Abstract
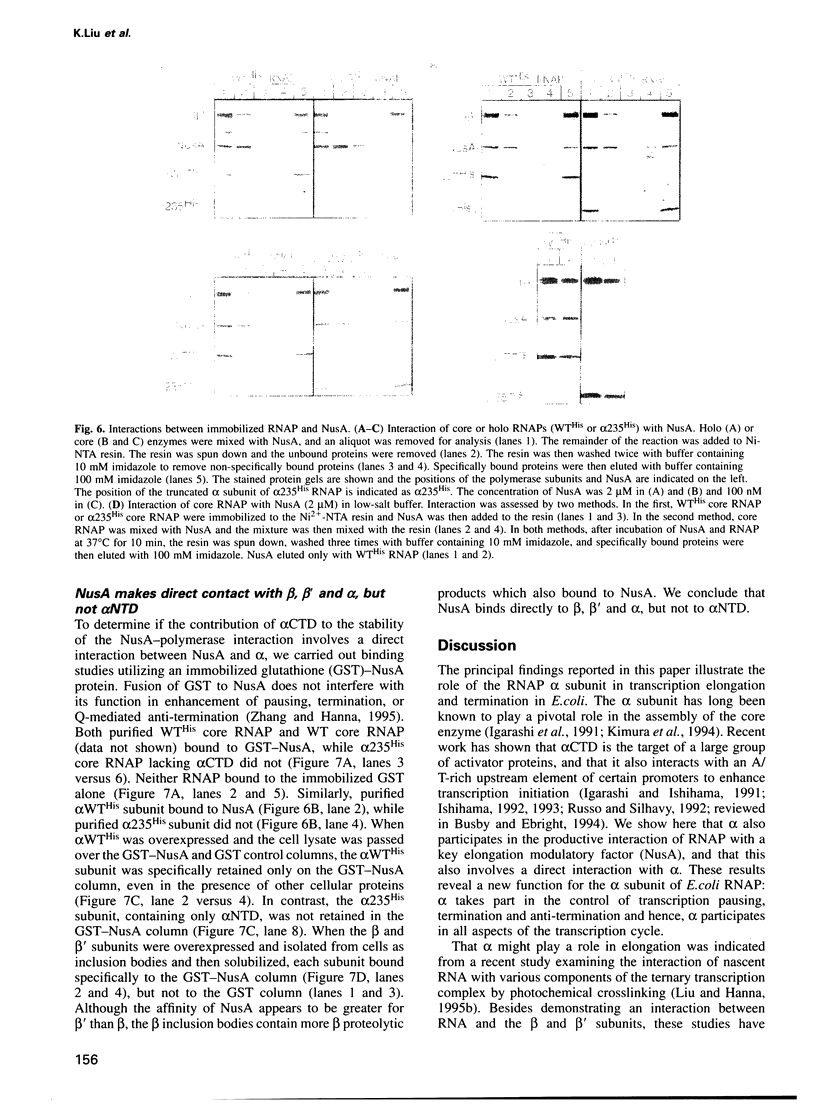
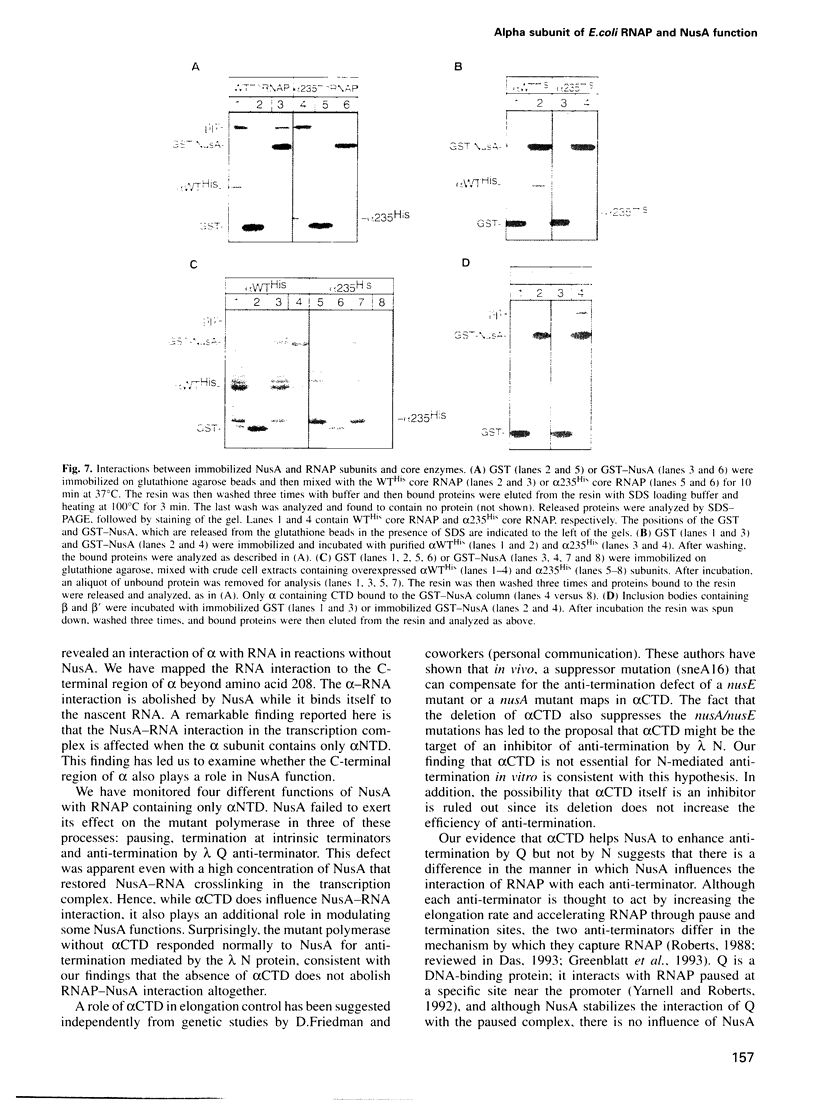
The alpha subunit (alpha) of RNA polymerase (RNAP) is critical for assembly of polymerase and positive control of transcription initiation in Escherichia coli. Here, we report that alpha also plays a role in transcription elongation, and this involves a direct interaction between alpha and NusA factor. During in vitro transcription without NusA, alpha interacts with the nascent RNA, as revealed by photocrosslinking. When NusA is present, RNA crosslinks to NusA rather than to alpha. We show that this NusA-RNA interaction is diminished during transcription with an RNAP mutant that lacks the C-terminus of alpha beyond amino acid 235, including the so-called alpha CTD. The absence of alpha CTD also affects NusA's ability to enhance transcription pausing, termination at intrinsic terminators and anti-termination by the phage lambda Q anti-terminator, but not anti-termination by the lambda N anti-terminator. NusA functions are not recovered even when transcription with mutant RNAP is done with excess NusA, a condition which does restore NusA-RNA crosslinking. By affinity chromatography, we show that NusA interacts directly with alpha, and also with beta and beta', but not with mutant alpha. Hence, alpha-NusA interaction is vital for the control of transcript elongation and termination.
Full text
PDF
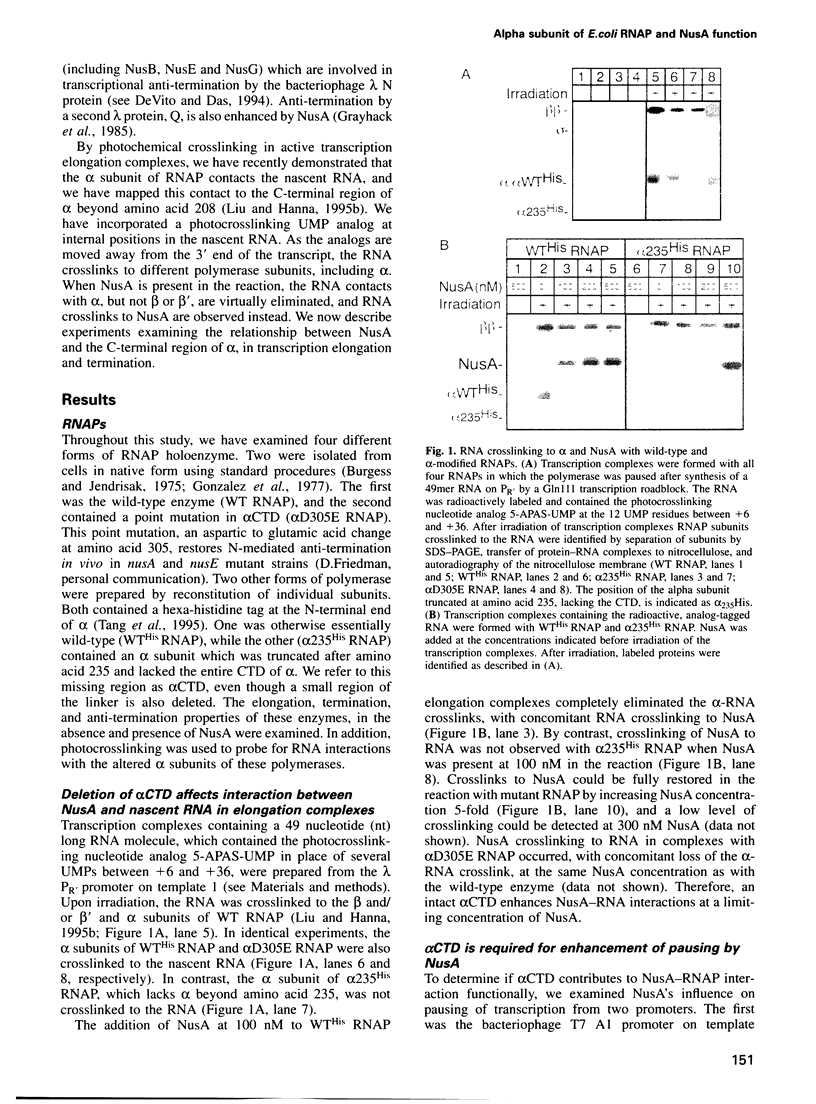

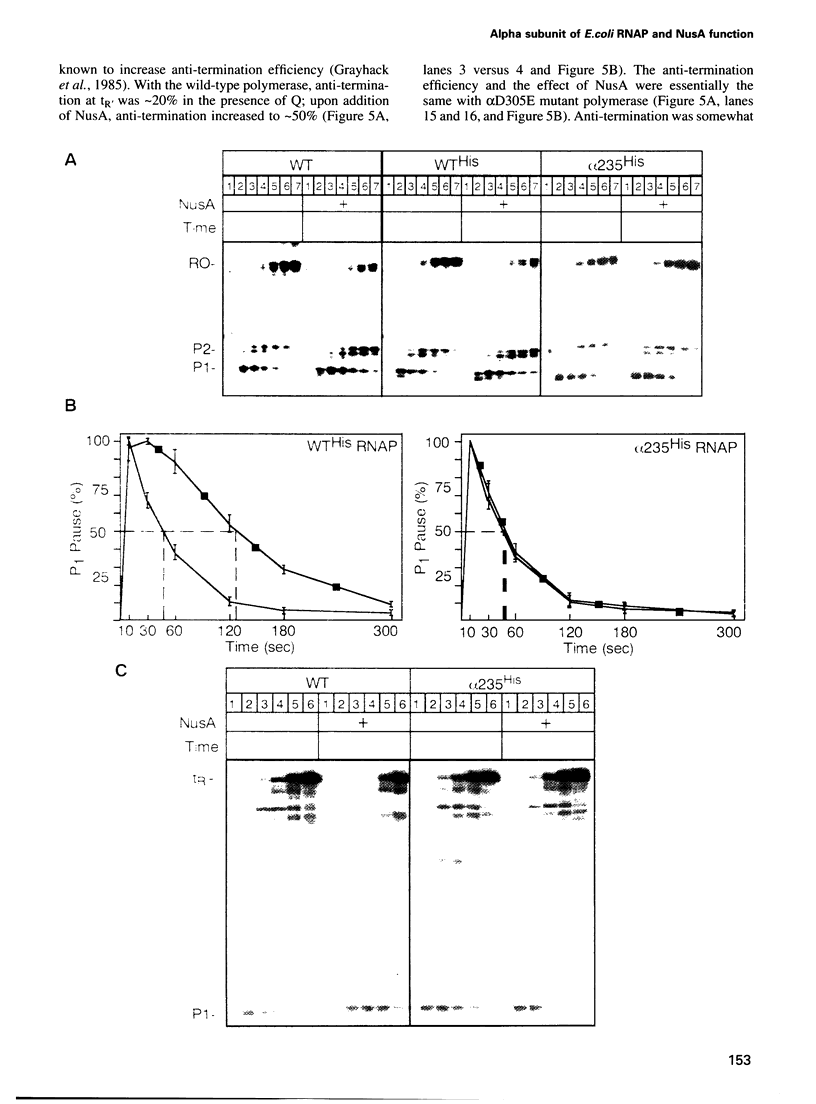
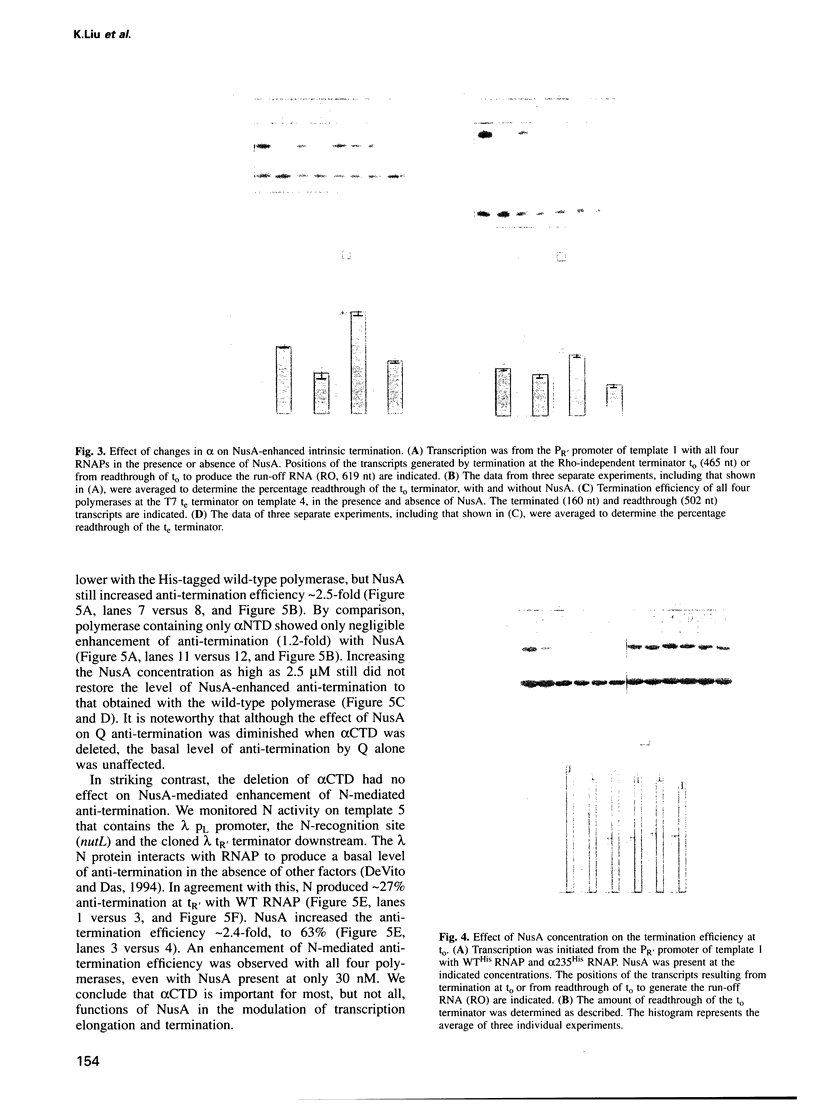
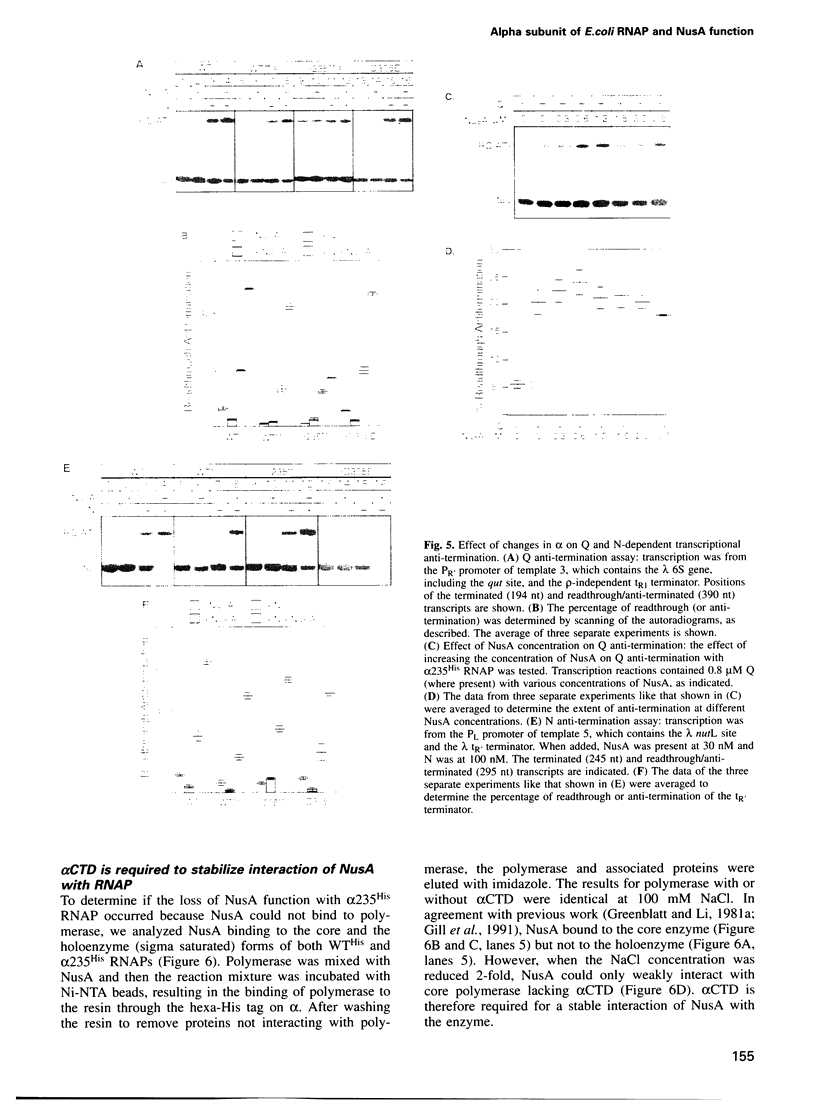




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blatter E. E., Ross W., Tang H., Gourse R. L., Ebright R. H. Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994 Sep 9;78(5):889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Bollag G., Kearney C. A., Molineux I., Chamberlin M. J. Tau factor from Escherichia coli mediates accurate and efficient termination of transcription at the bacteriophage T3 early termination site in vitro. J Mol Biol. 1987 Nov 5;198(1):43–49. doi: 10.1016/0022-2836(87)90456-6. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Burgess R. R. Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6168–6176. [PubMed] [Google Scholar]
- Busby S., Ebright R. H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994 Dec 2;79(5):743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Chan C. L., Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993 Sep 5;233(1):25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Garcia-Mena J., DeVito J., Wolska K., Das A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):4061–4065. doi: 10.1073/pnas.92.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ebright Y. W., Ebright R. H. Identification of the target of a transcription activator protein by protein-protein photocrosslinking. Science. 1994 Jul 1;265(5168):90–92. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- DeVito J., Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissinger S., Hanna M. M. RNA-protein interactions in a Nus A-containing Escherichia coli transcription complex paused at an RNA hairpin. J Mol Biol. 1991 May 5;219(1):11–25. doi: 10.1016/0022-2836(91)90853-x. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Greenblatt J., Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982 Jul;29(3):945–951. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Schauer A. T., Baumann M. R., Baron L. S., Adhya S. L. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B., Das A. nusB: a protein factor necessary for transcription antitermination in vitro by phage lambda N gene product. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6305–6309. doi: 10.1073/pnas.81.20.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C., Weitzel S. E., von Hippel P. H. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991 Jul 20;220(2):307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Grayhack E. J., Yang X. J., Lau L. F., Roberts J. W. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985 Aug;42(1):259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981 May;24(2):421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J Mol Biol. 1981 Mar 25;147(1):11–23. doi: 10.1016/0022-2836(81)90076-0. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., McLimont M., Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981 Jul 16;292(5820):215–220. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature. 1993 Jul 29;364(6436):401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991 Feb 1;192(2):262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hanna M. M., Dissinger S., Williams B. D., Colston J. E. Synthesis and characterization of 5-[(4-Azidophenacyl)thio]uridine 5'-triphosphate, a cleavable photo-cross-linking nucleotide analogue. Biochemistry. 1989 Jul 11;28(14):5814–5820. doi: 10.1021/bi00440a017. [DOI] [PubMed] [Google Scholar]
- Hanna M. M. Photoaffinity cross-linking methods for studying RNA-protein interactions. Methods Enzymol. 1989;180:383–409. doi: 10.1016/0076-6879(89)80113-2. [DOI] [PubMed] [Google Scholar]
- He B., Riggs D. L., Hanna M. M. Preparation of probe-modified RNA with 5-mercapto-UTP for analysis of protein-RNA interactions. Nucleic Acids Res. 1995 Apr 11;23(7):1231–1238. doi: 10.1093/nar/23.7.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Ishihama A. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991 Jun 14;65(6):1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993 May;175(9):2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Role of the RNA polymerase alpha subunit in transcription activation. Mol Microbiol. 1992 Nov;6(22):3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- Ito K., Egawa K., Nakamura Y. Genetic interaction between the beta' subunit of RNA polymerase and the arginine-rich domain of Escherichia coli nusA protein. J Bacteriol. 1991 Feb;173(4):1492–1501. doi: 10.1128/jb.173.4.1492-1501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Cashel M., Friedman D. I., Nakamura Y., Walter W. A., Gross C. A. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J Mol Biol. 1988 Nov 20;204(2):247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Chamberlin M. J. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981 Mar 25;256(6):2777–2786. [PubMed] [Google Scholar]
- Kimura M., Fujita N., Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase. Deletion analysis of the amino-terminal assembly domain. J Mol Biol. 1994 Sep 16;242(2):107–115. doi: 10.1006/jmbi.1994.1562. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Chamberlin M. J. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981 Dec;27(3 Pt 2):523–531. doi: 10.1016/0092-8674(81)90394-9. [DOI] [PubMed] [Google Scholar]
- Landick R., Yanofsky C. Isolation and structural analysis of the Escherichia coli trp leader paused transcription complex. J Mol Biol. 1987 Jul 20;196(2):363–377. doi: 10.1016/0022-2836(87)90697-8. [DOI] [PubMed] [Google Scholar]
- Lee D. N., Phung L., Stewart J., Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J Biol Chem. 1990 Sep 5;265(25):15145–15153. [PubMed] [Google Scholar]
- Liu K., Hanna M. M. NusA contacts nascent RNA in Escherichia coli transcription complexes. J Mol Biol. 1995 Apr 7;247(4):547–558. doi: 10.1006/jmbi.1994.0161. [DOI] [PubMed] [Google Scholar]
- Liu K., Hanna M. M. NusA interferes with interactions between the nascent RNA and the C-terminal domain of the alpha subunit of RNA polymerase in Escherichia coli transcription complexes. Proc Natl Acad Sci U S A. 1995 May 23;92(11):5012–5016. doi: 10.1073/pnas.92.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Nudler E., Kashlev M., Nikiforov V., Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995 May 5;81(3):351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- Pavco P. A., Steege D. A. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990 Jun 15;265(17):9960–9969. [PubMed] [Google Scholar]
- Ring B. Z., Roberts J. W. Function of a nontranscribed DNA strand site in transcription elongation. Cell. 1994 Jul 29;78(2):317–324. doi: 10.1016/0092-8674(94)90300-x. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Phage lambda and the regulation of transcription termination. Cell. 1988 Jan 15;52(1):5–6. doi: 10.1016/0092-8674(88)90523-5. [DOI] [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Russo F. D., Silhavy T. J. Alpha: the Cinderella subunit of RNA polymerase. J Biol Chem. 1992 Jul 25;267(21):14515–14518. [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry. 1984 Jan 17;23(2):197–203. doi: 10.1021/bi00297a004. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987 Jun 20;195(4):809–818. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- Sparkowski J., Das A. Simultaneous gain and loss of functions caused by a single amino acid substitution in the beta subunit of Escherichia coli RNA polymerase: suppression of nusA and rho mutations and conditional lethality. Genetics. 1992 Mar;130(3):411–428. doi: 10.1093/genetics/130.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Severinov K., Goldfarb A., Ebright R. H. Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A., Burgessrr Cyclic re-use of the RNA polymerase sigma factor. Nature. 1969 May 10;222(5193):537–540. doi: 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- Wang D., Meier T. I., Chan C. L., Feng G., Lee D. N., Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995 May 5;81(3):341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- Whalen W. A., Das A. Action of an RNA site at a distance: role of the nut genetic signal in transcription antitermination by phage-lambda N gene product. New Biol. 1990 Nov;2(11):975–991. [PubMed] [Google Scholar]
- Yang X. J., Hart C. M., Grayhack E. J., Roberts J. W. Transcription antitermination by phage lambda gene Q protein requires a DNA segment spanning the RNA start site. Genes Dev. 1987 May;1(3):217–226. doi: 10.1101/gad.1.3.217. [DOI] [PubMed] [Google Scholar]
- Yang X. J., Roberts J. W. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5301–5305. doi: 10.1073/pnas.86.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell W. S., Roberts J. W. The phage lambda gene Q transcription antiterminator binds DNA in the late gene promoter as it modifies RNA polymerase. Cell. 1992 Jun 26;69(7):1181–1189. doi: 10.1016/0092-8674(92)90639-t. [DOI] [PubMed] [Google Scholar]
- Zaychikov E., Denissova L., Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20: "jumping" of RNA polymerase and asymmetric expansion and contraction of the "transcription bubble". Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1739–1743. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hanna M. M. Expression and functional characterization of Escherichia coli NusA and lambda Q as glutathione S-transferase fusion proteins. Protein Expr Purif. 1995 Oct;6(5):625–631. doi: 10.1006/prep.1995.1082. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Hanna M. M. NusA changes the conformation of Escherichia coli RNA polymerase at the binding site for the 3' end of the nascent RNA. J Bacteriol. 1994 Mar;176(6):1787–1789. doi: 10.1128/jb.176.6.1787-1789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Fujita N., Igarashi K., Ishihama A. Mapping the cAMP receptor protein contact site on the alpha subunit of Escherichia coli RNA polymerase. Mol Microbiol. 1992 Sep;6(18):2599–2605. doi: 10.1111/j.1365-2958.1992.tb01437.x. [DOI] [PubMed] [Google Scholar]












