Abstract
Mutations in Isocitrate dehydrogenase 1 (IDH1) and IDH2 are among the most common genetic alterations in intrahepatic cholangiocarcinoma (IHCC), a deadly liver cancer1–5. Mutant IDH proteins in IHCC and other malignancies acquire an abnormal enzymatic activity allowing them to convert alpha-ketoglutarate (αKG) to 2-hydroxyglutarate (2HG), which inhibits the activity of multiple αKG-dependent dioxygenases, and results in alterations in cell differentiation, survival, and extracellular matrix maturation6–10. However, the molecular pathways by which IDH mutations lead to tumour formation remain unclear. Here we show that mutant IDH blocks liver progenitor cells from undergoing hepatocyte differentiation through the production of 2HG and suppression of HNF4α, a master regulator of hepatocyte identity and quiescence. Correspondingly, genetically engineered mouse models (GEMMs) expressing mutant IDH in the adult liver show aberrant response to hepatic injury, characterized by HNF4α silencing, impaired hepatocyte differentiation and markedly elevated levels of cell proliferation. Moreover, mutant IDH and activated Kras, genetic alterations that co-exist in a subset of human IHCCs4,5, cooperate to drive the expansion of liver progenitor cells, development of premalignant biliary lesions, and progression to metastatic IHCC. These studies provide a functional link between IDH mutations, hepatic cell fate, and IHCC pathogenesis, and present a novel GEMM of IDH-driven malignancy.
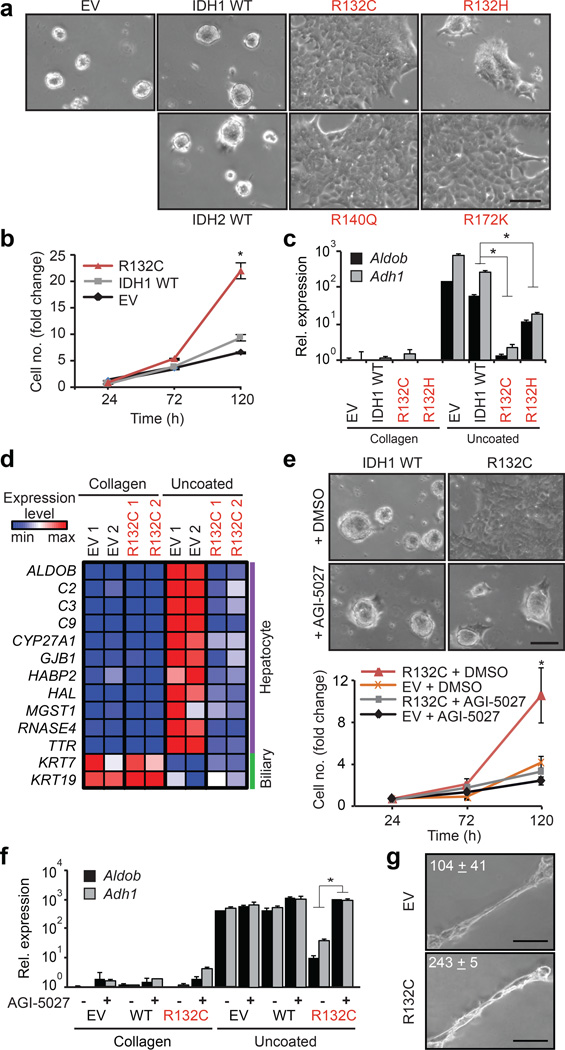
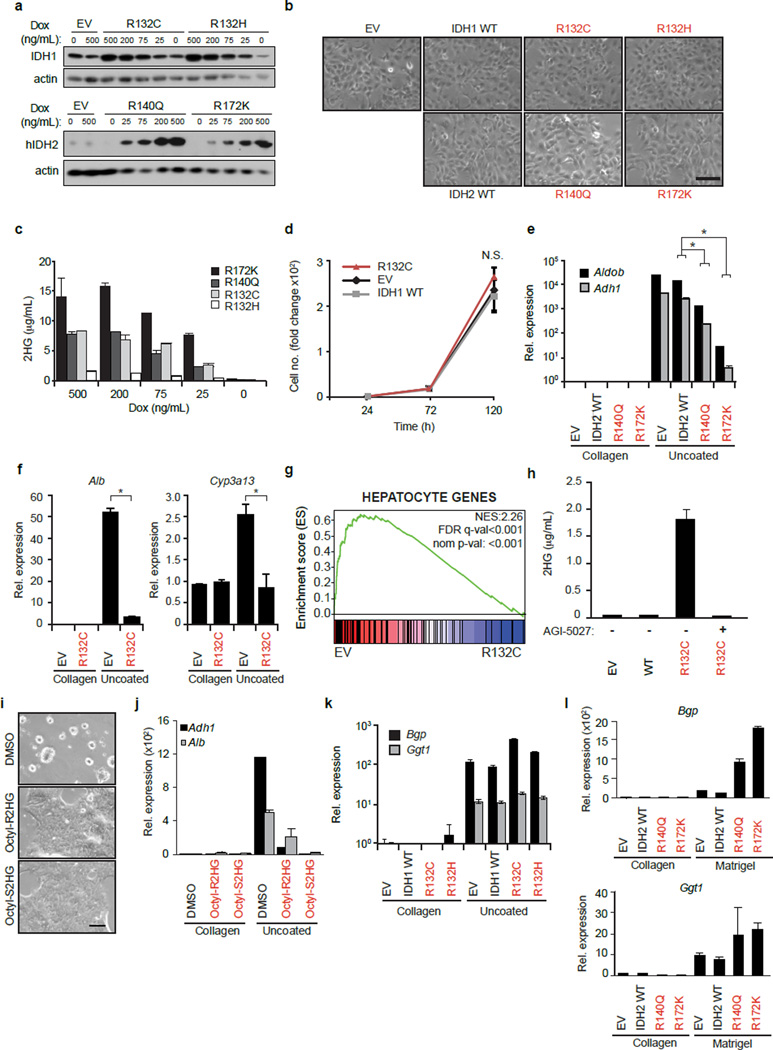
Gain-of-function IDH1/IDH2 mutations occur in ∼25% of IHCCs1,3–5 but have not been identified in hepatocellular carcinomas (http://www.sanger.ac.uk/cosmic) — liver malignancies that exhibit bile duct and hepatocyte differentiation, respectively. To examine the role of IDH mutations in liver tumourigenesis we isolated mouse hepatoblasts (HBs), which are embryonic progenitors that give rise to hepatocytes and bile duct cells and show correspondence to adult liver progenitors11,12. HBs expressing mutant IDH1 (R132C, R132H) or IDH2 (R140Q, R172K) produced increased 2HG, but exhibited morphology and proliferation rates indistinguishable from vector and IDH wild type (WT) controls (Extended Data Fig. 1a–d). However, unlike control HBs, which underwent hepatocyte differentiation when transferred from collagen-coated plates to uncoated plates13, forming hepatocyte clusters, decreasing proliferation, and activating a large program of hepatocyte-specific genes including Adh1 and Aldob, IDH mutant cells were refractory to differentiation (Fig. 1a–d, Extended Data Fig. 1e–g). IDH1-R132C and IDH2-R172K caused the most pronounced effects, correlating with relative 2HG levels. Treatment of R132C-expressing HBs with ML309 (AGI-5027), a specific inhibitor of mutant IDH114, attenuated 2HG production (Extended Data Fig. 1h) and restored hepatocyte differentiation (Fig. 1e–f). Conversely, differentiation of WT HBs was counteracted by (R)- or (S)-2HG octyl-esters (Extended Data Fig. 1i–j). In contrast to the complete inhibition of hepatocyte differentiation, mutant IDH did not impair biliary differentiation of HBs in matrigel (Fig. 1g, Extended Data Fig. 1k–l). Thus, mutant IDH specifically blocks hepatocyte lineage progression through 2HG production.
Figure 1. IDH mutant alleles block hepatocyte differentiation.
a–d. Hepatoblasts (HBs) expressing empty vector (EV) or the indicated IDH alleles at physiologic levels (see Extended Data Fig. 1a), were transferred to uncoated plates to induce hepatocyte differentiation. a, Hepatocyte sphere formation. b, proliferation, c, hepatocyte marker expression (qRT-PCR). d, Heat map of hepatocyte and biliary gene expression.
e,f. HB cells treated with 2.5µM AGI-5027 or DMSO vehicle. e, hepatocyte sphere formation (upper panel), proliferation (lower panel). f, hepatocyte marker expression.
g. HB cells in matrigel assessed for biliary differentiation. Tubular structures/6 cm dish ± std. are quantified. *P<0.05. Scale bars, 100 µm.
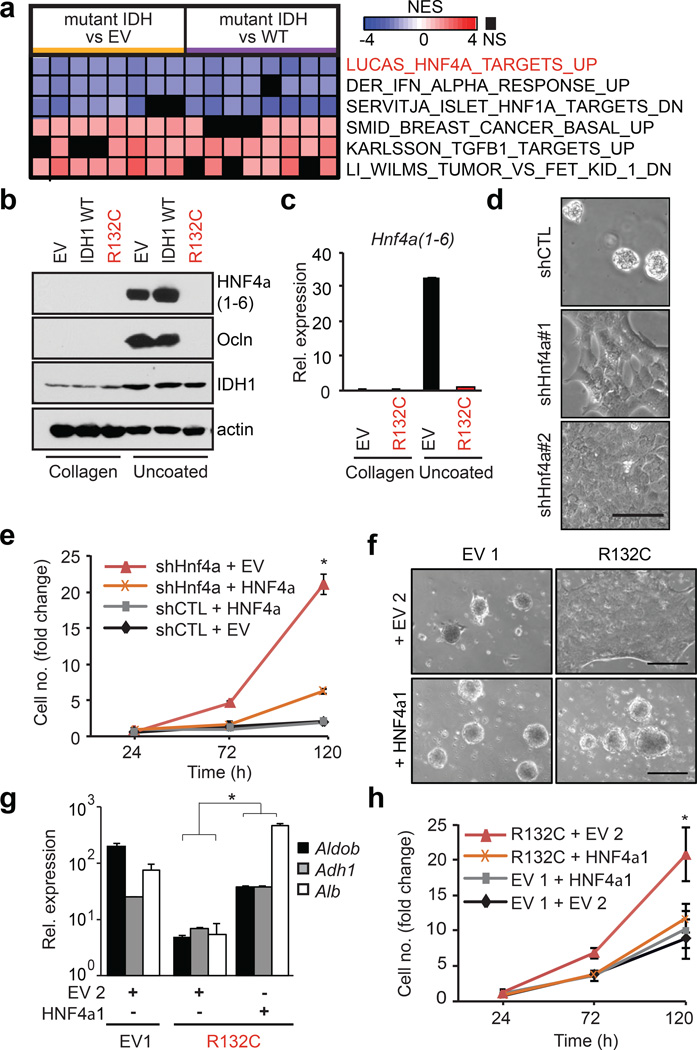
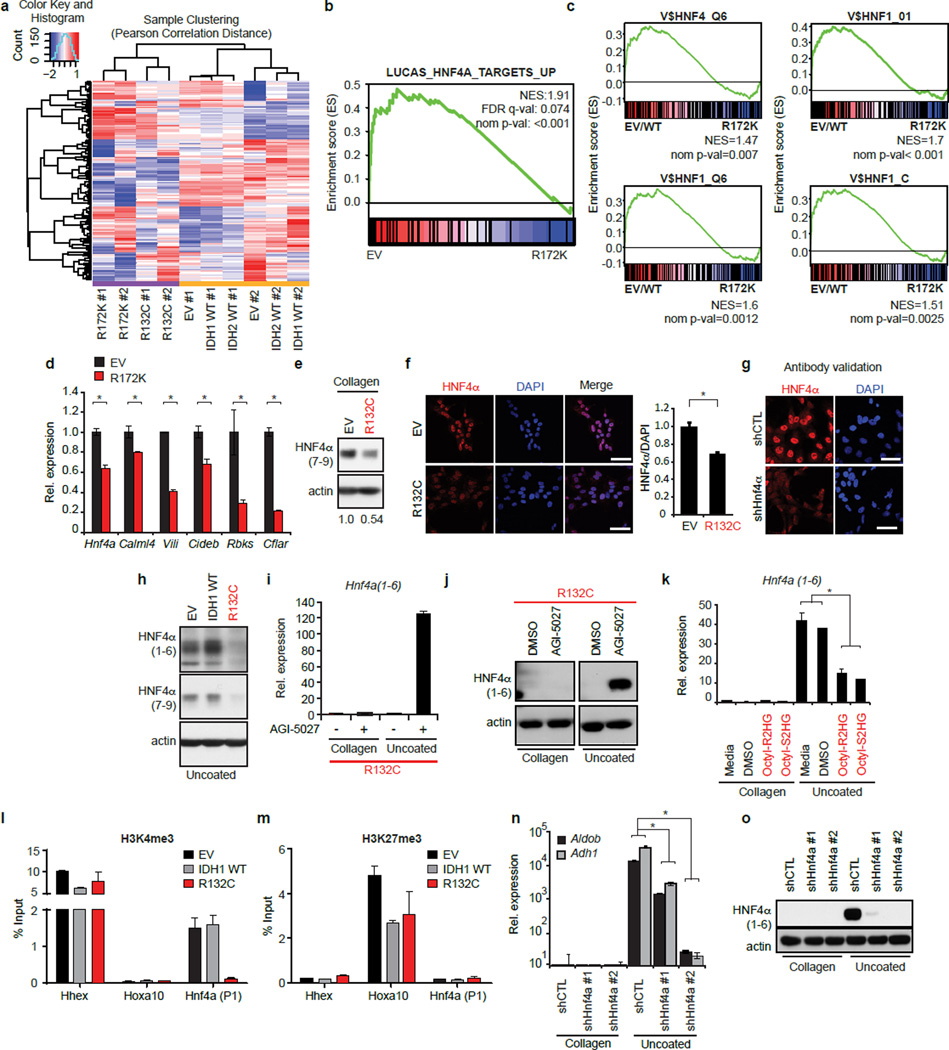
To uncover the molecular program underlying these defects, we examined the effect of mutant IDH1/2 on the transcriptome of HBs grown on collagen. Transcriptional profiles of mutant IDH 1 and 2 clustered together and Gene Set Enrichment Analysis (GSEA) demonstrated reduced expression of targets of HNF4α — a master transcriptional regulator of hepatocyte differentiation12 — and of HNF1α which acts downstream of HNF4α15. Moreover, canonical HNF4α and HNF1α binding sites were strongly enriched at the promoters of differentially expressed genes (Fig. 2a, Extended Data Fig. 2a–c).
Figure 2. Mutant IDH blocks hepatocyte differentiation by silencing HNF4α.
a. Heat map of GSEA showing top-ranked gene-sets distinguishing IDH1-R132C or IDH2-R172K from WT or EV control HBs (pairwise analysis; replicates for each condition; see Methods). NES = Normalized enrichment score.
b,c. HB cells analyzed by (b) immunoblot, and (c) qRT-PCR.
d,e. Analysis of WT HBs expressing the indicated shRNAs (uncoated plates). d, Hepatocyte sphere formation. e, Proliferation of shRNA-expressing HB cells co-expressing EV or shRNA-resistant Hnf4a1 cDNA.
f–h. Control and R132C–expressing HBs co-expressing vector control (EV2) or HNF4α, grown on uncoated plates. f, hepatocyte sphere formation, g, hepatocyte gene expression, h, proliferation. *P<0.05, Scale bar, 100µm (d), 250µm (f).
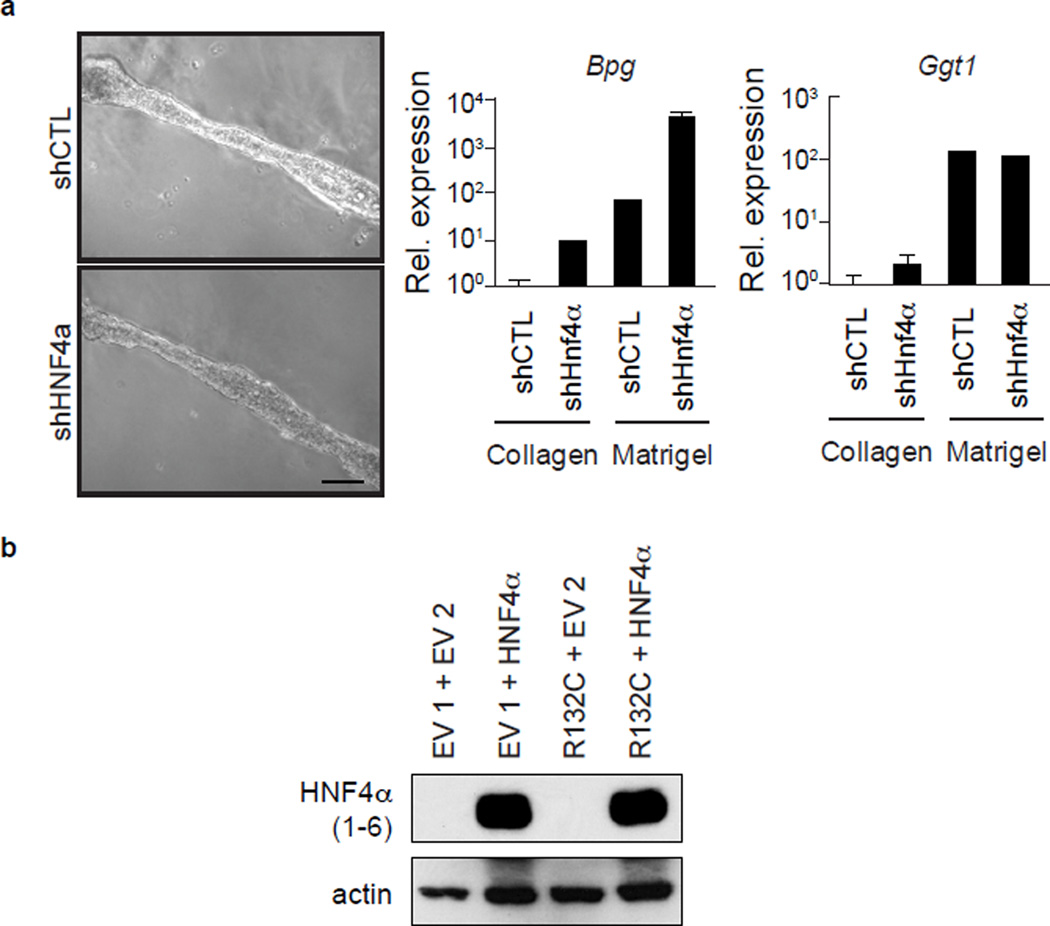
HNF4α has multiple isoforms expressed from separate promoters. The P2 promoter (encoding Hnf4a7-9) is active in hepatoblasts and gradually extinguished in adult hepatocytes, whereas the P1 promoter (encoding Hnf4a1-6) is hepatocyte-specific16. Hnf4a7-9 mRNA and protein were reduced in IDH-mutant HBs, as was expression of HNF4α targets (Extended Data Fig. 2d–g). Moreover, under hepatocyte differentiation conditions, mutant IDH completely inhibited the pronounced induction of HNF4α1-6 and its target OCLN that is observed in control cells (Fig. 2b–c, Extended Data Fig. 2h). Mutant IDH or octyl-2HG treatment blocked Hnf4a1-6 mRNA induction, whereas AGI-5027 restored Hnfa1-6 levels in R132C-expressing cells (Extended Data Fig. 2i–k). Histone H3 lysine-4 trimethylation (H3K4Me3) is associated with active transcription and was specifically reduced at the P1 promoter in R132C HBs, consistent with the observed silencing of Hnf4a1-6, whereas the repressive marks, H3K27me3 and H3K9me3, were unaffected (Extended Data Fig. 2l–m and data not shown). Significantly, HNF4α knockdown impaired hepatocyte differentiation of WT HBs without inhibiting biliary differentiation, whereas ectopic HNF4α expression rescued differentiation of IDH-mutant cells (Fig. 2d–h, Extended Data Fig. 2n–o, 3a–b). Thus, mutant IDH alters the epigenetic state of the P1 promoter — through targeting either direct regulators of the locus or more upstream factors — and prevents induction Hnf4a1-6, thereby blocking hepatocyte lineage progression. Notably, HNF4α and HNF1α have critical anti-proliferative and tumour suppressor functions in the adult liver17–20, suggesting the relevance of this pathway to mutant IDH-mediated tumourigenesis and prompting us to extend our studies in vivo.
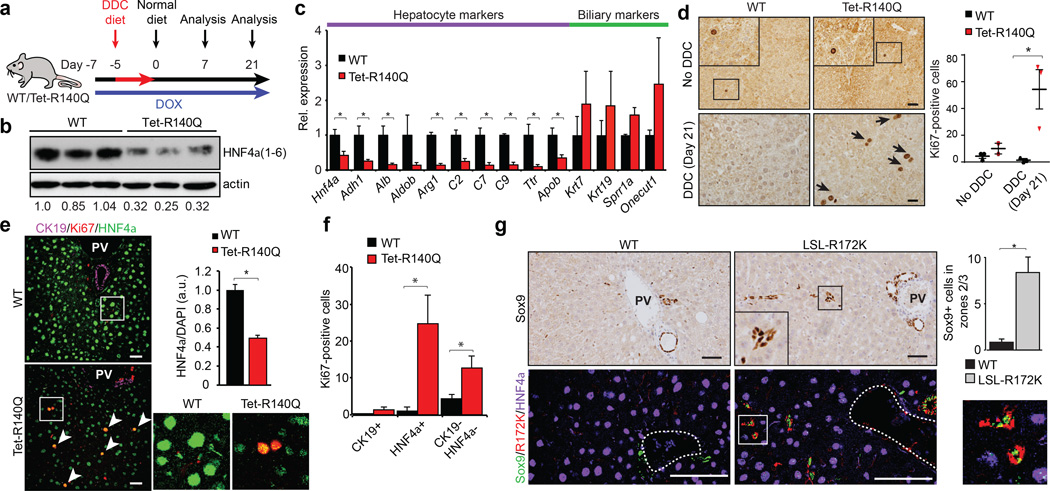
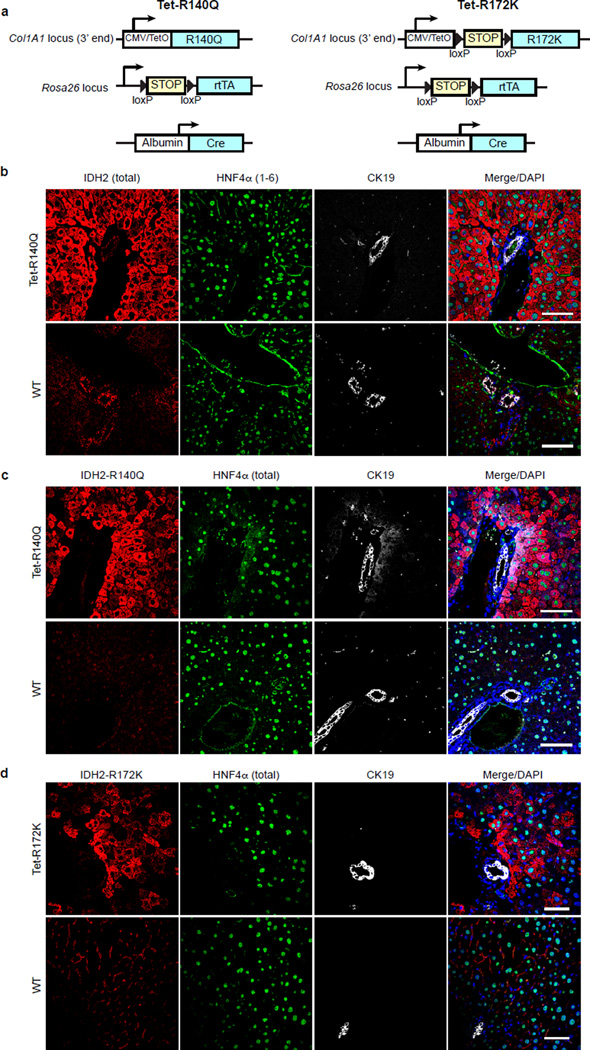
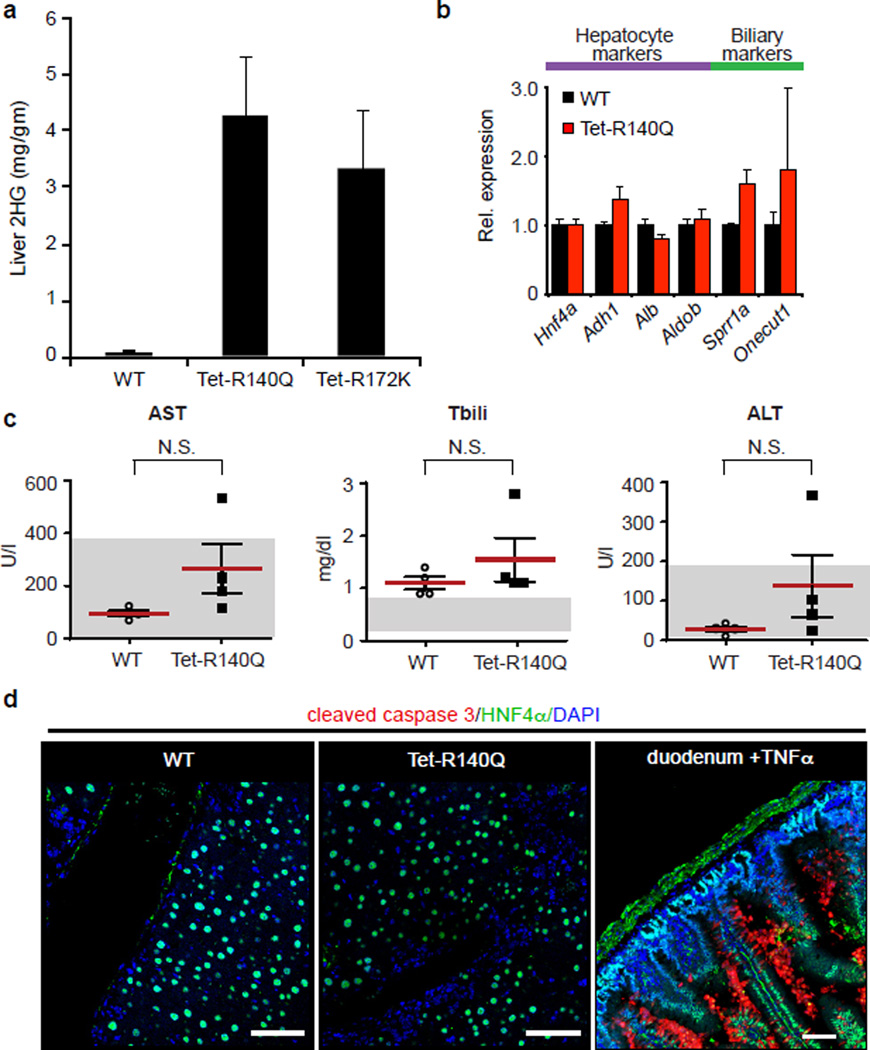
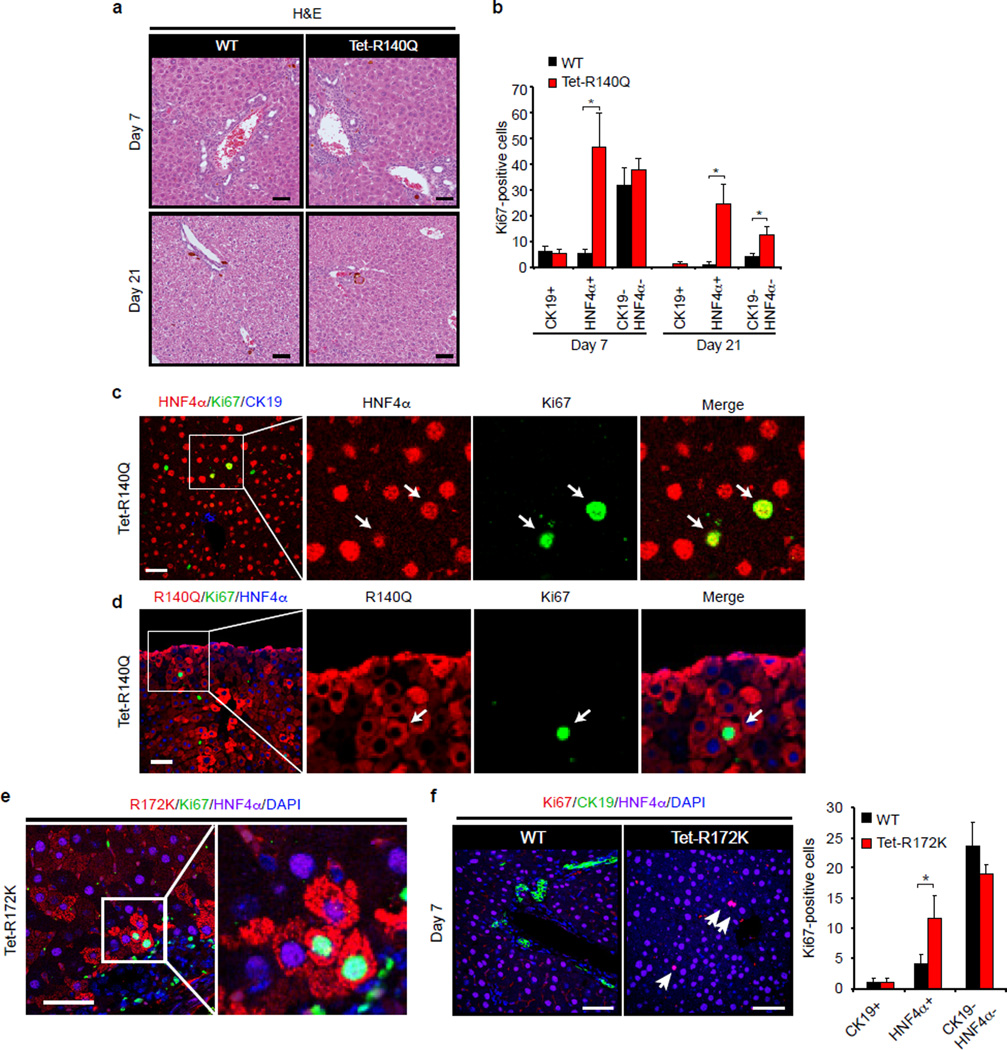
We generated transgenic mice with doxycycline-inducible expression of IDH2-R140Q or IDH2-R172K (Tet-R140Q, Tet-R172K strains) specifically in adult hepatocytes — R140Q was detected in virtually all hepatocytes and R172K showed more scattered expression, and liver 2HG levels were elevated (Extended Data Fig. 4a–d, 5a). Since mutant IDH blocks liver progenitors from undergoing hepatocyte differentiation in vitro, we sought to address whether it acts analogously in vivo to specifically override differentiation from a progenitor cell state, or conversely, whether it broadly alters homeostasis of mature hepatocytes. Although normally quiescent, the liver has extensive regenerative capacity following injury involving replication of mature hepatocyte and biliary cells, or activation of bipotential progenitors (oval cells) that may arise from either lineage11,21. In the absence of injury, Tet-R140Q mice were healthy up to 48 weeks, and had normal liver histology, marker expression, proliferation, and liver function (Fig. 3d, Extended Data Fig. 5b, and data not shown). By contrast, pronounced defects in restoration of hepatocyte differentiation were observed in mice fed a diet containing 3,5-diethoxycarbonyl-1,4-dihydrocollidin (DDC) for 5 days then switched to normal diet for 3 weeks (Fig. 3a), a protocol causing hepatocyte cell death and transient oval cell activation21,22. Hepatocyte markers including HNF4α were downregulated 3–10-fold, while biliary markers were unchanged, and proliferation was increased >40-fold relative to WT controls (Fig. 3b–d). Despite this depletion of mature hepatocytes, no changes were seen in parameters of liver function (Extended Data Fig. 5c–d and data not shown), consistent with the persistence of hepatocytes surviving short-term DDC treatment and the established capacity of reduced hepatocyte numbers to maintain normal physiology.
Figure 3. Mutant IDH inhibits hepatocyte differentiation and quiescence of liver progenitors.
a. Schematic of DDC study in Tet-R140Q (Tet-R140Q, Alb-Cre, Rosa26-LSL-rtTA) and WT littermate controls (Alb-Cre, Rosa26-LSL-rtTA).
b–f.Livers at day 21. b, Immunoblot (HNF4α1–6:actin is quantified). c, qRT-PCR. d, Ki-67 staining. Chart: Ki-67+ cells/20 high-powered fields. e,f, IF analysis. Graph: mean fluorescence intensity of HNF4α:DAPI (125 cells/group were scored). Inset: high power views of boxed regions. f, Quantification of Ki-67+ cells co-staining for the indicated markers (N = 3 mice/group, 5 high-powered fields/mouse).
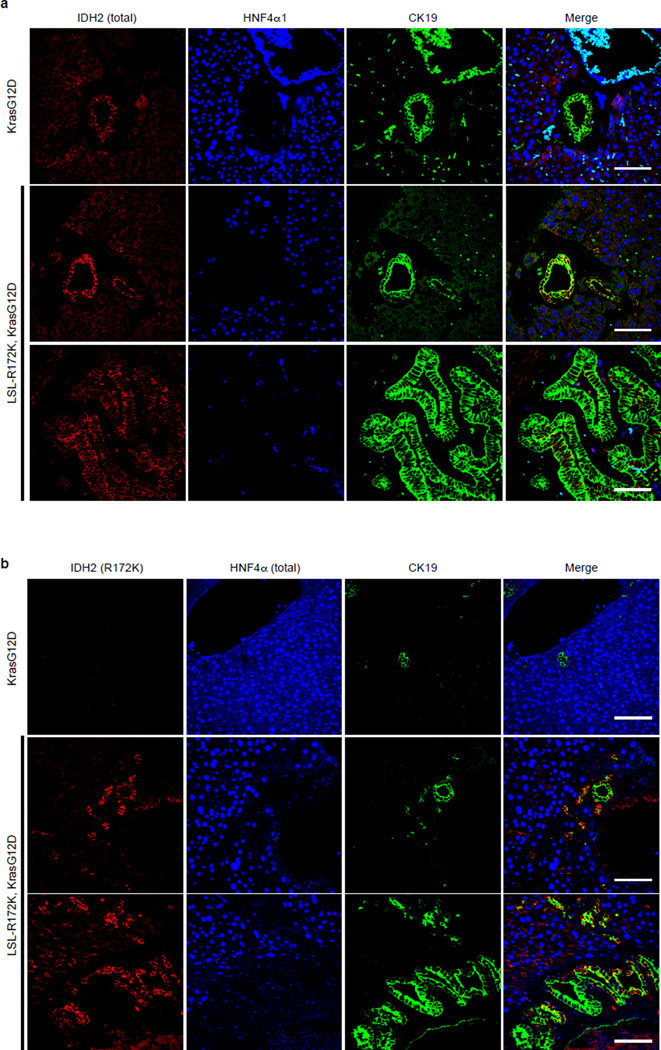
g. IHC (top) and IF (bottom) of WT and LSL-R172K livers at 20 months. Note accumulation Sox9+ cells located >25µm away from bile duct or portal structures (dashed-line), which express IDH2-R172K and lack HNF4α. Inset: higher magnification. Chart: quantification. N = 3 mice/group; 4 high-powered images/mouse were scored. PV=Portal vein.
Error bars, ±s.d. (c) and ±s.e.m. (d, f, g); Scale bars, 20 µm (d) and 50 µm (e, g).
Serial analyses of WT and R140Q livers revealed comparable numbers of proliferating periductal HNF4α−/CK19− oval cells at 1 week, and resolution of this population after 3 weeks (Extended Data Fig. 6a–b, and Fig. 3e–f). However, R140Q livers exhibited prominent induction of proliferating non-periductal cells with hepatocyte morphology but reduced or absent HNF4α expression (HNF4α+ and HNF4α−/CK19−), which persisted after 3 weeks (Fig. 3e–f, Extended Data Fig. 6a–d). Similar but more tempered phenotypes were seen in Tet-R172K mice, consistent with focal transgene expression (Extended Data Fig. 6e–f). Thus, mutant IDH specifically blocks restoration of hepatocyte differentiation following acute liver injury, leading to aberrant proliferation in the hepatic parenchyma. As HNF4α levels increase in liver progenitors undergoing hepatocyte commitment23,24, these proliferating HNF4α-low/absent cells appear to be committed progenitors (derived from de-differentiated hepatocytes or oval cells) whose differentiation to hepatocytes is subverted by mutant IDH2. The transgene was not expressed in Sox9+ oval cells, precluding assessment of effects on these earlier progenitors.
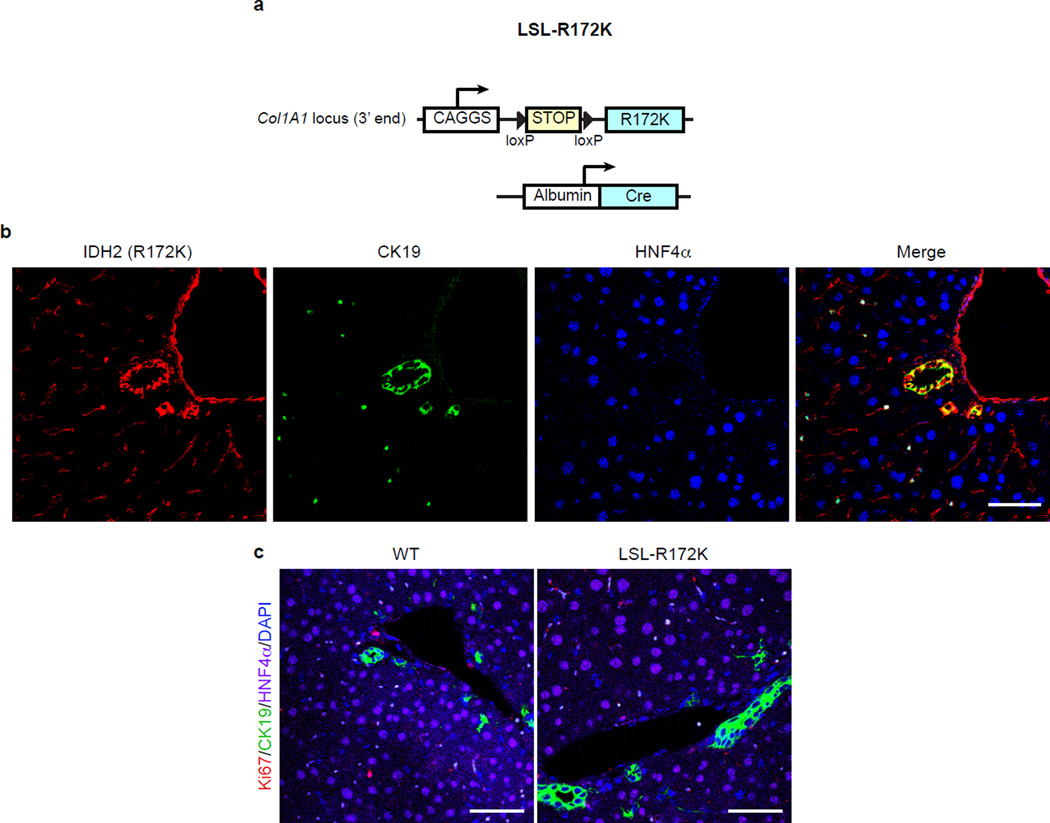
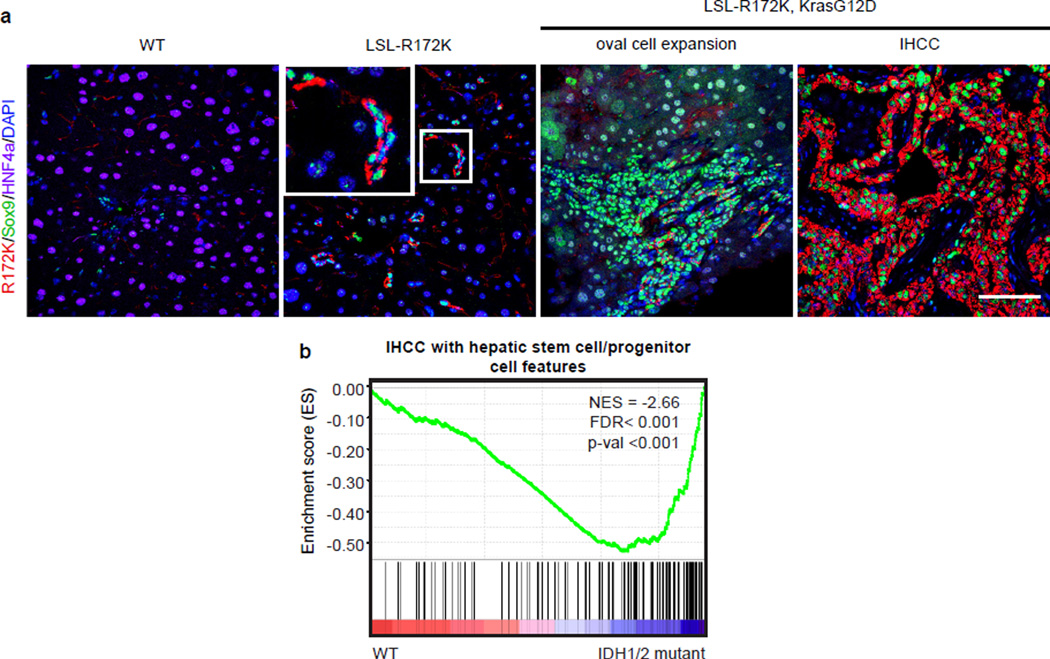
We developed an additional transgenic strain (LSL-R172K) expressing human IDH2-R172K in Sox9+ biliary cells but not in hepatocytes (Fig. 3g, Extended Data Fig. 7a–b). These animals displayed normal liver histology, gene expression, and proliferation at 3 months (Extended Data Fig. 7c and data not shown). However, by 20 months of age there was pronounced accumulation of Hnf4α−/Sox9+ oval cells expressing R172K >25µm away from any bile duct or portal structure (zones 2/3) (Fig. 3g). Collectively, our results suggest that mutant IDH2 abrogates differentiation of adult progenitors (activated spontaneously during aging or by injury), specifically blocking hepatocyte lineage progression.
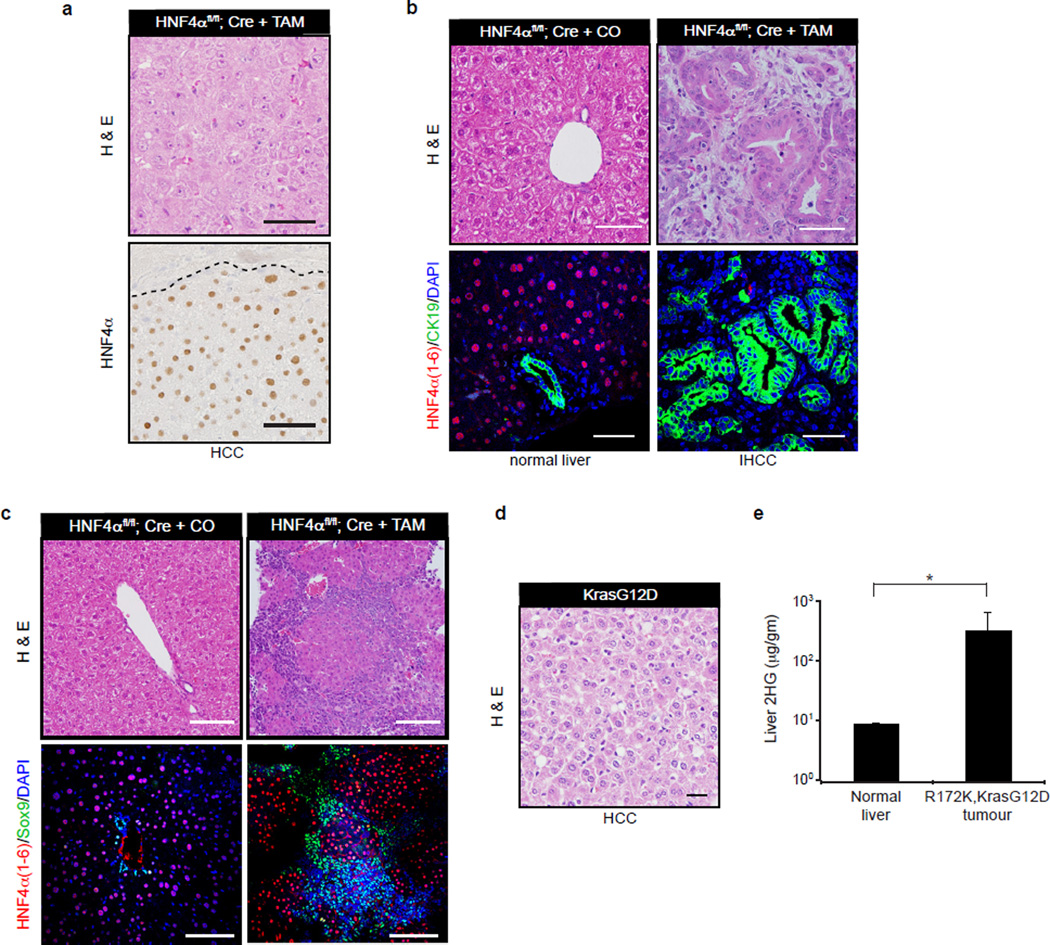
These findings are concordant with our observations in HB cells and suggest that failed HNF4α induction could contribute to the liver phenotypes caused by mutant IDH in vivo. In this regard, acute HNF4α deletion in hepatocytes is reported to provoke hepatocyte differentiation defects and, upon diethylnitrosamine (DEN) treatment, oval cell accumulation and formation of tumours showing hepatocellular carcinoma (HCC) and IHCC morphology20. To expand upon potential parallels with IDH-mutant mice, we characterized these phenotypes in further depth. Significantly, the HCC lesions in DEN-treated HNF4a conditional KO mice were uniformly HNF4α+, indicating that they arose from cells that escaped deletion of the locus (Extended Data Fig. 8a). By contrast, Sox9+ oval cells and CK19+ IHCCs were HNF4α-negative and were never observed in DEN-treated controls (Extended Data Fig. 8b–c). Thus, HNF4α ablation in DEN-treated livers drives progenitor expansion and progression specifically to IHCC. These findings establish Hnf4a as an IHCC tumor suppressor and are consistent with HNF4α acting downstream of mutant IDH in liver growth control.
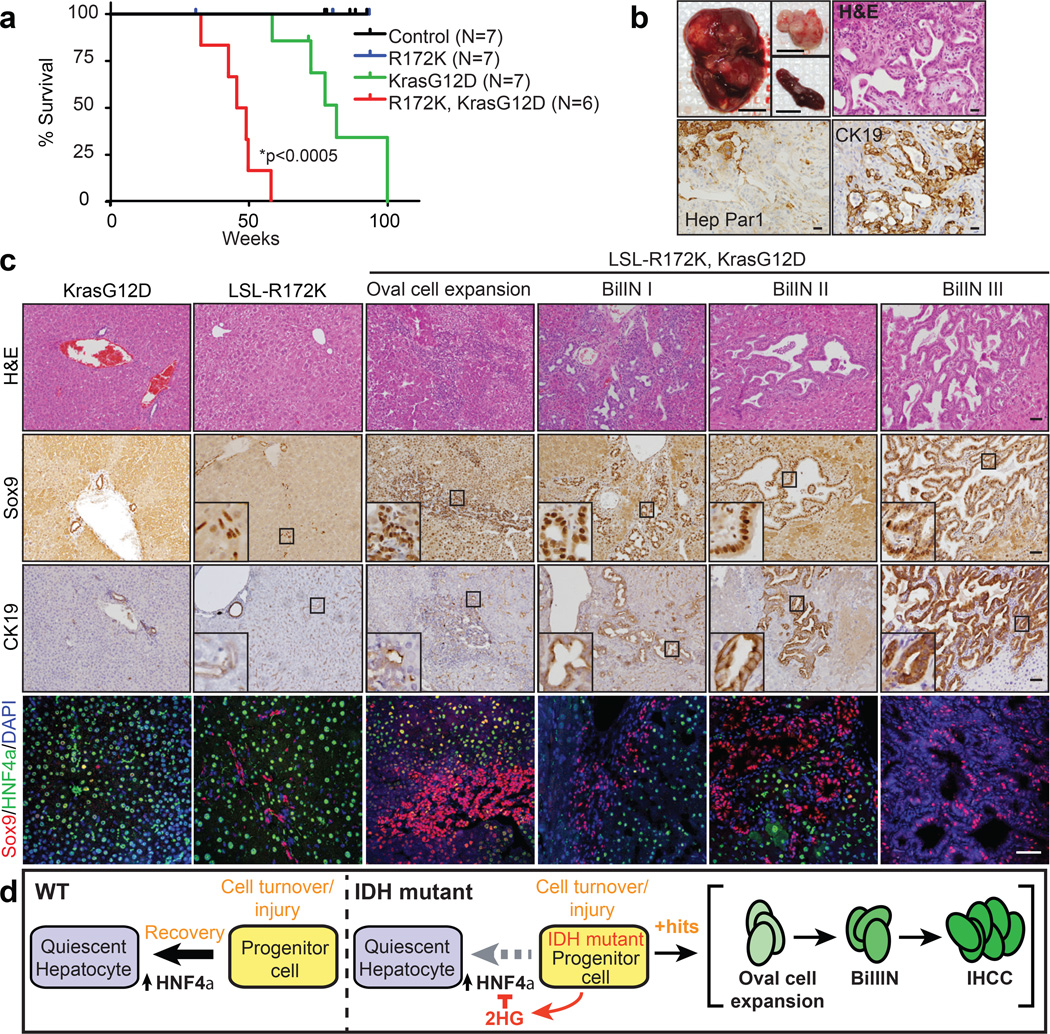
The capacity of mutant IDH to silence Hnf4a and impair differentiation would be expected to confer sensitivity to transformation by additional oncogenic lesions. In this regard, IDH and KRAS mutations exist concurrently in human IHCC4,5 and AML25. Previously, we showed that KrasG12D expression in mouse liver causes mixed IHCC/HCC with long latency, a phenotype accelerated by p53 deletion26. Intercrossing LSL-R172K, KrasG12D and Alb-Cre mice revealed dramatic oncogenic cooperation, with 6/6 Alb-Cre; LSL-R172K; KrasG12D animals developing poor body condition and palpable liver tumours between 33 and 58 weeks (mean 47.3 weeks; Fig. 4a). Multifocal liver masses with splenic invasion and peritoneal metastases were observed and demonstrated to be IHCC by histopathologic analysis, CK19 staining, and lack of reactivity for the hepatocyte/HCC marker, Hep Par1 (Fig. 4b). By contrast, only 1/7 Alb-Cre; KrasG12D mice sustained a tumour by 70 weeks (mean survival = 81.6 weeks), and solely HCC were detected (Extended Data Fig. 8d). Tumour 2HG levels in R172K mice were comparable to those in IDH-mutant human IHCC1 (Extended Data Fig. 8e).
Figure 4. Mutant IDH cooperates with KrasG12D to drive liver progenitor cell expansion and multi-step IHCC pathogenesis.
a. Kaplan-Meier analysis showing time until signs of illness necessitated euthanasia. All animals euthanized had liver tumours.
b. Upper left: Representative Alb-Cre;LSL-R172K;KrasG12D tumor, and peritoneal and spleen metastases (insets). Upper right: H&E staining showing IHCC histology. Lower panels: The tumour is Hep Par1- and CK19+ while adjacent hepatocytes stain Hep Par1+ and CK19-.
c.The livers of Alb-Cre;LSL-R172K;KrasG12D animals exhibit oval cell expansion and increasing grades of BilIN, which stain Sox9+. CK19 levels increase with higher grade lesions. IF analysis reveals focal accumulation Sox9+ oval cells in Alb-Cre:LSL-R172K livers and pronounced oval cell expansion in Alb-Cre;LSL-R172K; KrasG12D livers.
d. Model for mutant IDH in cholangiocarcinoma pathogenesis. Scale bars, 1cm (b, upper left), 50µm (b-c).
In humans, IHCC is thought to develop from precursor lesions, including biliary intraepithelial neoplasias (BilIN)2. Aberrant oval cell proliferation also precedes liver cancer development in murine models27. Notably, in all Alb-Cre; LSL-R172K; KrasG12D mice analyzed (N=6), the adjacent liver exhibited oval cell expansion and a graded series of BilIN-like lesions, which were Sox9+/CK19+, and expressed the IDH2-R172K transgene at physiologic levels (Fig. 4c, Extended Data Fig. 9a–b, 10a). Importantly, neither Alb-Cre; KrasG12D nor Alb-Cre; KrasG12D; p53Lox/+ livers26 showed oval cell expansion, and BilIN were found in only 2/8 Alb-Cre; KrasG12D; p53Lox/+ mice. Thus, IDH2-R172K and KrasG12D cooperatively incite activation of hepatic progenitors and multistage IHCC progression.
In summary, we present a novel GEMM driven by mutant IDH that exhibits the hallmarks of human IHCC. Mutant IDH blocks hepatocyte differentiation from progenitors in vitro, and regulates hepatic regeneration, progenitor cell expansion, and IHCC pathogenesis in vivo. Collectively, our findings are consistent with a model whereby mutant IDH subverts the HNF4α-mediated hepatocyte differentiation/quiescence program in proliferating hepatocytes or bipotential progenitors, creating a persistent pre-neoplastic state primed for transformation by additional oncogenic mutations, and leading to adenocarcinoma specifically with a biliary phenotype (Fig. 4d). While lineage-tracing studies are required to fully define the impact of IDH mutations on different liver cell types, this model predicts that IDH-mutant IHCC may have progenitor-like features, and that IDH mutations occur early in disease pathogenesis as observed in glioblastoma and AML28,29. Accordingly GSEA revealed that IDH-mutant tumors have strong enrichment of a hepatic stem cell expression signature (Extended Data Fig. 10b).
Defining the biologic functions of mutant IDH is of immediate significance as IHCCs are resistant to current treatment. Our GEMM provides an autochthonous context for ‘co-clinical trials’ to help determine the best way to deploy IDH inhibitors and identify biomarkers of response, thereby informing clinical decision-making in this deadly disease.
METHODS SUMMARY
Cell culture
HB cells were prepared from WT mice at embryonic day 14 and subjected to hepatocyte and bile duct differentiation as described13. For proliferation assays, cells were plated in duplicate on collagen-coated or uncoated 6 well plates. Cells were trypsinized and counted by trypan-blue exclusion.
Mice
Mice were housed in pathogen-free animal facilities. Studies were approved by the Subcommittee on Research Animal Care at Mass. General Hospital (protocol 2005N000148). The Tet-R140Q, Tet-R172K, and LSL-R172K transgenic mice and other strains are presented in the Methods Section. For DDC experiments, mice were maintained on 200 µg/mL doxycycline-containing water. Mice were fed with 0.1% DDC-containing chow (F4643, Bio-serv, Frenchtown, NJ) from days 2–7. Mice were then switched to normal chow until harvest. DEN experiments were performed as described20.
Statistics
Results are expressed as mean ±s.d. unless otherwise specified. Significance was analyzed using 2-tailed Student’s t test. A p-value of less than 0.05 was considered statistically significant.
METHODS
Cell culture
HB cells were prepared from WT mice at embryonic day 14 and immortalized by plating at clonal density13. HBs were maintained in HB media [DMEM/F-12 (Gibco Life Technologies, Grand Island NY) containing 10% fetal bovine serum, 1% penicillin-streptomycin, 50ng/mL epidermal growth factor, 30 ng/mL insulin-like growth factor II (PeproTech, Rocky Hill, NJ), 10 µg/mL insulin (Roche, Mannheim, Germany)] on plates coated with rat tail collagen (BD Biosciences, Bedford, MA) in a humidified atmosphere with 5% CO2 at 37°C. For IDH1 experiments, culture media was supplemented with 25 ng/mL of doxycycline (d9891, Sigma-Aldrich). For hepatocyte differentiation assays, 5×106 HB cells were cultured on uncoated 10 cm tissue culture dishes in HB cell medium for up to 5 days. Pictures were taken 2–5 days after plating. For bile duct differentiation (matrigel) assays, 0.5 mL of Basement Membrane Matrix (BD) was coated onto 6 cm tissue culture dishes and allowed to set for 1h. Then 0.5×106 HB cells were added to the plate in HB cell media supplemented with 100 ng/mL recombinant mouse hepatocyte growth factor (HGF) (R&D Systems, Minneapolis, MN). Tubules were counted after 24hrs. For gene expression analysis, cells were isolated after 10 days. For proliferation assays, cells were plated in duplicate on collagen-coated 6 well dishes (1×104/well) or uncoated 6 well plates (1×105/well) in culture medium. Adherent and non-adherent cells were harvested, trypsinized and counted by trypan-blue exclusion using a Countess Automated Cell Counter (Invitrogen) 24, 72 or 120 hrs later. Negative mycoplasma contamination status of all cell lines and primary cells used in the study was established using LookOut Mycoplasma PCR Kit (Sigma, MP0035).
AGI-5027
AGI-5027 (ML309) is a small molecule (phenyl-glycine scaffold) inhibitor of IDH1 R132C and R132H (IC50=30 nm and 68 nm respectively) that does not affect WT IDH1 activity (IC50 >36 µM and was developed by NIH Chemical Genomics Center and Agios Pharmaceuticals14.
Mice
Mice were housed in pathogen-free animal facilities. All experiments were conducted under protocol 2005N000148 approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital. Mice were maintained on mixed 129SV/C57Bl/6 background. Data presented include both male and female mice. All mice included in the survival analysis were euthanized when criteria for disease burden were reached.
Albumin-Cre transgenic mice
This strain30 harbors a transgene in which the mouse albumin enhancer/promoter drives Cre recombinase. Cre activity is inefficient at birth, but leads to progressive deletion of floxed sequences in all liver lineages by 4–6 weeks of age.
Rosa26-LSL-rtTA31 mice
In this strain, the doxycycline-responsive reverse tetracycline transactivator (rtTA) is knocked into the transcriptional start site at the ubiquitously expressed Rosa26 locus and is preceded by a lox-stop-lox (LSL) casette.
KrasG12D mice32
This allele consists of a mutant KrasG12D allele knocked into the endogenous Kras locus, preceded by an LSL cassette.
IDH mutant transgenic mouse models
To generate transgenic mice, the R140Q and R172K mutations were introduced into human IDH2 cDNA (Origene, Rockville, MD # SC319226, accession# NM_002168) using the Quickchange kit (Agilent Lexington, MA #200521) per the manufacturer’s instructions. Two different transgenic targeting approaches were used.
Doxycycline inducible models
To generate the doxycycline-inducible Tet-R140Q and Tet-R172K strains (Extended Data Fig. 4), IDH2-R140Q cDNA was cloned into the pBS31 Prime transgenic targeting vector (Thermo Scientific Open Biosystems, #MES4487). A modified version of pBS31 containing a floxed stop cassette was used for the IDH2-R172K mice. The targeting vectors were co-electroporated into C2 mouse embryonic stem cells (Thermo Scientific Open Biosystems, #MES4305) with a plasmid expressing FLPE recombinase (pCAGGS-FlpE, Thermo Scientific Open Biosystems, Waltham MA, #MES4488) as described33. This system involves single copy integration of the transgene downstream of the Collagen-1 locus, and within the liver, results in expression in hepatocytes but not in bile ducts33. ES cells were screened for integration of the transgene by PCR and correctly targeted ES cells were injected into C57Bl/6 blastocysts. Chimeras were crossed to the Albumin-Cre strain30, and offspring were successively crossed with Rosa26-LSL-rtTA31, and LSL-KrasG12D mice32. We confirmed the expected expression of the R140Q (Extended Data Figure. 4b–c) and R172K transgenes (Extended Data Fig. 4d) in the adult hepatocytes and lack of expression in bile ducts. Stably expressed model: To generate mice with a conditionally active IDH2-R172K allele (LSL-R172K), human IDH2-R172K was cloned into a modified version of the PGK-ATG-Frt vector (Open Biosystems, Waltham MA, #MES4490) containing the CAGGS promoter for stable expression (Extended Data Fig 7a)34. Gene targeting and chimeric mouse generation was as above. This strain was successively crossed with Albumin-Cre and LSL-KrasG12D animals. This transgene was specifically expressed in bile ducts and oval cells (Fig. 3g and Extended Data Fig. 7b). We did not observe developmental anomalies in these mice, although the inefficiency of recombination induced by the Alb-Cre strain in neonates and the restricted activity of the transgene in this system precludes definitive assessment of potential effects of mutant IDH2 on the embryonic liver.
DDC treatment
For DDC experiments, 4-week old mice were treated with 200 µg/mL doxycycline-containing water. 2 days later, mice were fed with 0.1% DDC-containing chow (F4643, Bio-serv, Frenchtown, NJ) for 5 days. The mice were then switched to normal chow and maintained on doxycycline-containing water until harvest 1 or 3 weeks later. Randomization was done according to genotype and blinding was applied during histological analysis. No mice were excluded from the analysis. Sample sizes were chosen to achieve statistical significance yet use the fewest animals possible to minimize suffering.
Statistics
Results are expressed as mean ±s.d. between two technical replicates, unless otherwise specified. Significance was analyzed using 2-tailed Student’s t test. A p-value of less than 0.05 was considered statistically significant.
Plasmids
Human wild-type IDH1 cDNA (Accession# NM_005896) was obtained from Origene and subcloned into pRetro-Puro using EcoRI and BamHI fragments. Human wild-type IDH2, R140Q and R172K cDNA (Origene) were subcloned into pRetro-puro and pMSCV-blast (derived by replacing the puromycin resistance gene from pMSCV-puro with a blasticidin resistance gene) (both from Clontech, Mountain View CA) using EcoRI and XhoI. Human IDH1 R132C was generated from wild-type IDH1 using QuikChange Lightning kit (Agilent Technologies, Santa Clara, CA) and primers 5’-GATGGGTAAAACC TATCATCATAGGTTGTCATGCTTATGGGGATCAATAC-3’ for sense and 5’-GTATTGATCCCCATAAGCATGACAACCTATGATGATAGGTTTTACCCATC-3’ for antisense. pLVX-Tet-On was obtained from Clontech (Mountain View, CA). Murine Hnf4a was obtained from Addgene (Cambridge, MA) and cloned into pRetro-blast (derived by replacing the puromycin resistance with blasticidin) as a BglII-EcoRI fragment. Lentiviral (pLKO.1) Hnf4α shRNA vectors TRCN0000026154, target sequence: 5’-CGACAATGTGTGGTAGACAAA-3’ and TRCN0000026216, target sequence: 5’-GCAGATTGATGACAATGAATA-3’ were used as shHnf4a#1 and shHnf4a#2, respectively, and were obtained from the Broad Institute TRC shRNA library. pLKO.1 shRNA with target sequence 5’-GCAAGCTGACCCTGAAGTTCAT-3’ was used as negative control shRNA. Hnf4α resistant to shHnf4a#2 was generated using QuikChange Lightning kit and primers 5’-GCCCTTCCAAGAGCTGCAGA TCGACGATAACGAATATGCCTGCCTCAAAGCC-3’ for sense and 5’-GGCTTTGAGGCAGGCATATTCGTTATCGTCGATCTGCAGCTCTTGGAAGGGC-3’ for antisense.
Immunoblot analysis
Cell extracts were prepared in 1x RIPA buffer (150mM NaCl, 1% IGEPAL, 0.1%SDS, 50mM Tris, 0.5% DOC) supplemented with a protease inhibitor cocktail (Complete, Roche Applied Science, Indianapolis, IN) and phosphatase inhibitors (Phosphatase Inhibitor Cocktail Sets I and II, Calbiochem, San Diego, CA) and quantified by BCA Protein Assay (Thermo Scientific, Rockford, IL). 30 µg protein was resolved on 9% SDS-PAGE gels and transferred onto PVDF membranes (GE Healthcare Life Sciences, Pittsburgh, PA). Membranes were blocked in TBS with 5% non-fat milk and 0.1% Tween and probed with anti-IDH1 polyclonal antibody (3997S, Cell Signaling, Danvers, MA), anti-IDH2 polyclonal antibody (ab84726, Abcam, Cambridge, MA), anti-occludin (71-1500, Invitrogen), anti-HNF4α (1-6) monoclonal antibody (ab41898, Abcam), anti-HNF4α(1–6)/NP2A1 monoclonal antibody (PP-K9218-00, R&D Systems, Minneapolis, MN), anti-HNF4α(7–9) monoclonal antibody (PP-H6939-00, R&D Systems), or anti-βactin monoclonal antibody (A5316, Sigma-Aldrich, St. Louis, MO) as loading control. Bound proteins were detected with horseradish-peroxidase-conjugated secondary antibodies (Vector Biolaboratories, Burlingame, CA) and SuperSignal West Pico Luminol/Enhancer Solution (Thermo Scientific).
RNA analysis
Total cellular RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA synthesis was performed with the QuantiTect Reverse Transcription Kit (Qiagen) with 1µg total RNA. Sense and antisense for amplification of target genes included: Alb (5’-CATGCCAAATTAGTGCAGGA-3’ and 5’-GCTGGGGTTGTCATCTTTGT-3’); Aldob (5’-TGTCTGGAGGTATGAGTGAGG-3’ and 5’-CTGGGTTGCCTTCTTGTTTGC-3’); Adh1 (5’-GTGACTTGTGTGAAACCAGGT-3’ and 5’-GCTACAAAAGTTGCTTTCCGGG-3’); Ggt1 (5’-CTTGTGCGAGGTGTTCTGC-3’ and 5’-GCATAGTCACCGTCTCTCCTT-3’); Bgp (5’-CAAAAGGAGGCCTCTCAGAT-3’ and 5’-GCTGAGGGTTTGTGCTCTGT-3’); Hnf4α(1–6) (5’-GGTAGGGGAGAATGCGACTC-3’ and 5’-AAACTCCAGGGTGGTGTAGG-3’); Foxm1 (5’-ATCGCTACTTGACATTGGACCA-3’ and 5’-GATTGGGTCGTTTCTGCTGTG-3’); Krt19 (5’-TGCTGGATGAGCTGACTCTG-3’ and 5’-AATCCACCTCCACACTGACC-3’); Onecut1 (5’-GGCAACGTGAGCGGTAGTTT-3’ and 5’-TTGCTGGGAGTTGTGAATGCT-3’); Arg1 (5’-TTGGGTGGATGCTCACACTG-3’ and 5’-GTACACGATGTCTTTGGCAGA-3’); C2 (5’-GGAACCCATTTGCCGACAG-3’ and 5’-GGCCCAAACTTTTTGTCAGAAG-3’); C7 (5’-GCAGGAAAGTGTTCAGCGG-3’ and 5’-CATGACCGTAAGTATTCCGTGAG-3’); C9 (5’-CATGCCGTGACCGAGTAGC-3’ and 5’-TCACAGAGTCCGTTGTAGAACT-3’); Krt7 (5’-CACCCGGAATGAGATTGCG-3’ and 5’-GCACGCTGGTTCTTCAAGGT-3’); Sprr1a (5’-TTGTGCCCCCAAAACCAAG-3’ and 5’-GGCTCTGGTGCCTTAGGTTG-3’); Ttr (5’-CACCAAATCGTACTGGAAGACA-3’ and 5’-GTCGTTGGCTGTGAAAACCAC-3’); ApoB (5’-TGGCTCTGATCCCAAATCCCT-3’ and 5’-GCTGCTCCTTGGCAGTATTAAA-3’); Cyp3a13 (5’-GACCACAAGCAGTGCTCTTTC-3’ and 5’-GCAGGGTATCATAGGTGGCAG-3’).
Gene expression profiling and gene-set enrichment analysis
Gene expression of HBs expressing IDH1 WT, IDH1 R132C, IDH2 WT, R172K and empty vector controls (N=2 cultures for each condition) grown on collagen-coated plates, as well as IDH1 R132C and empty vector controls grown on uncoated plates, was evaluated using Affymetrix Mouse 430Av2 DNA microarrays (DFCI Microarray core facility). Raw expression values were processed and normalized using RMA in the R Bioconductor package. This dataset is available in GEO (GSE57002). Differentially expressed genes in IDH1 R132C and IDH2 R172K expressing HBs were identified using the signal-to-noise metric. Gene Set Enrichment Analysis (GSEA) of the expression data was used to uncover the molecular program accounting for the differentiation defects. GSEA was performed using two libraries from version 3.1 of the molecular signature database (http://www.broadinstitute.org/gsea/msigdb/index.jsp): the c2 curated gene sets from online pathway databases, PubMed publications, knowledge of domain experts and the c3 motif gene sets. Pairwise GSEA analysis such as shown in Fig. 2a was performed by creating ranked lists of genes using the log2 ratio of mutant IDH samples to vector or WT control and running GSEA in the ranked list mode. The heat map in Fig. 2a presents the gene-sets that distinguish IDH1 R132C or IDH2 R172K as compared to cells expressing WT IDH1 or IDH2 or EV. Each column represents pairwise analysis of HB cells expressing mutant IDH (IDH1 R132C or IDH2 R172K) with either EV (left columns) or WT IDH (right columns) control cells for two independent replicates of each sample. All other results were obtained using GSEA to rank genes according to signal-to-noise ratio between mutant IDH samples and controls. Statistical significance for GSEA results were obtained from permuting gene sets (1000 permutations). Gene sets of ‘Hepatocyte Genes’ for the GSEA results shown in Extended Data Fig. 1g were created from expression data from Shin, et al. 201137. The expression data was downloaded from GEO (GSE28891) in series matrix form and loaded into R. Undifferentiated, differentiated hepatocytes, and primary hepatocyte samples were selected from GSE28891 (4 samples of each type for a total of 12). Within that sample subset, the normalized signal intensities were thresholded to a minimum value of 10 and features were filtered out if they had less than 3 fold difference between minimum and maximum values or had absolute differences less than 50 between minimum and maximum values (leaving 21277 features out of 37908). A t-test was performed between the undifferentiated and the combined differentiated and primary hepatocyte samples and p-values from the t-test were corrected for multiple hypothesis testing using the Benjamini-Hochberg correction (in R using the Bioconductor multtest package). This left 206 features up and 407 down in the hepatocyte direction with an adjusted BH p-value < 0.01 which are represented by 133 and 311 unique gene symbols respectively. 96 of these genes were represented on the murine Affymetrix array. The feature list was made into a gene set labeled as ‘Hepatocyte Genes’ and is shown in Supplementary Table 1.
Chromatin Immunoprecipitation
5×106 EV, IDH1 WT or R132C-expressing HBs were transferred from collagen-coated to uncoated plates for 5 days and fixed in 1% formaldehyde for 10 minutes at 37°C. Cross-linking was terminated by the addition of glycine at a final concentration of 125mM for 5min. Cells were then harvested and washed twice with ice-cold PBS. Chromatin extraction, DNA sonication, and immunoprecipitation were performed using a Chromatin Immunoprecipitation (ChIP) Assay Kit (17-295, Millipore, Temecula, CA) and ChIPAb+ Trimethyl-Histone H3 (Lys4) (17-614, Millipore) or Tri-Methyl-Histone H3 (Lys4) Antibody (Cell Signaling, #9727S) validated antibody. Sense and antisense for amplification of target genomic regions included: Hhex (5’-GAAGGAGCCTGACCCTTTCC-3’ and 5’-ATCAGCAGCGTGCACTACTC-3’); Hoxa10 (5’-AACAGTAAAGCTTCGCCGGA-3’ and 5’-AGTTCAGAAGGTCAGCCTGC-3’); Hnf4a P1 (5’-AACATGGCCCTATCTTCGGG-3’ and 5’-AAACGCACACCGCTATGTTG-3’).
Histology and Immunostaining
Tissue samples were fixed overnight in 4% buffered formaldehyde, and then embedded in paraffin and sectioned (5 µm thickness) by the DF/HCC Research Pathology Core. Hematoxylin & eosin staining was performed using standard methods. For immunohistochemistry, unstained slides were baked at 55°C overnight, deparaffinized in xylenes (2 treatments, 6 min each), rehydrated sequentially in ethanol (5 min in 100%, 3 min in 95%, 3 min in 75%, and 3 min in 40%), and washed for 5 min in 0.3% Triton X-100/PBS (PBST) and 3 min in water. For antigen unmasking, specimens were cooked in a 2100 Antigen Retriever (Aptum Biologics Ltd, Southampton, UK) in 1X Antigen Unmasking Solution, Citric Acid Based (H-3300, Vector Laboratories), rinsed 3 times with PBST, incubated for 10 min with 1% H2O2 at room temperature to block endogenous peroxidase activity, washed 3 times with PBST, and blocked with 5% goat serum in PBST for 1h. Primary antibodies were diluted in blocking solution as follows: anti-R172K (26163, New East Biosciences) 1:100, anti-R140Q (26165 New East Biosciences) 1:100, anti-IDH2 (NBP2-22166, Novus Biologicals) 1:100, anti-HNF4α monoclonal antibody (PP-K9218-00, R&D Systems) 1:250, anti-CK19 (Troma3) 1:100; anti-Sox9 (ab5535, Millipore), 1:300; or anti-Ki67 (ab15580, Abcam), 1:100 and incubated with the tissue sections at 4°C overnight. Specimens were then washed 3 times for 3 min each in PBST and incubated with biotinylated secondary antibody (Vector Laboratories) in blocking solution for 1h at room temperature. Then, specimens were washed 3 times in PBST and treated with ABC reagent (Vectastain ABC kit #PK-6100) for 30 min, followed by 3 washes for 3 min each. Finally, slides were stained for peroxidase for 3 min with the DAB (Di-amine-benzidine) substrate kit (SK-4100, Vector Laboratories), washed with water and counterstained with hematoxylin. Stained slides were photographed with an Olympus DP72 microscope. For Immunofluorescence studies of tissue specimens, antigen unmasking was performed in 1X Antigen Retrieval Buffer, Tris-EDTA Buffer, pH 9.0 (ab93684, Abcam), rinsed briefly with dH2O, washed for 5min in PBS, followed by 5 min in 0.05% Tween-20/TBS (TBST). Specimens were then blocked using 0.5% blocking reagent (FP1020, PerkinElmer) diluted in TBS (TNB) for 1h and incubated in primary antibodies at 4°C overnight in TNB, followed by 3 min washes in TBST and 1h incubation in secondary antibodies (1:400) diluted in TNB. Finally, specimens were rinsed 3 × 5 min in TBST and mounted using mounting media with DAPI (H-1500, Vectashield Vector Laboratories). For cell culture staining, samples were fixed in 4% paraformaldehyde (Fisher Scientific) in PBS for 20 min followed by washing four times in PBS. Residual paraformaldehyde was removed with three 5-min washes with 100 mM glycine in PBS. Cells were permeabilized with 0.3% Triton X-100/PBS and blocked for 30 min with blocking buffer (5% bovine serum albumin, 0.2% Triton X-100, 0.05% Tween 20, PBS). Coverslips were incubated with primary (1:100) and secondary (1:1000) antibodies diluted in blocking buffer for 1 h and 40 min, respectively, at room temperature, and nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Coverslips were mounted with immu-mount (Thermo-Shandon, Pittsburgh, PA). The following primary antibodies were used: rabbit anti-Sox9 (1:500; Millipore AB5535); goat anti-HNF4α (SC6556, 1:250; Santa Cruz). Secondary antibodies were conjugated to Alexa-488, 555 or 647 (Life Technologies). Confocal images were captured by a scanning laser confocal microscope (Nikon Eclipse Ti; Nikon) using 40× or 60× oil lenses and captured using NIS-Elements software (Nikon). Images were processed using ImageJ and Adobe Photoshop CS4. For quantification of signal intensity ratios, channel intensities of individual cells were selected from at least 60 cells per sample from at 3- 5 images using the Measure RGB plugin of ImageJ and exported into Excel for further analysis.
2HG production and measurement
2HG-octyl esters were prepared from commercially available 2HG lactones (CAS# 21461-84-7 (S-) and 53558-93-3 (R-)). Esterification of lactone with 1-octanol and EDCI (CAS# 25952-53-8), followed by opening of the lactone with lithium hydroxide in tetrahydrofuran/water, yielded the 2HG-octyl esters (free carboxylic acid) upon neutralization and chromatographic purification. The sodium salts of both 2HG-octyl esters are available from Toronto Research Chemicals. LC-MS/MS analysis was performed using an AB Sciex 4000 (Framingham, MA) operating in negative electrospray mode. MRM data were acquired for each compound using the following transitions: 2HG (146.9/128.8 amu), 13C5-2HG (151.9/133.8 amu) and 3HMG (160.9/98.9 amu). Chromatographic separation was performed using an ion-exchange column (Bio-Rad Fast Acid Analysis, 9µm, 7.8 mm X 100 mm; Bio-Rad). The flow rate was 1ml/min of 0.1 % formic acid in water with a total run time of 4 minutes. Samples were prepared as follows: Tissues were weighed out and homogenized in appropriate volumes of PBS using Tissue Lyser (Qiagen). 10µl of homogenate were extracted by adding 200µl of methanol with 200 ng/ml 3HMG as internal standard. Cell pellets containing 1×106 cells each were resuspended in specified volumes of 80:20 MeOH:water, centrifuged for 10 minutes at 14,000 rpm. 30µl of supernatant were extracted by adding 170µl of methanol with 200 ng/ml 13C5-2HG as internal standard. All sample types were then vortexed, centrifuged at 4000 rpm at 5°C and 150µl of supernatant transferred to a clean 96-well plate. The samples were dried down and reconstituted in 200µl 0.1% formic acid in water, and 10 µl injected on a column.
Human samples and genomic studies
An initial cohort of 149 formalin-fixed, paraffin-embedded (FFPE) samples was obtained from ICC patients resected between 1995 and 2007 at three centers from the HCC Genomic Consortium: IRCCS Istituto Nazionale Tumori (Milan), Mount Sinai School of Medicine (New York), and Hospital Clinic (Barcelona). Pathological diagnosis of ICC was confirmed by two independent liver pathologists (MS and ST). DNA and RNA extraction from human tissue, whole-genome gene-expression profiling (DASL assay Illumina), and the Infinium FFPE restoration kit (Illumina) are extensively described elsewhere36. The study protocol was approved at each center’s Institutional Review Board. A total of 127 samples had available data for both gene expression and DNA copy number changes and were analyzed for molecular characterization to define molecular-based classes. IDH1 and 2 mutations occurring at Arginine residue 132 and 172 respectively were analyzed by PCR technique using primers 5’-GTGGCACGGTCTTCAGAGA-3’ (forward primer) and 5’- TGCTTAATGGGTGTAGATACCAA-3’ (reverse primer), and 5’-GCCCACACATTTGCACTCTA-3’ (forward primer) and 5’-AAGGAAAGCCACGAGACAGA-3’ (reverse primer). The PCR amplifications were performed in a volume of 25µL reaction mixture containing 1.5mM MgCl2, 0.2mM of each dNTP, 0.125mM of each primer and 1U of Platinum Taq DNA Polymerase (Invitrogen). PCR products were purified using the Qiaquick PCR purification kit (Qiagen) and sequenced using an Applied Biosystems 3700 DNA sequencer (ABI PRISM® 3730XL; Applied Biosystems). Good quality sanger sequencing data for both IDH1 and IDH2 was obtained for 107 ICC DNA samples and has been included in all the subsequent analyses.
Human Gene Set Enrichment Analysis
Genes, molecular pathways, and gene expression signatures associated with the classes were evaluated using Gene Set Enrichment Analysis (GSEA) for Molecular Signature Database gene sets (MSigDB, www.broadinstitute.org/msigdb). Data analysis was conducted using the GenePattern Analytical Toolkit37, whereas correlation with clinico-pathological parameters was performed with SPSS software® (version 18) as previously described36.
Extended Data
Extended Data Figure 1. Impact of mutant IDH1 and IDH2 on HB cell differentiation.
a. Primary HB cells were engineered to express the indicated human IDH alleles or empty vector (EV) under a Dox-inducible system. Lysates from HB cells cultured in the presence of increasing Dox concentrations were analyzed by immunoblot using an antibody that recognizes both murine and human IDH1 (Upper panel) or with an antibody specific to human IDH2 (lower panel); actin is the loading control. Note that 25 ng/ml Dox induces physiologic levels of IDH1 expression and was used in the experiments shown in Fig. 1 and 2 of the main text.
b. Photomicrographs of cells grown for 2 days on collagen-coated plates in the presence of 25ng/mL Dox.
c. HB cells cultured in the presence of increasing Dox concentrations were analyzed by LC-MS for levels of intracellular 2HG.
d. Growth curve of HBs cultivated on collagen-coated dishes.
e, f. HB cells from (a) were grown on collagen-coated or uncoated plates and analyzed for expression of hepatocyte markers by qRT-PCR.
g. Enrichment plot showing downregulation of a hepatocyte gene set (Gene Expression Omnibus, GSE28892) in R132C-expressing cells. NES = Normalized Enrichment Score. FDR = False discovery rate.
h. 2HG levels in HB cells expressing the indicated IDH alleles or EV and treated with AGI-5027 (+) or DMSO vehicle (-).
i, j. WT HBs were treated with 500µm octyl-(R) or (S) enantiomers of 2HG or DMSO vehicle, and tested for hepatocyte differentiation upon transfer to uncoated plates by hepatocyte sphere formation (i) or qRT-PCR (j).
k, l. HB cells expressing the indicated alleles were tested for biliary differentiation upon transfer to matrigel as assessed by qRT-PCR for the induction of biliary makers Bgp and Ggt1.
Scale bars, 100µm. Error bars indicate ±s.d. between technical duplicates. * Student’s t-test P<0.05. Data are representative of at least 2 independent experiments.
Extended Data Figure 2. Mutant IDH represses the HNF4α-mediated hepatocyte differentiation program.
a–c. Gene expression profiling of HB cells expressing the indicated alleles and grown on collagen. (a) Clustering analysis, (b) enrichment plot (GSEA) showing downregulation of HNF4α targets in the IDH mutant cells relative to controls, (c) GSEA plots using the collection of cis regulatory elements from the Molecular Signatures Database (C3 collection) reveals strong downregulation of genes containing consensus HNF4α and HNF1α binding sites in the IDH mutant HB cultures.
d. qRT-PCR showing expression of Hnf4a and its target genes in HB cells expressing either EV or IDH2 R172K and grown on collagen-coated plates.
e. HNF4α(7–9) expression (immunoblot) in HB cells grown on collagen-coated plates (quantification of HNF4α:actin is indicated). Data are representative of 2 independent experiments.
f, g. Immunofluorescence (IF) staining for HNF4α (using an antibody that detects all isoforms) in HB cells expressing EV or IDH1 R132C and grown on collagen (f). The chart shows quantification of the ratio of the HNF4α and DAPI signals. The specificity of the antibody was demonstrated by the diminished staining in cells with knockdown of endogenous HNF4α (g).
h. Immunoblot showing that R132C suppresses HNF4α(1–6) and HNF4α(7–9) isoforms on uncoated plates.
i, j. AGI-5027 restores Hnf4a(1–6) induction in R132C-expressing HBs as shown by qRT-PCR (i) and immunoblot (j). DMSO=vehicle.
k. WT HBs were treated with 500µm octyl-(R) or -(S) enantiomers of 2HG, DMSO vehicle, or media alone. Hnf4a1–6 levels were determined by qRT-PCR.
l, m. Cultures of HB cells expressing the indicated alleles were grown on collagen or transferred to uncoated plates for 5 days and subjected to chromatin immunoprecipitation (ChIP) for H3K4me3 (l) or H3K27me3 (m). Enrichment for the promoter regions of Hhex (highly expressed gene in HB cells), Hoxa10 (transcriptionally silent in HB cells) and P1 Hnf4a was measured by qPCR.
n, o. Analysis of WT HBs expressing shRNA control (shCTL) or targeting different HNF4a sequences (shHnf4a#1, shHnf4a#2): (n) hepatocyte marker expression (qRT-PCR) (o) HNF4a immunoblot.
Error bars indicate ±s.e.m for (f) and ±s.d. for (d, i, k-n) between technical duplicates. Scale bars indicate 50 µm. Student’s t-test * P<0.05.
Extended Data Figure 3. HNF4 α is dispensable for biliary differentiation.
a. WT HB cells expressing shRNA control (shCTL) or targeting HNF4α were tested for ability to undergo biliary differentiation upon transfer to matrigel as assessed by tubule formation 24h after transfer (left) and qRT-PCR for induction of the biliary markers Bgp and Ggt1 10 days after transfer to matrigel (right).
b. Immunoblot showing ectopic expression of HNF4α1 in HB cells. Error bars indicate ±s.d. between technical replicates. Scale bar indicates 100µm.
Extended Data Figure 4. Analysis of GEM model with hepatocyte-specific mutant IDH expression.
a. Schematic of Dox-inducible IDH2 mutant alleles (See Methods for details). Mice harboring mutant human IDH2 alleles were crossed with Alb-Cre and LSL-rtTA strains for liver-specific expression.
b–d. Characterization of expression pattern of mutant IDH2 in Tet-IDH2 R140Q and Tet-R172K GEM models compared to control WT mice, after 4 weeks of doxycycline supplementation reveals hepatocyte-specific expression, consistent with previous models using this transgenic targeting system40. (b) IF analysis of Tet-IDH2 R140Q and control WT livers using an antibody that detects both endogenous and transgenic IDH2 expression. Note that transgenic IDH2 R140Q is expressed in the hepatocytes which stain for HNF4α(1–6), but not in the bile ducts which are marked by CK19. (c) IF analysis of Tet-IDH2 R140Q and control WT livers using an antibody that is specific to IDH2 R140Q, showing an identical pattern of transgene expression.
(d) IF analysis of Tet-IDH2 R172K and control WT livers using an antibody that is specific to IDH2 R172K. This allele is also expressed in the hepatocytes, but shows more focal expression compared to R140Q. In (b) and (c) an antibody to total HNF4α was used to label hepatocytes. Scale bars = 50µm.
Extended Data Figure 5. Characterization GEM model with hepatocyte-specific mutant IDH expression in the presence or absence of liver injury.
a. Measurement of 2HG levels in liver lysates from IDH2 mutant and control mice treated with Dox for 1 month.
b. Uninjured WT and IDH2 R140Q livers exhibit comparable expression of the hepatocyte markers, Hnf4a, Adh1, Alb, and Aldob and biliary markers Sprr1a and Onecut1 by qRT-PCR. c-d. Tet-R140Q mice and littermate controls (WT) receiving Dox were fed a DDC-containing diet for 5 days before being switched to normal chow for 1 week (See main text, Fig. 3a for schematic). Mutant IDH2 does not provoke liver injury as reflected by (c) comparable levels of serum AST, Tbili and ALT, and (d) absence of cleaved caspase 3 staining in Tet-R140Q compared to WT mice. Duodenum from a TNFα-treated mouse was used as a positive control for cleaved caspase-3 staining. Scale bars = 50µm.
Extended Data Figure 6. Characterization of response to liver injury in GEM model with hepatocyte-specific mutant IDH expression.
Tet-R140Q or Tet-R172K mice and littermate controls (WT) receiving Dox were fed a DDC-containing diet for 5 days before being switched to normal chow for 7 or 21 days (See main text, Fig. 3a for schematic).
a. Representative H&E images of livers from WT and Tet-R140Q mice analyzed at day 7 and 21.
b. Quantification of Ki67-positive cells for indicated markers as shown in main text, Fig. 3e (N = 3 mice per group, from at least 5 high-powered field per mouse scored). Error bars, ±s.e.m. Data from Fig. 3f (day 21) is reproduced here for comparison.
c, d.IF analysis of IDH2 mutant livers showing that Ki-67 co-localizes with HNF4α (c) and R140Q-expressing cells (d). As shown in main text, Fig. 3e, these cells express lower levels of HNF4α compared to WT hepatocytes.
e, f. Tet-R172K mice and littermate controls (WT) receiving Dox were fed a DDC-containing diet for 5 days before being switched to normal chow for 1 week. IF analysis of IDH2 mutant livers shows that Ki-67 co-localizes with HNF4α and R172K-expressing cells (e) and greater numbers of Ki-67+,HNF4α+ cells in Tet-R172K compared to WT mice (f).
Error bars indicate ±s.e.m. between 3 mice; Scale bars, 50µm. Student’s t-test * P<0.05.
Extended Data Figure 7. Characterization of LSL-IDH2 R172K GEMM.
a. Schematic of LSL-IDH2 R172K GEMM (See Methods for details). Mice harboring the LSL-IDH2 R172K allele were crossed with the Alb-Cre strain to target expression to the liver.
b. IF analysis revealed specific expression of IDH2 R172K in CK19+ biliary cells, whereas hepatocytes were negative.
c. IF analysis showing comparable low levels of proliferation (Ki-67) in LSL-R172K and littermate control livers at 3 months of age. Scale bars = 50µm.
Extended Data Figure 8. Impact HNF4 α ablation on IHCC pathogenesis in vivo.
a–c. Albumin-CreERT2; Hnf4afl/fl (HNF4αfl/fl,Cre) mice were treated with diethylnitrosamine (DEN), on post-natal day 15, and subsequently administered Tamoxifen (TAM) or corn oil (CO) control at 8 months of age as described20. DEN is activated to its carcinogenic form by cytochrome P450 (CYP) enzymes, including CYP2E1, specifically in hepatocytes38,39. Livers were harvested two months later and analyzed by H&E, IF and IHC. In Hnf4afl/fl,Cre + TAM livers, HCC arises from HNF4α+ cells that have escaped HNF4α ablation (a), while CK19+ IHCC stains negative for HNF4α (b, right panel). Normal liver from control corn oil-treated livers is shown in (b, left panel).
c. Representative IHC (top) and IF images (bottom) show marked expansion of Sox9+ oval cells in TAM-injected mice. Surrounding these oval cells are HNF4α+ hepatocytes which escaped Hnf4a ablation in TAM-injected mice.
d. H&E stain confirming that the liver tumour arising in a KrasG12D mouse is an HCC.
e. Measurement of 2HG in liver tumours from Alb-Cre; LSL-R172K; KrasG12D mice compared to normal liver from control mice (N=2). Scale bars, 50 µm. Error bars, ±s.d. * Student’s t-test P<0.05.
Extended Data Figure 9. Expression of mutant IDH2 in the LSL R172K GEM model.
a, b. IF analysis to characterize expression of IDH2 in the liver of LSL-R172K; KrasG12Dcompound mice. (a) Staining with an antibody that recognizes both endogenous WT and mutant IDH2 shows that endogenous IDH2 is expressed in bile duct in the non-diseased liver from KrasG12D mice (top panels). In LSL-R172K; KrasG12D mice, total IDH2 staining is at near endogenous levels in the normal bile ducts (middle panels) and in the BilIN lesions (bottom panels). (b) IF using an antibody specific to IDH2-R172K showing specific expression of the R172K transgene in CK19+ bile ducts (middle row) and BilINs (bottom row) in LSL-R172K, KrasG12D animals. Scale bars = 50µm.
Extended Data Figure 10. Stem cell features in IDH mutant murine and human IHCC pathogenesis.
a. IDH2 R172K expression using mutant-specific antibodies in livers from LSL-R172K and KrasG12D, R172K compound mice showing specific expression of the R172K transgene in Sox9+ oval cells and IHCC. Scale bars = 50µm.
b. Normalized enrichment plot of a cohort of 127 human IHCC samples genotyped for IDH status, showing that the subset of tumors with IDH1/IDH2 mutations exhibits enrichment of a gene signature40 that identifies IHCCs with hepatic stem cell/progenitor features.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Mostoslavsky, L. Ellison, A. Kimmelman, and members of the Bardeesy lab for valuable input. We also thank S. Thorgeirsson and J. Andersen for sharing unpublished datasets. This work was supported by grants from the NIH (R01CA136567-02 and P50CA1270003) and TargetCancer Foundation to N.B. N.B. holds the Gallagher Endowed Chair in Gastrointestinal Cancer Research at Massachusetts General Hospital. S.K.S. is the recipient of a Cholangiocarcinoma Foundation/Conquer Cancer Foundation of ASCO Young Investigator Award, and American Cancer Society Postdoctoral Fellowship (PF-13-294-01-TBG). C.A.P is the recipient of a CIHR postdoctoral fellowship. N.B., J.M.L, and D.S are members of the Samuel Waxman Cancer Research Foundation Institute Without Walls. J.M.L. and D.S. are supported by the Asociación Española para el Estudio del Cáncer
Footnotes
AUTHOR CONTRIBUTIONS
S.K.S. and C.A.P. contributed equally to the study. S.K.S., C.A.P., K.S.G., M.S.N. and S.G. carried out the experiments involving hepatoblasts and mouse models. K.R. and S.R. performed computational analysis on gene expression data. J.F. performed IHC on tissue sections. E.A.A. and K.W. assisted with the generation of the IDH mutant mice. V.D. analyzed the histology from the murine liver specimens. C.W. and U.A. carried out the experiments involving the HNF4a KO mice. D.S., H.C., O.M., and J.M.L. performed the GSEA analysis on human IHCC samples. A.X.Z., A.F.H., and C.R.F. were involved in the study design. K.Y., K.S., J.T., J.P.M., and C.G. developed and provided the AGI-5027 compound and measured 2HG in our samples. S.K.S., C.A.P., and N.B. designed the experiments and wrote the paper. N.B. supervised the studies. All authors discussed the results and commented on the manuscript.
AUTHOR INFORMATION
K.Y., K.S., J.T., J.P.M., and C.G. are employees of Agios Pharmaceuticals. All other authors have no financial conflict of interest.
REFERENCES
- 1.Borger DR, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol. 2010;28:3531–3540. doi: 10.1200/JCO.2009.27.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014 doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voss JS, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum Pathol. 2013;44:1216–1222. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol. 2013;3:485–513. doi: 10.1002/cphy.c120014. [DOI] [PubMed] [Google Scholar]
- 12.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Strick-Marchand H, Weiss MC. Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology. 2002;36:794–804. doi: 10.1053/jhep.2002.36123. [DOI] [PubMed] [Google Scholar]
- 14.Davis MI, et al. Biochemical, Cellular and Biophysical Characterization of a Potent Inhibitor of Mutant Isocitrate Dehydrogenase IDH1. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.511030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo CJ, et al. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 16.Torres-Padilla ME, Fougere-Deschatrette C, Weiss MC. Expression of HNF4alpha isoforms in mouse liver development is regulated by sequential promoter usage and constitutive 3’ end splicing. Mech Dev. 2001;109:183–193. doi: 10.1016/s0925-4773(01)00521-4. [DOI] [PubMed] [Google Scholar]
- 17.Bluteau O, et al. Bi-allelic inactivation of TCF1 in hepatic adenomas. Nat Genet. 2002;32:312–315. doi: 10.1038/ng1001. [DOI] [PubMed] [Google Scholar]
- 18.Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. The Journal of biological chemistry. 2012;287:7345–7356. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Servitja JM, et al. Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol. 2009;29:2945–2959. doi: 10.1128/MCB.01389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walesky C, et al. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013;57:2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanger K, et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes & development. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preisegger KH, et al. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- 23.Malato Y, et al. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin S, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes & development. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Dell MR, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer research. 2012;72:1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avruch J, Zhou D, Fitamant J, Bardeesy N. Mst1/2 signalling to Yap: gatekeeper for liver size and tumour development. Br J Cancer. 2011;104:24–32. doi: 10.1038/sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tefferi A, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic-or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 30.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 31.Belteki G, et al. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Dell MR, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer research. 2012;72:1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 34.Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin S, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes & development. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sia D, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
EXTENDED DATA FIGURE LEGEND REFERENCES
- 38.Kang JS, Wanibuchi H, Morimura K, Gonzalez FJ, Fukushima S. Role of CYP2E1 in diethylnitrosamine-induced hepatocarcinogenesis in vivo. Cancer research. 2007;67:11141–11146. doi: 10.1158/0008-5472.CAN-07-1369. [DOI] [PubMed] [Google Scholar]
- 39.Verna L, Whysner J, Williams GM. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacology & therapeutics. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 40.Oishi N, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.