales, and levels of within-population variance. Estimation of variance components was done in R using the lmer function where the responses were the phenotypic traits and population was a random effect.
Genetic divergence
DNA was extracted from the fin samples using the Wizard® SV 96 Genomic DNA Purification System (Promega) kit. DNA concentration was then measured using a Nanodrop spectrophotometer and diluted to 4 ng·μl−1. To first identify candidate chromosomal regions that might be linked to phenotypic differences between lake and stream populations, we pooled the DNA of all individuals within a given population (12 pools) and screened for genetic variation at 192 microsatellite loci distributed across the stickleback genome (Peichel et al. 2001). From these screens, we selected twelve loci that showed little overlap in allele size distributions within at least two of the six population pairs (see Gow et al. 2006 for further explanation and rationale of this method). We then confirmed these hypothesized differences by genotyping eight individuals from each population at those loci. Six loci (Stn45, Stn168, Stn232, Stn246, Stn321, Stn386; Appendix S3) were found to have little overlap in allele size distributions and were then genotyped on all the individuals for all populations. Chromosome regions singled out in this type of scan are likely under divergent selection (Stinchcombe and Hoekstra 2008), but this method will not detect all such regions, particularly because of the relatively small number of loci (192). As a result, we consider this a first step to quantifying the genetic basis of divergence in lake-stream stickleback.
For ease of presentation, we refer to the above loci as “selected loci,” while remembering that they are probably not themselves under selection but rather could be linked to a chromosomal region under selection. We next established a null expectation for genetic divergence by screening all individuals at six microsatellite loci (Stn34, Stn67, Stn87, Stn159, Stn199, Stn234; Appendix S3) not tightly linked to any known quantitative trait locus (QTL) and thus likely to experience relatively weak selection. For ease of presentation, we refer to these loci as “neutral loci”, while remembering that it remains possible they might be linked to as yet unknown genes or QTL subject to disruptive selection.
For both selected and neutral loci, we used FSTAT (Goudet 1995) to estimate allelic richness for each population. We then used GENEPOP V.4 (Rousset 2008) to test each locus within each population for deviations from Hardy–Weinberg equilibrium (heterozygote deficiency), and to test each locus pair within each population for deviations from linkage disequilibrium. Using Genetix (Belkhir et al. 2004), we estimated average heterozygosity of the neutral loci in each population and the overall average for each watershed. Also, between the lake and stream populations within each watershed, we estimated the average θ (multi-locus Fst) across all neutral loci (Weir and Cockerham 1984). As a final conservative screen for those loci most likely under the influence of divergent selection between lake and stream populations, we examined whether θ for a given “selected” locus in a given watershed fell outside the 95 % CI for the average θ at neutral loci within that watershed. Despite the abundance of Fst outlier tests in the literature, the small number of loci used in this study precludes methods with more complex neutral models, such as Dfdist (Beaumont and Balding 2004).
Ecological, morphological, genetic correlations
The above analyses yielded, for each watershed, estimates of lake-stream divergence based on Pst for each of six morphological traits (RelW1, gill raker length and number, dorsal and pelvic spine length, and plate number) and Fst for each of 12 loci. To estimate a comparable ecological divergence metric, we estimated “Est” within each watershed as the proportion of variance in three ecological variables (proportion of limnetic prey, δ13C, and trophic position) attributable to differences between lake and stream environments. As traditionally used, these variance ratios (Pst, Fst, and here also Est) do not indicate the direction of divergence. We therefore added a sign to Pst and Est to indicate whether the direction of divergence within a given lake-stream pair was the same as (positive) or the opposite of (negative) the typical direction of divergence across watersheds. For example, gill rakers are generally more numerous in lake than stream fish, and so Pst values for pairs diverging in that (versus the other) direction would be positive (versus negative).
We next calculated the correlation across watersheds (N = 6 data points for each correlation) between each measure of ecological divergence and each measure of morphological divergence (18 correlations), each measure of ecological divergence and each measure of genetic divergence (36 correlations), and each measure of morphological divergence and each measure of genetic divergence (72 correlations). With 126 correlations to consider, each with relatively low power, tests of statistical significance are questionable and corrections for multiple comparisons are not feasible. We therefore treat these correlations not as hypothesis tests, but rather as an investigative tool to identify promising associations that could be the subject of future research. We suggest that a reasonable way to select these associations is to consider those where at least half of the variance in one could be explained by the other (r2 = 0.5, |r| = 0.707107). For completeness, we do also report individual P values for these correlations.
RESULTS
Ecological divergence
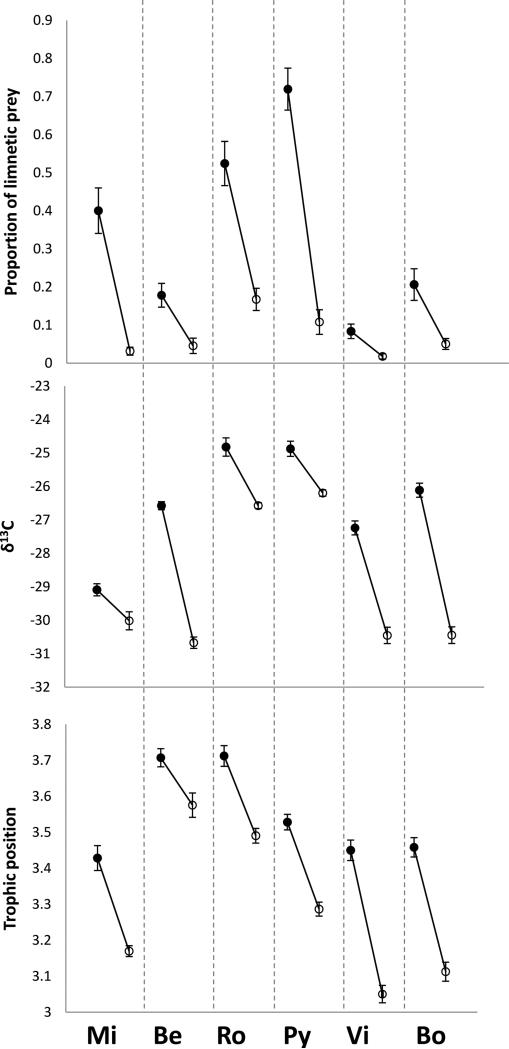
All ecological variables differed among watersheds and between lake and stream habitats, with a significant interaction between watershed and habitat (Table 1; Fig. 3). In each case, at least 50% of the total variation could be explained jointly by these three terms, with the strongest contributions from the watershed and habitat main effects (Table 1). Relative to stream stickleback, lake stickleback ate more limnetic prey, had lower δ13C, and had a higher trophic position. The interaction term was not as strong because lake-stream divergence in these ecological variables was in the same direction in all watersheds (Table 1; Fig. 3). Instead, any deviations from convergent patterns of divergence were the result of variation among watersheds in the magnitude of lake-stream differences.
Table 1.
Watershed, habitat, and interaction effects on ecological and morphological variables in general linear models. For morphological traits correlated with body size (gill raker length and dorsal and pelvic spine lengths), results are for allometrically standardized trait sizes. Estimates of variance explained are the proportion of the total variance based on sums of squares (SS): SS effect / SS total.
| Variables | Watershed | Habitat | Interaction | Total Var. explained | |
|---|---|---|---|---|---|
| Proportion of limnetic prey | F | F5.449 = 7.829 | F1.449 = 9.257 | F5.449 = 4.012 | 0.48 |
| Var. Explained | 0.18 | 0.21 | 0.09 | ||
| P | < 0.001 | < 0.001 | < 0.001 | ||
| δ13C | F | F5.449 = 1085.06 | F1.449= 796.07 | F5.449 = 205.91 | 0.75 |
| Var. Explained | 0.39 | 0.28 | 0.07 | ||
| P | < 0.001 | < 0.001 | < 0.001 | ||
| Trophic position | F | F5.449 = 10.794 | F1.449 = 8.3 | F5.449 = 0.8666 | 0.63 |
| Var. Explained | 0.34 | 0.26 | 0.03 | ||
| P | < 0.001 | < 0.001 | < 0.001 | ||
| Relative warp 1a | F | F5.458 = 20.495 | F1.458 = 549.778 | F5.458 = 10.316 | 0.61 |
| Var. Explained | 0.09 | 0.47 | 0.04 | ||
| P | < 0.001 | < 0.001 | < 0.001 | ||
| Gill raker length | F | F5.458 = 9.106 | F1.458= 10.698 | F5.458 = 11.811 | 0.20 |
| Var. Explained | 0.08 | 0.02 | 0.10 | ||
| P | < 0.001 | 0.001 | < 0.001 | ||
| Gill raker number | F | F5.431= 13.709 | F1.431 = 259.986 | F5.431 = 19.128 | 0.50 |
| Var. Explained | 0.08 | 0.30 | 0.11 | ||
| P | < 0.001 | < 0.001 | < 0.001 | ||
| Pelvic spine length | F | F5.458 = 6.335 | F1.458 = 0.470 | F5.458 = 6.783 | 0.13 |
| Var. Explained | 0.06 | 0.00 | 0.06 | ||
| P | < 0.001 | 0.493 | < 0.001 | ||
| Dorsal spine length | F | F5.458 = 8.284 | F1.458 = 0.103 | F5.458 = 7.977 | 0.15 |
| Var. Explained | 0.08 | 0.00 | 0.07 | ||
| P | < 0.001 | 0.749 | < 0.001 | ||
| Plate number | F | F5.457 = 26.469 | F1.457 = 0.282 | F5.457 = 35.273 | 0.40 |
| Var. Explained | 0.17 | 0.00 | 0.23 | ||
| P | < 0.001 | 0.595 | < 0.001 | ||
When including centroid size as a covariate, the full model explains 71% of the variation and all terms were significant. The variance explained by the habitat and watershed terms did not change, but variation explained by the watershed*habitat interaction was reduced to 2.7%. In addition, centroid size explained 11% of the variation and the centroid size by watershed interaction explained 1% of the variation.
Figure 3.
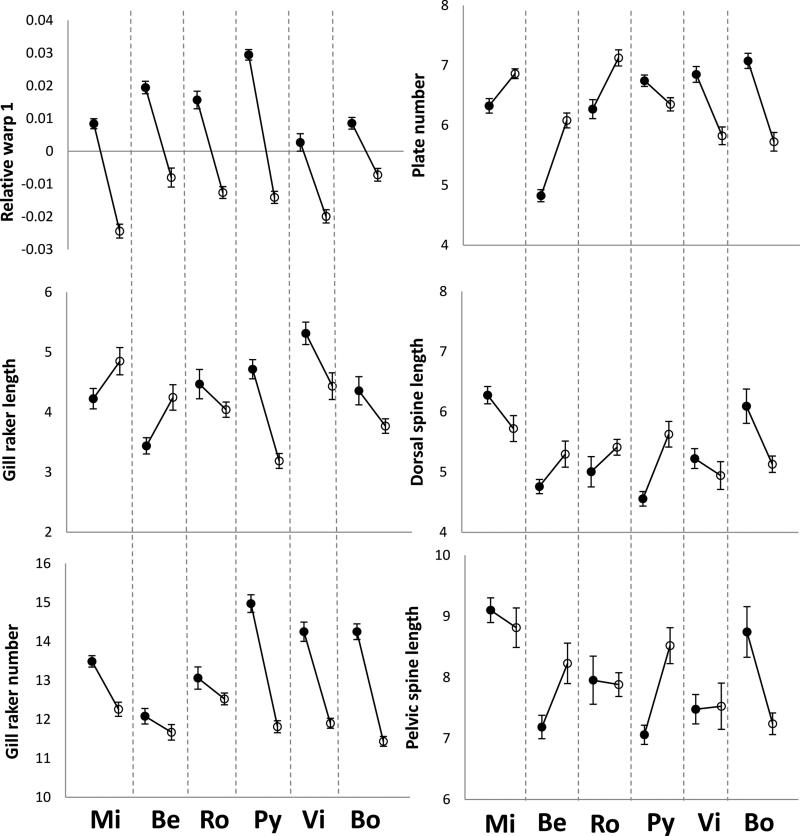
Means and standard errors of the ecological variables for each lake (filled circle) and stream (open circle) site in each watershed (Mi = Misty, Be = Beaver, Ro = Roberts, Py = Pye, Vi = Village Bay, Bo = Boot).
Although the diet and trophic position data are easily interpretable in the context of our earlier expectations, the δ13C data require further explanation. In particular, previous studies within lakes documented a lower δ13C for limnetic than benthic environments (France 1995; Bolnick et al. 2008; Snowberg and Bolnick 2008; Mathews et al. 2010), whereas we documented higher δ13C in lakes (more limnetic) as compared to streams (more benthic). The likely reason for this difference is that water flow increases the availability of CO2 to benthic algae, which may increase fractionation of 13CO2 (Calder and Parker 1973; Pardue et al. 1976; Finlay et al. 1999). In short, the direction of δ13C divergence in our study could be an effect of water velocity (Finlay et al. 1999) and/or food type (Bolnick et al. 2008), but we are unable to disentangle these alternatives at present. Thus, although we report results for δ13C, we do not make much of their interpretation.
Morphological divergence
Significant watershed, habitat, and watershed-by-habitat interactions were present in the multivariate analyses (Table 2). For the analysis combining univariate traits, the three predictor variables (watershed, habitat, and interaction) explained roughly similar amounts of variation. For the analysis using geometric morphometric body shape variables, the watershed term explained about twice as much of the variation as did the habitat term and the watershed-by-habitat interaction. Overall, much more of the total variation could be explained for geometric morphometric body shape than for the univariate traits. We next describe the results trait-by-trait, given that different traits had different expectations.
Table 2.
Results of multivariate analysis of variance (MANCOVA) for linear measurements (gill raker number and size, spines length and plate number: size standardized as needed), and total body shape (all relative warps and uniform components combined). Partial variance explained is η2 = SS effect / [SS effect + SS error] = 1- Wilk's lambda, as appropriate for multivariate analyses (Langerhans and DeWitt 2004). Partial variance explained can sum to greater than 100%. In a MANCOVA, η2 is also the effect size.
| Effects | Wilk's lambda | F | d.f. | P value | Partial variance explained (%) | |
|---|---|---|---|---|---|---|
| Linear measurements | Body (centroid) size | 0.923 | 7.46 | 5,448 | <0.001 | 7.7 |
| Habitat | 0.916 | 8.258 | 5,448 | <0.001 | 8.4 | |
| Watershed | 0.891 | 10.915 | 5,448 | <0.001 | 10.8 | |
| Habitat*Size | 0.915 | 8.341 | 5,448 | <0.001 | 8.5 | |
| Watershed*Size | 0.898 | 10.141 | 5,448 | <0.001 | 10.2 | |
| Habitat*Watershed | 0.911 | 8.78 | 5,448 | <0.001 | 8.9 | |
| Habitat*Watershed* Size | 0.909 | 8.919 | 5,448 | <0.001 | 9.1 | |
| Total body shape | Body (centroid) size | 0.537 | 13.4 | 28, 435 | <0.001 | 46.3 |
| Habitat | 0.751 | 5.154 | 28, 435 | <0.001 | 24.9 | |
| Watershed | 0.548 | 12.788 | 28, 435 | <0.001 | 45.1 | |
| Habitat*Size | 0.756 | 5.005 | 28, 435 | <0.001 | 24.3 | |
| Watershed*Size | 0.539 | 13.292 | 28, 435 | <0.001 | 46.1 | |
| Habitat*Watershed | 0.779 | 4.413 | 28, 435 | <0.001 | 22.1 | |
| Habitat*Watershed*Size | 0.776 | 4.472 | 28, 435 | <0.001 | 22.3 | |
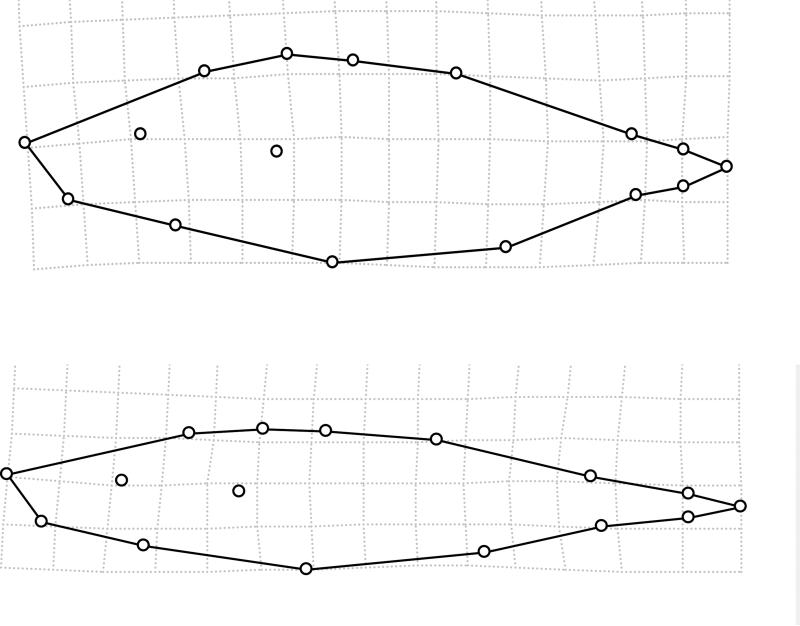
The first relative warp for body shape (RelW1) explained 27.9% of total body shape variance, and higher scores were associated with shallower bodies along the entire length of the body (Fig. 4). This relative warp was very similar to that extracted in other studies of lake-stream stickleback (e.g., Sharpe et al. 2008; Berner et al. 2009, 2010b; 2011). The other relative warps explained considerably less of the variation (RelW2 = 15.5%, RelW3 = 11.7%, and RelW4 = 9.3%) and did not have clear functional interpretations. These relative warps are not analyzed further. RelW1 differed strongly among watersheds and between lake and stream habitats, with a significant interaction (Table 1). The full model explained 61% of the variance, and habitat was by far the most important effect (47% of the total variance). Specifically, lake fish have shallower bodies than do stream fish across and within all watersheds (Table 3; Figs. 4 and 5). The small interaction effect (4%) is the result of differences among systems only in the magnitude of lake-stream divergence. For instance, divergence in body depth was greatest in the Pye system and least in the Boot system (Table 3; Fig. 5).
Figure 4.
Variation in body depth captured by relative warp 1, with a lower score (more stream-like) shown on top and a higher score (more lake-like) shown on the bottom. The illustrated deformations represent the maximum (Pye Lake) and minimum (Village Bay Inlet Stream) population averages (see Fig. 5).
Table 3.
Pst for morphological traits and Est for ecological variables between lake and stream populations within each watershed. Negative signs indicate divergence opposite to the most common direction between lakes and streams.
| Watershed | Proportion limnetic prey |
δ13C | Trophic position |
Relative warp 1 |
Gill raker length |
Gill raker number |
Pelvic spine length |
Dorsal spine length |
Plate number |
|---|---|---|---|---|---|---|---|---|---|
| Misty | 0.299 | 0.089 | 0.375 | 0.682 | −0.049 | 0.297 | 0.00 | −0.047 | −0.136 |
| Beaver | 0.128 | 0.838 | 0.105 | 0.451 | −0.111 | 0.016 | 0.08 | 0.049 | −0.443 |
| Roberts | 0.284 | 0.333 | 0.338 | 0.500 | 0.019 | 0.027 | 0.00 | 0.014 | −0.173 |
| Pye | 0.531 | 0.278 | 0.477 | 0.797 | 0.412 | 0.652 | 0.185 | 0.185 | 0.073 |
| Village Bay | 0.108 | 0.557 | 0.589 | 0.356 | 0.093 | 0.467 | 0.00 | 0.000 | 0.245 |
| Boots | 0.129 | 0.688 | 0.509 | 0.302 | 0.047 | 0.635 | −0.112 | −0.094 | 0.356 |
Figure 5.
Means and standard errors of the morphological traits for each lake (filled circle) and stream (open circle) population in each watershed (Mi = Misty, Be = Beaver, Ro = Roberts, Py = Pye, Vi = Village Bay, Bo = Boot).
Gill raker traits differed among watersheds and between lake and stream habitats, with a significant interaction in each case (Tables 1 and 3; Fig. 5). These terms explained much more of the total variation for gill raker number than for gill raker length, possibly because the latter tends to be one of the most variable traits within populations (Berner et al. 2008, 2010a). On average, lake fish had more and longer gill rakers than did stream fish, with the direction of divergence always being the same for gill raker number but not for gill raker length (Table 3; Fig. 5). Thus, for gill raker number, convergence in lake-stream divergence was high and the interaction term was the result of variation in the magnitude of lake-stream divergence (highest for Pye, Village, and Boot; Table 3; Fig. 5). For gill raker length, however, convergence was very weak, and the significant interaction term reflected differences among watersheds in both the magnitude and direction of divergence (Table 3; Fig. 5).
The three armor traits differed among watersheds, but consistent differences were not evident between lake and stream habitats, and the lake by habitat interaction was significant in each case (Table 1). Not much of the total variation could be explained by these terms for pelvic or dorsal spine lengths, but 40% could be explained for lateral plate number (Table 1). The importance of watershed and the interaction were roughly equivalent for each of the traits. Convergence was thus very low for armor traits, with the direction of lake-stream divergence frequently differing among watersheds (Table 3; Fig. 5).
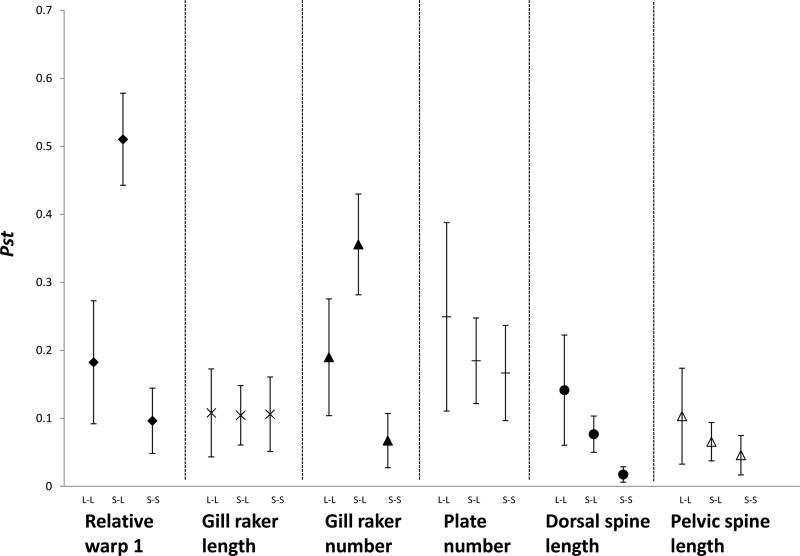
The magnitude (here ignoring the direction) of phenotypic divergence can be qualitatively compared among traits and contrasts (lake-lake, lake-stream, stream-stream) by reference to average pair-wise Pst values (Table 3; Fig. 6). First, gill raker number and RelW1 showed the greatest lake-stream divergence: 2.8 times higher than lake-lake and 5.3 times higher than stream-stream divergence. Second, armor traits showed relatively high lake-lake divergence, whereas gill raker length showed similarly low divergence in all three contrasts. Third, stream-stream divergence for each trait was lower than lake-lake or lake-stream divergence. Overall, then, morphological traits that show the highest level of convergence in lake-stream divergence (gill raker number and RelW1, as described above) also showed the greatest overall magnitude of lake-stream divergence.
Figure 6.
Averages of all pairwise Pst (and 95 % CI) values between lake-lake (L-L), stream-lake (S-L) and stream-stream (S-S) comparisons.
Genetic divergence
Average lake-stream Fst at the six “neutral” loci ranged from 0.045 (Roberts) to 0.192 (Beaver), with an average of 0.108 (Table 4). By contrast, average lake-stream Fst at the “selected” loci identified by our scan (Stn45, Stn168, Stn232, Stn246, Stn321, Stn386) ranged from 0.071 (Village Bay) to 0.395 (Boot), with an average of 0.218 (Table 4). The six neutral markers only rarely showed evidence of departures from Hardy-Weinberg equilibrium: 9 out of 76 possible tests were individually significant, involving four different loci in six different populations. Similarly, only 17 out of 180 tests for linkage disequilibrium were individually significant, and these were not associated with particular locus pairs or populations. By contrast, 29 out of 76 possible tests for departures from Hardy-Weinberg equilibrium were individually significant for the selected loci, further supporting the ongoing action of selection. In particular, Stn321 and Stn246 showed strong departures from Hardy-Weinberg equilibrium in 8 and 9 populations, respectively. Similar to neutral loci, however, only 18 of 180 tests for linkage disequilibrium were individually significant for the selected loci, which is expected given that these loci are on different linkage groups (Appendix S3).
Table 4.
Fst for each “selected” locus (see text) between lake and stream populations within each watershed. Average values and 95% confidence intervals (CI) are shown for both selected and neutral loci. Also indicated are Fst values that are higher (in bold) or lower (in italics) than the 95% CI from Fst values based on neutral loci.
| Selected locus | Misty | Beaver | Roberts | Pye | Village Bay | Boot |
|---|---|---|---|---|---|---|
| Stn168 | 0.476 | 0.337 | 0.084 | 0.149 | 0.048 | 0.306 |
| Stn232 | 0.388 | 0.202 | 0.026 | 0.180 | 0.027 | 0.285 |
| Stn246 | 0.172 | 0.223 | 0.152 | 0.184 | 0.029 | 0.595 |
| Stn321 | 0.323 | 0.356 | 0.084 | 0.664 | 0.250 | 0.662 |
| Stn386 | 0.137 | 0.162 | 0.078 | 0.155 | 0.071 | 0.471 |
| Stn45 | 0.152 | 0.106 | 0.124 | 0.137 | 0.026 | 0.062 |
| Average selected | 0.275 | 0.225 | 0.095 | 0.246 | 0.071 | 0.395 |
| 95% CI | 0.182-0.38 | 0.158-0.297 | 0.061-0.127 | 0.15-0.417 | 0.031-0.141 | 0.214-0.553 |
| Average neutral | 0.121 | 0.192 | 0.045 | 0.069 | 0.046 | 0.178 |
| 95% CI | 0.059 - 0.181 | 0.083 - 0.331 | 0.029 - 0.063 | 0.041 - 0.098 | 0.018 - 0.084 | 0.080 - 0.302 |
Comparisons of Fst at selected loci to the distribution of Fst at neutral loci (Table 4) yielded several key observations. First, two of the selected loci exhibited high Fst in most watersheds: Stn321 in all six watersheds and Stn168 in five watersheds. The other selected loci appeared to be under divergent selection in only two or three watersheds each. Second, some watersheds (especially Pye) showed evidence of divergent selection at multiple loci, whereas others showed evidence of divergent selection at only one or two loci (i.e. Village Bay or Beaver). Note, however, that this approach to confirming divergent selection is limited in situations where neutral-locus Fst values are already very high (e.g., Beaver).
Ecology-morphology-genetic correlations
As noted earlier, the correlations identified here do not represent hypothesis testing but rather hypothesis generation. Five ecology-morphology correlations were identified (Table 5; Appendix S4). First, stickleback from watersheds with greater lake-stream divergence in diet (PLP) also showed greater lake-stream divergence in body depth (RelW1) and gill raker number. Second, stickleback from watersheds with greater lake-stream divergence in δ13C showed lower divergence in body depth (RelW1), although this is difficult to interpret in light of the previously mentioned effects of stream flow on δ13C values. Finally, stickleback from watersheds with greater lake-stream divergence in trophic position showed greater divergence in gill raker number and lateral plate number. Three morphology-genetic correlations were identified. Two of these correlations involved selected loci. First, stickleback from watersheds with greater lake-stream divergence in Stn321 showed greater divergence in gill raker number. Second, stickleback from watersheds with greater lake-stream divergence in Stn45 showed greater divergence in body depth (RelW1). Four ecology-genetic correlations were identified, and one of these involved a selected locus. Specifically, stickleback from watersheds with greater lake-stream divergence in diet (PLP) had greater divergence in Stn45.
Table 5.
The strongest observed correlations between ecological, morphological, and genetic variables. The coefficients calculated take into account the direction of lake-stream divergence in each system, as indicated in Table 3. Locus names with an asterisk are expected to be neutral.
| Pairs of variables | Coefficient | P value | ||
|---|---|---|---|---|
| Ecology vs. Morphology | Trophic position | Plate number | 0.936 | 0.0059 |
| Proportion limnetic prey | Relative warp 1 | 0.922 | 0.0089 | |
| Trophic position | Gill raker number | 0.810 | 0.0504 | |
| δ13C | Relative warp 1 | −0.746 | 0.0883 | |
| Proportion limnetic prey | Gill raker length | 0.738 | 0.0937 | |
| Morphology vs. Genetics | Stn45 | Relative warp 1 | 0.830 | 0.0409 |
| Stn199* | Plate number | −0.791 | 0.0609 | |
| Stn321 | Gill raker number | 0.782 | 0.0662 | |
| Ecology vs. Genetics | Stn67* | Trophic position | −0.846 | 0.0339 |
| Stn199* | Trophic position | −0.813 | 0.0491 | |
| Stn95* | Trophic position | −0.732 | 0.0981 | |
| Stn45 | Proportion of limnetic prey | 0.718 | 0.1077 | |
DISCUSSION
We quantified the extent to which divergence between lake and stream stickleback within watersheds was convergent (or parallel) versus non-convergent (or non-parallel) across those watersheds. Ecological variables related to foraging (diet and stable isotopes) generally showed a reasonable degree of convergence, with more benthic diets in streams than in lakes: 21-28% of the total diet variation among all individuals could be explain by the lake-stream contrast. Morphological traits showed varying degrees of convergence, ranging from very low (~0% by the above index for armor traits) to very high (47% for body depth, RelW1), and some the non-convergence could be explained by ecological variables. (Although these patterns were based on wild-caught fish, previous work suggests they have a strong genetic basis – see Methods.) Finally, we found varying degrees of similarity in lake-stream divergence at genetic markers, some of which was associated with divergence in morphological traits.
Convergent traits
Several lines of evidence indicate that lake-stream divergence for body depth and gill raker number are strongly convergent. First, lake-stream divergence was in the same direction in all watersheds (Table 3; Fig. 5). Second, the habitat term explained a substantial amount of the total variation among all individuals: 47% for body depth and 30% for gill raker number. Third, the habitat term was considerably more important than the watershed or interaction terms (Table 1). The strong convergence shown by these traits is consistent with previous work on lake and stream populations, and is thought to reflect natural selection related to foraging mode and diet (Reimchen et al. 1985; Lavin and McPhail 1993; Walker 1997; Hendry and Taylor 2004; Berner et al. 2008, 2009; Aguirre 2009). Our analysis further supports this interpretation because lake-stream divergence in both body depth and gill raker number was positively associated with lake-stream divergence in diet as assayed by stomach contents and stable isotopes (Tables 3 and 5).
Despite generally high convergence for these two traits, some non-convergence was still present and calls for explanation. In particular, both the watershed and interaction terms were highly significant (4-11% of the total variation), and at least 39% of the variation remains unexplained (Table 1). The interaction term reflects differences among watersheds in the extent to which stream fish have fewer gill rakers and deeper bodies than do lake fish, and these differences are likely the result of differences in the strength of divergent selection related to diet. Supporting this contetion, lake-stream divergence in body depth was closely associated with lake-stream divergence in the proportion of limnetic prey (r = 0.922) and lake-stream divergence in gill raker number was closely associated with lake-stream divergence in trophic position (r = 0.810). These associations parallel those reported in other studies of lake-stream divergence in stickleback (Berner et al. 2008), benthic-limnetic divergence in stickleback (Schluter and McPhail 1992), and benthic-limnetic divergence in other fishes (Landry and Bernatchez 2010). As for the unexplained variation, this likely arises because both habitats, but particularly lakes, have both limnetic and benthic resources, which maintains variation in foraging traits through disruptive selection and individual specialization (Berner et al. 2008; Snowberg and Bolnick 2008; Bolnick and Paull 2009; Berner et al. 2010a). In short, the non-convergence for generally convergent traits can be explained by variation in the ecological factors thought to influence divergent selection.
Although we have not yet deployed the full power of genomic tools available for stickleback, our initial scan suggests some interesting patterns of variation at the genetic level. Specifically, some marker loci showed high divergence in multiple lake-stream pairs, whereas others showed high divergence in only a few pairs (Table 4). In the first case, Stn321 was considerably more divergent than neutral loci in all six lake-stream pairs, and divergence at this locus has also been recorded for another lake-stream pair (Bolnick et al. 2009). In addition, Stn168 was highly divergent in five watersheds. In the second case, shared patterns of genetic divergence were less evident for Stn246 (three watersheds) and Stn232, Stn45, and Stn386 (two watersheds each). These results suggest that divergent selection between lakes and streams drives divergence in some similar and some different genetic regions. Note, however, that our analysis targeted only loci showing signs of high lake-stream divergence in at least two watersheds (see Methods).
Some of the above genetic markers are likely associated with the traits we measured because (1) some are linked to QTL identified in other studies (Appendix S3), and (2) some showed levels of lake-stream divergence that were associated with lake-stream divergence in morphological traits (Table 5). Most notably, Stn45 has been associated with QTL for body shape (Albert et al. 2008), and its divergence here appeared to be positively correlated with divergence in body depth (r = 0.830) and diet (r = 0.718). (The latter correlation was probably indirect, reflecting the association of diet with body depth and body depth with the marker.) In addition, Stn321 has been associated with QTL for lateral plate number and body shape (Colosimo et al. 2004; Albert et al. 2008), and its divergence hear appeared to be positively correlated with divergence in gill raker number (r = 0.782). Whether or not this last result reflects a novel genetic association, pleiotropy, or simply coincidence remains to be seen, but it does suggest the value of rapid genetic screens such as ours for suggesting novel associations between genotype and phenotype. Our results also highlight more generally the fact that markers identified in linkage mapping studies of particular population pairs are sometimes associated with divergence between population pairs in other contexts (see also Hohenlohe et al. 2010). That the same QTL appear to be important in multiple environmental contrasts suggests that adaptation in general at least sometimes draws on a shared genetic tool kit.
Larger-scale genomic studies are required to comprehensively assess associations between genetic and phenotypic divergence in the lake-stream stickleback system. Any such associations could arise in two ways: (1) differences in genetic architecture could cause phenotypic divergence to deviate from that expected solely under divergent selection; or (2) the strength of divergent selection jointly determines the extent to which both phenotypic and genetic divergence proceeds. The first (bias) possibility invokes the “genetic explanation for phenotypic non-convergence” mentioned in the Introduction. The second (no bias) possibility is that phenotypic divergence proceeds as dictated by selection, and genetic divergence occurs to the corresponding extent. We cannot conclusively separate these two alternatives, but suggest the second is more likely given that morphological divergence was so predictable based solely on divergence in the relevant ecological variables.
Non-convergent traits
Lake-stream divergence in armor traits and gill raker length showed very low convergence: the habitat term explained essentially none of the variation (Table 1) and the direction of lake-stream divergence differed among watersheds (Fig. 5). For armor traits, differences among populations generally arise owing to differences in selective factors such as predation (Hagen and Gilbertson 1972; Gross 1977; Reimchen 1994; Marchinko and Schluter 2007; Marchinko 2009) and ionic concentration (Giles 1983; Reimchen and Nosil 2006). Presumably reflecting the same factors, we too found strong among-population variation in armor traits. This variation was sometimes manifest as divergence between lake and stream populations within a watershed – but not in a consistent direction or magnitude across watersheds. This result confirms and extends the previous observation of Hendry and Taylor (2004) that “relative to stream fish, lake fish have shorter pelvic spines and more lateral plates in the Mayer and Drizzle watersheds (Moodie 1972; Reimchen et al. 1985) but longer pelvic spines and fewer lateral plates in the Misty watershed (Lavin and McPhail 1993; Hendry et al. 2002).” We suggest that selection from predators does sometimes differ between lakes and streams – just not in a consistent fashion. In short, strong trait convergence is only expected when selective regimes are very similar.
To test the above hypothesis, we would ideally quantify the difference in predation between lakes and streams and relate this to differences in armor traits. This test is not yet possible, owing to the difficulty of accurately assessing predator-based selection in nature. Interestingly, however, lake-stream divergence in one armor trait (lateral plates) appeared positively correlated with lake-stream divergence in trophic position (r = 0.936). Perhaps this association arose because trophic position reflects ecological niche (e.g., benthic versus limnetic) and different ecological niches cause different exposure to predators. For instance, Reimchen et al. (2008) documented associations between trophic position and plate number within lakes. Another possibility is that the different foraging environments require different aspects of swimming performance (Hendry et al. 2011), which is influenced by plate number (Bergstrom 2002; Hendry et al. 2011).
In contrast to body depth and gill raker number, we found no suggestive associations between lake-stream divergence in armor traits and lake-stream divergence in putatively selected genetic markers: i.e., markers showing lake-stream divergence in multiple watersheds (see Methods). We did, however, find a suggestive but puzzling negative association between lake-stream divergence at a putatively neutral marker (Stn199) and lake-stream divergence in lateral plate number (r = −0.791). Perhaps this is just a spurious association resulting from the large number of tests – or perhaps Stn199 is linked to an undiscovered QTL. Additional work will be needed to test this possibility. As discussed above for convergent traits, any such association would likely reflect the action of selection, rather than the “genetic explanation for non-convergence” – because divergence in both lateral plates and Stn199 was seemingly associated with ecological divergence (trophic position; Table 5).
Non-convergence was not surprising for armor traits, but it might be for gill raker length. Lake stickleback have more limnetic diets than do stream stickleback, and so might be expected to have longer gill rakers (Bentzen and McPhail 1984; Schluter and McPhail 1992; Bolnick 2004; Berner et al. 2008; Matthews et al. 2010; Berner et al. 2010b). In the present study, however, the opposite was true for the Misty and Beaver watersheds (Fig. 5), as was also found in previous samples from these watersheds (Berner et al. 2008) and from other lake-stream pairs (Berner et al. 2010b). More research is clearly needed on how selection acts on gill raker length in stickleback.
Conclusions
Convergence (or parallelism) was strong for body depth and gill raker number: 47% and 30% of the total phenotypic variation among all individuals could be explained by the simple lake-stream habitat classification (Table 1), and divergence was always in the same direction. These patterns are likely genetic (see Methods), which implies that selection has played a predominant role in the shaping the morphological patterns. We can therefore conclude that when selection is strong and consistently divergent, the relevant traits diverge in a consistent and predictable fashion. Most studies stop with the above positive assertion, but it is just as important to understand aspects of non-convergence. First, convergence was not overwhelming even for the above traits. Instead, understanding the degree of lake-stream divergence required quantitative measures of relevant ecological variables: watersheds with greater lake-stream divergence in diets showed greater-lake stream divergence in body depth and gill raker number. Second, armor traits and gill raker length did not diverge consistently between lake and stream environments, and this variation could not be explained by the measured ecological variables. These traits are certainly sometimes under divergent selection, just not consistently so between lakes and streams. We can therefore infer that divergent selection often involves multiple ecological variables that do not always map easily onto a simple habitat contrast, such as lake versus stream, benthic versus limnetic, or high predation versus low predation.
We also provided an initial assessment of the extent to which lake-stream genetic divergence was similar across watersheds, and the extent to which genetic and morphological divergence were correlated. We found some instances of the same marker diverging in multiple watersheds, particularly for markers putatively associated with QTL for the traits we measured. However, we also found some evidence of markers diverging strongly in only some watersheds, suggesting that unique aspects of genetic divergence are involved in adaptive divergence in different watersheds. However, these conclusions are preliminary, owing to the need for a denser array of markers and a more objective screen for genomic divergence. We hope that our analysis, along with other recent studies (e.g., Berner et al. 2008; Landry and Bernatchez 2010; Rosenblum and Harmon 2011; Ożgo 2011), reinforces the value of quantifying the degree of trait convergence and non-convergence, and then exploring ecological and genetic correlates thereof.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J.S. Moore for his help in the field and Western Forest Products for accommodation in Port McNeil. We also thank Simon Barrette, Camille Macnaugthon, and Shahin Muttalib for their help in morphological measurements, and the entire Peichel lab for their help with the genetic analyses. Daniel Berner had many insightful comments on the manuscript and gave permission to use his stickleback drawings. Additional improvements were made based on comments y Mike Bell and five other referees. This work was financially supported by an FQRNT postdoctoral fellowship (RK), an NIH grant P50 HG002568 (CLP), a David and Lucille Packard Foundation fellowship (DIB), the Howard Hughes Medical Institute (DIB), and NSERC (APH).
BIBLIOGRAPHY
- Aguirre WE. Microgeographical diversification of threespine stickleback: body shape-habitat correlations in a small, ecologically diverse Alaskan drainage. Biol. J. Linn. Soc. 2009;98:139–151. [Google Scholar]
- Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Alfaro ME, Bolnick DI, Wainwright PC. Evolutionary dynamics of complex biomechanical systems: an example using the four-bar mechanism. Evolution. 2004;58:495–503. [PubMed] [Google Scholar]
- Alfaro ME, Bolnick DI, Wainwright PC. Evolutionary consequences of many-to-one mapping of jaw morphology to mechanics in labrid fishes. Am. Nat. 2005;165:E140–E154. doi: 10.1086/429564. [DOI] [PubMed] [Google Scholar]
- Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Arnold SJ. Morphology, performance and fitness. Am. Zool. 1983;23:347–361. [Google Scholar]
- Barton NH, Charlesworth B. Genetic revolutions, founder effects, and speciation. Annu. Rev. Ecol. Syst. 1984;15:133–164. [Google Scholar]
- Baumgartner JV, Bell MA, Weinberg PH. Body form differences between the Enos Lake species pair of threespine sticklebacks (Gasterosteus aculeatus complex). Can. J. Zool. 1988;66:467–474. [Google Scholar]
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Bell MA. Reduction and loss of the pelvic girdle in Gasterosteus (Pisces): a case of parallel evolution. Natur. Hist. Mus. Los Angeles Co. Contrib. Sci. 1974;257:1–36. [Google Scholar]
- Bell MA. Evolution of phenotypic diversity in the Gasterosteus aculeatus superspecies on the Pacific coast of North America. Syst. Zool. 1976;25:211–227. [Google Scholar]
- Bell MA. Interacting evolutionary constraints in pelvic reduction of threespine sticklebacks, Gasterosteus aculeatus (Pisces: Gasterosteidae). Biol. J. Linn. Soc. 1987;31:347–382. [Google Scholar]
- Bell MA, Francis RC, Havens AC. Pelvic reduction and its directional asymmetry in threespine sticklebacks from the Cook Inlet region, Alaska. Copeia. 1985;1985:437–444. [Google Scholar]
- Bell MA, Ortí G, Walker JA, Koenings JP. Evolution of pelvic reduction in threespine stickleback fish: a test of competing hypotheses. Evolution. 1993;47:906–914. doi: 10.1111/j.1558-5646.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Bell MA, Ortí G. Pelvic reduction in threespine stickleback from Cook Inlet lakes: geographical distribution and intrapopulation variation. Copeia. 1994;1994:314–325. [Google Scholar]
- Bell MA, Andrews CA. Evolutionary consequences of colonization of fresh water by primitively anadromous fishes. In: Streit B, Städler T, Lively CM, editors. Evolutionary ecology of freshwater animals. Current Trends and Case Studies; Birkhäuser, Basel, Switzerland: 1997. pp. 323–363. [Google Scholar]
- Bell MA, Foster SA. The evolutionary biology of the threespine stickleback. Oxford University Press; Oxford, UK: 1994. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations., Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. Université de Montpellier II; Montpellier (France): 2004. [Google Scholar]
- Bentzen P, McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): specialization for alternative trophic niches in the Enos Lake species pair. Can. J. Zool. 1984;62:2280–2286. [Google Scholar]
- Bergstrom CA. Fast-start swimming performance and reduction in lateral plate number in threespine stickleback. Can. J. Zool. 2002;80:207–213. [Google Scholar]
- Berner D, Adams DC, Grandchamp A-C, Hendry AP. Natural selection drives patterns of lake-stream divergence in stickleback foraging morphology. J. Evol. Biol. 2008;21:1653–1665. doi: 10.1111/j.1420-9101.2008.01583.x. [DOI] [PubMed] [Google Scholar]
- Berner D, Grandchamp A-C, Hendry AP. Variable progress toward ecological speciation in parapatry: stickleback across eight lake-stream transitions. Evolution. 2009;63:1740–1753. doi: 10.1111/j.1558-5646.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- Berner D, Stutz WE, Bolnick DI. Foraging trait (co)variances in stickleback evolve deterministically and do not predict trajectories of adaptive diversification. Evolution. 2010a;64:2265–2277. doi: 10.1111/j.1558-5646.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- Berner D, Roesti M, Hendry AP, Salzburger W. Constraints on speciation suggested by comparing lake-stream stickleback divergence across two continents. Mol. Ecol. 2010b;19:4963–4978. doi: 10.1111/j.1365-294X.2010.04858.x. [DOI] [PubMed] [Google Scholar]
- Berner D, Kaeuffer R, Grandchamp A-C, Raeymaekers JAM, Räsänen K, Hendry AP. Quantitative genetic inheritance of morphological divergence in a lake–stream stickleback ecotype pair: implications for reproductive isolation. J. Evol. Biol. 2011 doi: 10.1111/j.1420-9101.2011.02330.x. In Press. [DOI] [PubMed] [Google Scholar]
- Blake RW. Fish functional design and swimming performance. J. Fish Biol. 2004;65:1193–1222. [Google Scholar]
- Bolnick DI. Can intraspecific competition drive disruptive selection? An experimental test in natural populations of sticklebacks. Evolution. 2004;58:608–618. [PubMed] [Google Scholar]
- Bolnick DI, Nosil P. Natural selection in populations subject to a migration load. Evolution. 2007;61:2229–2243. doi: 10.1111/j.1558-5646.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Caldera EJ, Matthews B. Evidence for asymmetric migration load in a pair of ecologically divergent stickleback populations. Biol. J. Linn. Soc. 2008;94:273–287. [Google Scholar]
- Bolnick DI, Lau OL. Predictable patterns of disruptive selection in stickleback in postglacial lakes. Am. Nat. 2008;172:1–11. doi: 10.1086/587805. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Paull J. Diet similarity declines with morphological distance between conspecific individuals. Evol. Ecol. Res. 2009;11:1217–1233. [Google Scholar]
- Bolnick DI, Snowberg LK, Patenia C, Stutz WE, Ingram T, Lau OL. Phenotype-dependent native habitat preference facilitates divergence between parapatric lake and stream stickleback. Evolution. 2009;63:2004–2016. doi: 10.1111/j.1558-5646.2009.00699.x. [DOI] [PubMed] [Google Scholar]
- Calder JA, Parker PL. Geochemical implications of induced changes in c-13 fractionation by blue-green-algae. Geochim. Cosmochim. Acta. 1973;37:133–140. [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends Ecol. Evol. 2003;18:41–47. [Google Scholar]
- Cohan FM, Hoffmann AA. Uniform selection as a diversifying force in evolution - evidence from Drosophila. Am. Nat. 1989;134:613–637. [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2:635–641. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, Bell MA, Kimmel CB, Postlethwait JH. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. P. Natl. Acad. Sci. USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brito RA, Pletscher LS, Cheverud JM. The evolution of genetic architecture. I. Diversification of genetic backgrounds by genetic drift. Evolution. 2005;59:2333–2342. doi: 10.1554/05-151.1. [DOI] [PubMed] [Google Scholar]
- Endler JA. A predator's view of animal color patterns. Evol. Biol. 1978;11:319–364. [Google Scholar]
- Endler JA. Natural selection in the wild. Princeton University Press; Princeton, NJ.: 1986. [Google Scholar]
- Finlay JC, Power ME, Cabana G. Effects of water velocity on algal carbon isotope ratios: Implications for river food web studies. Limnol. Oceanogr. 1999;44:1198–1203. [Google Scholar]
- France RL. Differentiation between littoral and pelagic food webs in lakes using stable carbon isotopes. Limnol. Oceanogr. 1995;40:1310–1313. [Google Scholar]
- Francis RC, Baumgartner JV, Havens AC, Bell MA. Historical and ecological sources of variation among lake populations of threespine sticklebacks, Gasterosteus aculeatus, near Cook Inlet, Alaska. Can. J. Zool. 1986;64:2257–2265. [Google Scholar]
- Futuyma DJ. Evolutionary biology. Sinauer Associates; Sunderland, MA.: 1986. [Google Scholar]
- Giles N. The possible role of environmental calcium levels during the evolution of phenotypic diversity in Outer Hebridean populations of three-spined stickleback, Gasterosteus aculeatus. J. Zool. 1983;199:535–544. [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995;86:485–486. [Google Scholar]
- Gow JL, Peichel CL, Taylor EB. Contrasting hybridization rates between sympatric three-spined sticklebacks highlight the fragility of reproductive barriers between evolutionarily young species. Mol. Ecol. 2006;15:739–752. doi: 10.1111/j.1365-294X.2006.02825.x. [DOI] [PubMed] [Google Scholar]
- Grether GF, Millie DF, Bryant MJ, Reznick DN, Mayea W. Rain forest canopy cover, resource availability, and life history evolution in guppies. Ecology. 2001;82:1546–1559. [Google Scholar]
- Gross HP. Adaptive trends of environmentally sensitive traits in three-spined stickleback, Gasterosteus aculeatus L. Z. zool. Syst. Evolut.-forsch. 1977;15:252–278. [Google Scholar]
- Gross HP, Anderson JM. Geographic-variation in the gillrakers and diet of European threespine sticklebacks, Gasterosteus aculeatus. Copeia. 1984;1984:87–97. [Google Scholar]
- Hagen DW, Gilbertson LG. Geographic variation and environmental selection in Gasterosteus aculeatus L. in the Pacific Northwest, America. Evolution. 1972;26:32–51. doi: 10.1111/j.1558-5646.1972.tb00172.x. [DOI] [PubMed] [Google Scholar]
- Hagen DW, Gilbertson LG. Selective predation and the intensity of selection acting upon lateral plates of threespine sticklebacks. Heredity. 1973a;30:273–287. [Google Scholar]
- Hagen DW, Gilbertson LG. The genetics of plate morphs in freshwater threespine sticklebacks. Heredity. 1973b;31:75–84. [Google Scholar]
- Hatfield T. Genetic divergence in adaptive characters between sympatric species of stickleback. Am. Nat. 1997;149:1009–1029. doi: 10.1086/286036. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Taylor EB. How much of the variation in adaptive divergence can be explained by gene flow? - An evaluation using lake-stream stickleback pairs. Evolution. 2004;58:2319–2331. doi: 10.1111/j.0014-3820.2004.tb01606.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Bolnick DI, Berner D, Peichel CL. Along the speciation continuum in sticklebacks. J. Fish Biol. 2009;75:2000–2036. doi: 10.1111/j.1095-8649.2009.02419.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Taylor EB, McPhail JD. Adaptive divergence and the balance between selection and gene flow: lake and stream stickleback in the Misty system. Evolution. 2002;56:1199–1216. doi: 10.1111/j.0014-3820.2002.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Hudson K, Walker JA, Räsänen K, Chapman L. Genetic divergence in morphology-performance mapping between Misty Lake and inlet stickleback. J. Evol. Biol. 2011;24:23–35. doi: 10.1111/j.1420-9101.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e10000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram T, Stutz WE, Bolnick DI. Does intraspecific size variation in a predator affect its diet diversity and top-down control of prey? PLoS ONE. 2011;6:e20782. doi: 10.1371/journal.pone.0020782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irschick DJ, Meyers JJ, Husak JF, Le Galliard JF. How does selection operate on whole-organism functional performance capacities? A review and synthesis. Evol. Ecol. Res. 2008;10:177–196. [Google Scholar]
- Jones R, Culver DC, Kane TC. Are parallel morphologies of cave organisms the result of similar selection pressures? Evolution. 1992;46:353–365. doi: 10.1111/j.1558-5646.1992.tb02043.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Neutral theory of quantitative genetic variance in an island model with local extinction and colonization. Evolution. 1992;46:381–389. doi: 10.1111/j.1558-5646.1992.tb02046.x. [DOI] [PubMed] [Google Scholar]
- Landry L, Bernatchez L. Role of epibenthic resource opportunities in the parallel evolution of lake whitefish species pairs (Coregonus sp.). J. Evol. Biol. 2010;23:2602–2613. doi: 10.1111/j.1420-9101.2010.02121.x. [DOI] [PubMed] [Google Scholar]
- Langerhans RB, DeWitt TJ. Shared and unique features of evolutionary diversification. Am. Nat. 2004;164:335–349. doi: 10.1086/422857. [DOI] [PubMed] [Google Scholar]
- Lavin PA, McPhail JD. Parapatric lake and stream sticklebacks on northern Vancouver Island - disjunct distribution or parallel evolution. Can. J. Zool. 1993;71:11–17. [Google Scholar]
- Lindsey CC. Experimental study of meristic variation in a population of threespine sticklebacks, Gasterosteus aculeatus. Can. J. Zool. 1962;40:271–312. [Google Scholar]
- Logan JM, Jardine TD, Miller TJ, Bunn SE, Cunjak RA, Lutcavage ME. Lipid corrections in carbon and nitrogen stable isotope analyses: comparison of chemical extraction and modelling methods. J. Anim. Ecol. 2008;77:838–846. doi: 10.1111/j.1365-2656.2008.01394.x. [DOI] [PubMed] [Google Scholar]
- Losos JB. Lizards in an evolutionary tree: ecology and adaptive radiation of Anoles. University of California Press; Berkeley, CA.: 2009. [Google Scholar]
- Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65 doi: 10.1111/j.1558-5646.2011.01289.x. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- Marchinko KB. Predation's role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution. 2009;63:127–138. doi: 10.1111/j.1558-5646.2008.00529.x. [DOI] [PubMed] [Google Scholar]
- Marchinko KB, Schluter D. Parallel evolution by correlated response: Lateral plate reduction in threespine stickleback. Evolution. 2007;61:1084–1090. doi: 10.1111/j.1558-5646.2007.00103.x. [DOI] [PubMed] [Google Scholar]
- Matthews B, Marchinko KB, Bolnick DI, Mazumder A. Specialization of trophic position and habitat use by sticklebacks in an adaptive radiation. Ecology. 2010;91:1025–1034. doi: 10.1890/09-0235.1. [DOI] [PubMed] [Google Scholar]
- McKinnon JS, Rundle HD. Speciation in nature: the threespine stickleback model systems. Trends Ecol. Evol. 2002;17:480–488. [Google Scholar]
- McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): origin of the species pairs. Can. J. Zool. 1993;71:515–523. [Google Scholar]
- Millar NP, Reznick DN, Kinnison MT, Hendry AP. Disentangling the selective factors that act on male colour in wild guppies. Oikos. 2006;113:1–12. [Google Scholar]
- Moodie GEE. Predation, natural selection and adaptation in an unusual threespine stickleback. Heredity. 1972;28:155–167. [Google Scholar]
- Myhre F, Klepaker T. Body armour and lateral-plate reduction in freshwater three-spined stickleback Gasterosteus aculeatus: adaptations to a different buoyancy regime? J. Fish Biol. 2009;75:2062–2074. doi: 10.1111/j.1095-8649.2009.02404.x. [DOI] [PubMed] [Google Scholar]
- Newsome SD, del Rio CM, Bearhop S, Phillips DL. A niche for isotopic ecology. Front. Ecol. Environ. 2007;5:429–436. [Google Scholar]
- Ożgo M. Rapid evolution in unstable environments: a success story of the polymorphic land snail Cepaea nemoralis (Gastropoda: Pulmonata). Biol. J. Linn. Soc. 2011;102:251–262. [Google Scholar]
- Pardue JW, Scalan RS, Vanbaalen C, Parker PL. Maximum carbon isotope fractionation in photosynthesis by blue-green-algae and a green-alga. Geochim. Cosmochim. Acta. 1976;40:309–312. [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, et al. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Post DM. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology. 2002;83:703–718. [Google Scholar]
- Post DM, Layman CA, Arrington DA, Takimoto G, Quattrochi J, Montana CG. Getting to the fat of the matter: models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia. 2007;152:179–189. doi: 10.1007/s00442-006-0630-x. [DOI] [PubMed] [Google Scholar]
- Reimchen TE. Injuries on stickleback from attacks by a toothed predator (Oncorhynchus) and implications for the evolution of lateral plates. Evolution. 1992;46:1224–1230. doi: 10.1111/j.1558-5646.1992.tb00631.x. [DOI] [PubMed] [Google Scholar]
- Reimchen TE. Predators and morphological evolution in threespine stickleback. In: Bell MA, Foster SA, editors. The evolutionary biology of the threespine stickleback. Oxford University Press; Oxford, UK: 1994. pp. 240–273. [Google Scholar]
- Reimchen TE, Ingram T, Hansen SC. Assessing niche differences of sex, armour and asymmetry phenotypes using stable isotope analyses in Haida Gwaii sticklebacks. Behaviour. 2008;145:561–577. [Google Scholar]
- Reimchen TE, Nosil P. Replicated ecological landscapes and the evolution of morphological diversity among Gasterosteus populations from an archipelago on the west coast of Canada. Can. J. Zool. 2006;84:643–654. [Google Scholar]
- Reimchen TE, Stinson EM, Nelson JS. Multivariate differentiation of parapatric and allopatric populations of threespine stickleback in the Sangan River watershed, Queen Charlotte Islands. Can. J. Zool. 1985;63:2944–2951. [Google Scholar]
- Reist JD. An empirical-evaluation of coefficients used in residual and allometric adjustment of size covariation. Can. J. Zool. 1986;64:1363–1368. [Google Scholar]
- Reznick DN, Rodd FH, Cardenas M. Life-history evolution in guppies (Poecilia reticulata: Poeciliidae).4. Parallelism in life-history phenotypes. Am. Nat. 1996;147:319–338. [Google Scholar]
- Robinson BW. Trade offs in habitat-specific foraging efficiency and the nascent adaptive divergence of sticklebacks in lakes. Behaviour. 2000;137:865–888. [Google Scholar]
- Rohlf FJ. tpsRelw. 2005 http://life.bio.sunysb.edu/morph/
- Rohlf FJ. tpsDIG2. 2006 http://life.bio.sunysb.edu/morph/
- Romero A. The evolution of cave life. Am. Sci. 2011;99:144–151. [Google Scholar]
- Rousset F. GENEPOP ' 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Res. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Harmon LJ. “Same same but different”: replicated ecological speciation at White Sands. Evolution. 2011 doi: 10.1111/j.1558-5646.2010.01190.x. doi:10.1111/j.1558-5646.2010.01190.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50:1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS. Parallel evolution and inheritance of quantitative traits. Am. Nat. 2004;163:809–822. doi: 10.1086/383621. [DOI] [PubMed] [Google Scholar]
- Schluter D, McPhail JD. Ecological character displacement and speciation in sticklebacks. Am. Nat. 1992;140:85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Sharpe DMT, Rasanen K, Berner D, Hendry AP. Genetic and environmental contributions to the morphology of lake and stream stickleback: implications for gene flow and reproductive isolation. Evol. Ecol. Res. 2008;10:849–866. [Google Scholar]
- Simoes P, Santos J, Fragata I, Mueller LD, Rose MR, Matos M. How repeatable is adaptive evolution? The role of geographical origin and founder effects in laboratory adaptation. Evolution. 2008;62:1817–1829. doi: 10.1111/j.1558-5646.2008.00423.x. [DOI] [PubMed] [Google Scholar]
- Snowberg LK, Bolnick DI. Assortative mating by diet in a phenotypically unimodal but ecologically variable population of stickleback. Am. Nat. 2008;172:733–739. doi: 10.1086/591692. [DOI] [PubMed] [Google Scholar]
- Spitze K. Population-structure in Daphnia obtusa - quantitative genetic and allozymic variation. Genetics. 1993;135:367–374. doi: 10.1093/genetics/135.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoljaric MA, Reimchen TE. 10 000 years later: evolution of body shape in Haida Gwaii three-spined stickleback. J. Fish Biol. 2007;70:1484–1503. [Google Scholar]
- Steiner CC, Rompler H, Boettger LM, Schoneberg T, Hoekstra HE. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol. Biol. Evol. 2009;26:35–45. doi: 10.1093/molbev/msn218. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Svensson EI, Eroukhmanoff F, Friberg M. Effects of natural and sexual selection on adaptive population divergence and premating isolation in a damselfly. Evolution. 2006;60:1242–1253. [PubMed] [Google Scholar]
- Taylor EB, McPhail JD. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proc. R. Soc. Lond. B. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CE, Taylor EB, McPhail JD. Parallel evolution of lake-stream pairs of threespine sticklebacks (Gasterosteus) inferred from mitochondrial DNA variation. Evolution. 1997;51:1955–1965. doi: 10.1111/j.1558-5646.1997.tb05117.x. [DOI] [PubMed] [Google Scholar]
- Wainwright PC, Alfaro ME, Bolnick DI, Hulsey CD. Many-to-one mapping of form to function: a general principle of organismal design. Integr. Comp. Biol. 2005;45:256–262. doi: 10.1093/icb/45.2.256. [DOI] [PubMed] [Google Scholar]
- Wake DB, Wake MH, Specht CD. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science. 2011;331:1032–1035. doi: 10.1126/science.1188545. [DOI] [PubMed] [Google Scholar]
- Walker JA. Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L. (Gasterosteidae) body shape. Biol. J. Linn. Soc. 1997;61:3–50. [Google Scholar]
- Walker JA. A general model of functional constraints on phenotypic evolution. Am. Nat. 2007;170:681–689. doi: 10.1086/521957. [DOI] [PubMed] [Google Scholar]
- Webb PW. Body form, locomotion and foraging in aquatic vertebrates. Am. Zool. 1984;24:107–120. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population-structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Withler RE, McPhail JD. Genetic variability in freshwater and anadromous sticklebacks (Gasterosteus aculeatus) of southern British Columbia. Can. J. Zool. 1985;63:528–533. [Google Scholar]
- Wund MA, Baker JA, Clancy B, Golub JL, Foster SA. A test of the “flexible stem” model of evolution: ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. Am. Nat. 2008;172:449–462. doi: 10.1086/590966. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric morphometrics for biologists: a primer. Elsevier Academic Press; New York: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.