SUMMARY
Thousands of cis-elements in genomes are predicted to have vital functions. While conservation, activity in surrogate assays, polymorphisms, and disease mutations provide functional clues, deletion from endogenous loci constitutes the gold-standard test. A GATA-2-binding, Gata2 intronic cis-element (+9.5) required for hematopoietic stem cell genesis in mice is mutated in a human immunodeficiency syndrome. As +9.5 is the only cis-element known to mediate stem cell genesis, we devised a strategy to identify functionally comparable enhancers (“+9.5-like”) genome-wide. Gene editing revealed +9.5-like activity to mediate GATA-2 occupancy, chromatin opening, and transcriptional activation. A +9.5-like element resided in Samd14, which encodes a protein of unknown function. Samd14 increased hematopoietic progenitor levels/activity, promoted signaling by a pathway vital for hematopoietic stem/progenitor cell regulation (Stem Cell Factor/c-Kit), and c-Kit rescued Samd14 loss-of-function phenotypes. Thus, the hematopoietic stem/progenitor cell cistrome revealed a mediator of a signaling pathway that has broad importance for stem/progenitor cell biology.
INTRODUCTION
The ease of accessing genome sequences, “epigenetic” maps, and a plethora of bioinformatic tools have catalyzed efforts to translate nucleotide sequence into functional principles. Perhaps the most rudimentary problem involves identifying small DNA sequences that constitute cis-regulatory elements, primary determinants of gene expression and therefore cellular phenotypes. This problem may seem quite tractable, given chromatin immunoprecipitation (ChIP) for acquiring snapshots of protein binding to chromatin and gene editing technologies. However, only a subset of the thousands of a given cis-element are occupied in cells. Integrating factor co-occupancy, evolutionary conservation, and chromatin environment increases the fidelity of predictions of cis-element occupancy. These parameters do not invariably predict importance, however, as cis-elements bound by multiple factors have been deleted from a genome with little to no consequence (Bender et al., 2000; Sanalkumar et al., 2014; Snow et al., 2011). Occupancy measured by ChIP may reflect factor trapping at sites where they do not function, redundancy, or actions not measurable by existing assays. Sifting through cis-element ensembles to identify functional elements remains challenging.
Dissecting mammalian genome function ushered in cis-element “encyclopedias” (Yue et al., 2014) presumed to harbor a treasure-trove of regulatory content. As intrinsic and environmental mechanisms mold chromatin structure and confer plasticity in specialized contexts, it is crucial to address genome science problems with biologically robust systems. Given lineage relationships between hematopoietic stem/progenitor cells and progeny, and regenerative biology/medicine significance, the hematopoietic system is instructive as a model to discover mechanisms governing fundamental processes, including cell fate determination and gene regulation (Orkin and Zon, 2008; Rieger and Schroeder, 2012).
A single protein, GATA-2, governs hematopoietic stem cell (HSC) genesis from hemogenic endothelium in the aorta gonad mesonephros (AGM) region of the embryo and development of the hematopoietic system (de Pater et al., 2013; Tsai et al., 1994). GATA-2 also controls proliferation/survival of hematopoietic progenitors (Tsai and Orkin, 1997). In hemogenic endothelium, GATA-2 instigates a complex genetic network (Gao et al., 2013), including the regulator of hematopoiesis/leukemogenesis Runx1 (Wang et al., 1996). Since reduced GATA-2 expression/activity causes primary immunodeficiency, myelodysplastic syndrome, and myeloid leukemia (Dickinson et al., 2014; Spinner et al., 2013), the integrity of GATA-2-dependent genetic networks must be maintained. Many questions remain unanswered regarding the composition and dynamics of GATA-2 target gene ensembles. Given the caveats of extrapolating chromatin binding to function, traversing this divide will benefit from new approaches.
We described a cis-element essential for GATA-2 function (Gao et al., 2013; Johnson et al., 2012), which provides a unique opportunity to elucidate GATA-2 mechanisms genome-wide. Deletion of an intronic sequence 9.5 kb downstream of the Gata2 transcription start site (CATCTG-8bp-AGATAA), reduces Gata2 expression and abolishes the capacity of hemogenic endothelium to generate HSCs in the AGM, thereby causing anemia and embryonic lethality (Gao et al., 2013; Johnson et al., 2012). This contrasts with deletions of other Gata2 cis-elements bearing chromatin attributes that imply importance, but lack essential functions (Sanalkumar et al., 2014; Snow et al., 2011). The +9.5 conforms to an E-box-spacer-GATA composite element (Grass et al., 2006; Wozniak et al., 2007), originally reported to mediate assembly of a complex containing GATA-1 and Scl/TAL1 transcription factors and the coactivators Ldb1 and Lmo2 (Wadman et al., 1997). GATA-2 occupies a small fraction of these genomic elements (Fujiwara et al., 2009; Wozniak et al., 2008).
Herein, we leveraged +9.5 structure/function to establish an ensemble of GATA-2-regulated cis-elements termed the “hematopoietic stem/progenitor cell (HSPC) cistrome”. We envisioned that this cistrome would reveal new GATA factor-dependent pathways that control HSPC genesis/function and would constitute a resource for dissecting mechanisms governing the function of an abundant class of cis-elements in a genome – GATA motifs. We devised a strategy to identify GATA-2-regulated cis-elements, based on sequence/attributes shared with the +9.5. A “+9.5-like” cis-element resided in Samd14, encoding a sterile alpha motif domain protein of unknown function. Samd14 has sequence homology to neurabin-2 (Allen et al., 1997), which opposes β-arrestin-mediated suppression of G-protein coupled receptor (GPCR) signaling (Wang et al., 2004). GATA-2 upregulated Samd14 expression, which promoted stem cell factor (SCF)/c-Kit signaling and hematopoietic progenitor function. This mechanism exemplifies the unique biological/mechanistic content that can be mined from the cistrome and how our strategy can guide the traversal from genetic sequence and epigenetic signatures to new modes of cell regulation.
RESULTS
GATA-2-Regulated Stem/Progenitor Cell Cistrome
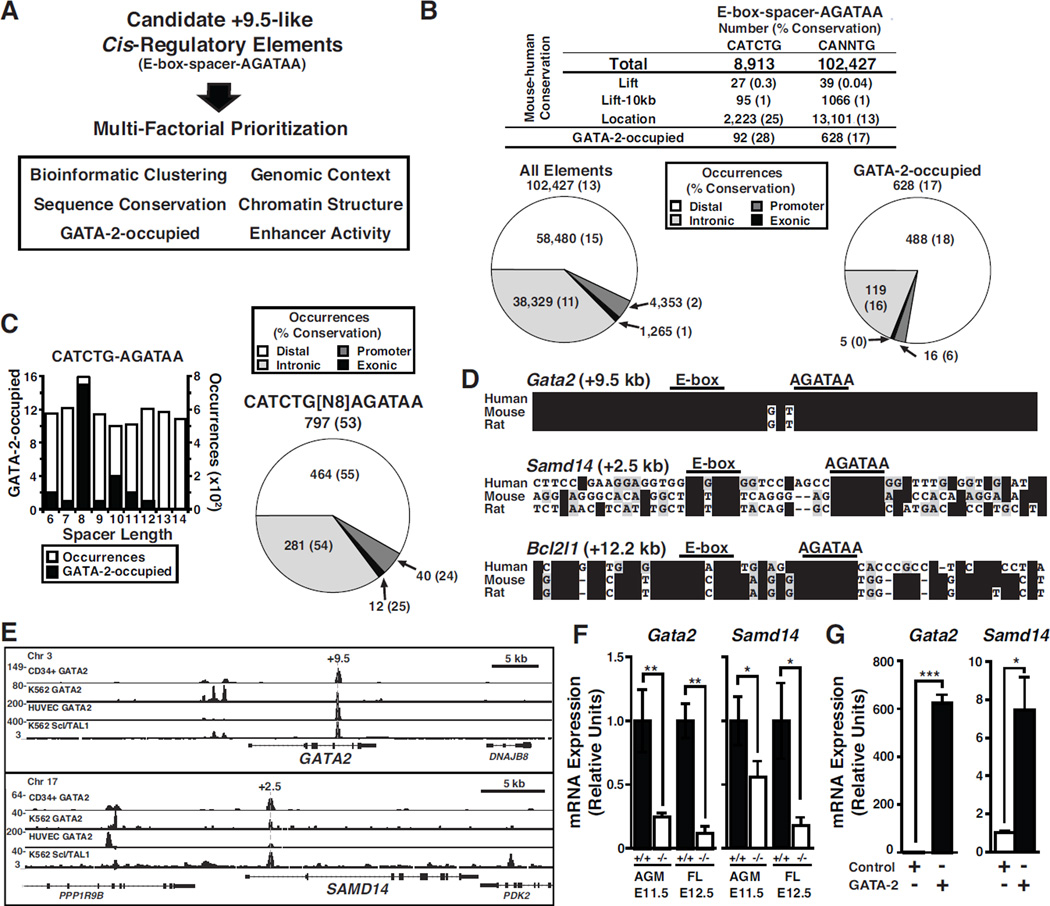
To discover an ensemble of E-box-GATA composite cis-elements resembling the Gata2 +9.5, we used multiple parameters to identify and analyze candidate sequences (Figure 1A). The human genome contains 102,427 occurrences of CANNTG, followed by a 6–14 bp spacer and AGATAA. This number drops 11.5-fold to 8,913 when CATCTG, is considered. Only small percentages (0.4% for CANNTG-(N6–14)-AGATAA and 0.3% CATCTG-(N6–14)-AGATAA) of sequences are conserved between the human and mouse genomes using the standard lift-over utility of the UCSC Genome Browser. To apply a broader definition of conservation, we annotated these elements as distal, promoter, intronic, and exonic, relative to known genes and assessed whether these elements exhibited location-based conservation between human and mouse. This comparison revealed that 13% and 25% of the human CANNTG-(N6–14)-AGATAA and CATCTG-(N6–14)-AGATAA elements are conserved in mouse, respectively (Figure 1B). We integrated GATA-2 occupancy data from CD34+ bone marrow cells (Beck et al., 2013) and observed that 17% and 28% of the GATA-2-occupied CANNTG-(N6–14)-AGATAA and CATCTG-(N6–14)-AGATAA elements were conserved. The conserved elements were located in diverse contexts and not predominantly at promoters (Figure 1B).
Figure 1. GATA-2-regulated Hematopoietic Stem/Progenitor Cell Cistrome.
(A) Gata2 +9.5 sequence and molecular attributes used to prioritize a “+9.5-like” element cohort. (B) Human to mouse conservation analysis of composite elements with CANNTG or CATCTG motifs by genome lift-over position or annotated location. Pie charts depict the location of human composite elements at distal, intronic, promoter and exonic. Pie chart values represent the number of elements in each location, and percent conserved in mouse (parentheses). left, all elements; right, GATA-2-occupied elements. (C) GATA-2-occupancy in murine Lin− progenitor cells at sites containing CATCTG-(N)x-AGATAA and variable spacer lengths. The pie chart depicts the location of 797 mouse elements with CATCTG-(N8)-AGATAA, and the percent conserved in human (parentheses). (D) Gata2 +9.5, Samd14 +2.5 and Bcl2l1 +12.2 conservation. (E) ChIP-seq of human CD34+ bone marrow, K562, and HUVEC cells at GATA2 and SAMD14 (GEO accessions: GSE18829, GSE29531). (F) Gata2 and Samd14 mRNA expression in E11.5 AGM and E12.5 fetal liver from the +9.5 mutant mouse (Johnson et al., 2012). (G) Gata2 and Samd14 mRNA expression in fetal liver cells infected with control (empty vector) or GATA-2-expressing retrovirus. Statistical significance: mean +/− SEM; *, p<0.05; **, p<0.01; ***, p<0.001.
We devised a multi-factorial strategy to prioritize the elements with the goal to identify enhancers functionally resembling +9.5. GATA-2 occupancy was overrepresented (p = 5.4 × 10−9) at composite elements containing 8 base pair spacers in lineage-negative (Lin−) mouse bone marrow hematopoietic progenitors (Figure 1C). Prioritization involving only composite elements with CATCTG-(N8)-AGATAA yielded 797 (excluding +9.5) in the mouse genome, which we considered to be candidate enhancers involved in HSPC genesis/function (Table S1). While these elements were similarly distributed throughout the genome, more than half (53%) showed location-based conservation in humans (Figure 1C).
We reasoned that elements sharing factor occupancy and histone modification patterns with the +9.5 may functionally resemble the +9.5. We compiled mouse ChIP-seq datasets from hematopoietic/erythroid cell lines (Wu et al., 2011), HPC-7 cells (Wilson et al., 2010), G1ME cells (Dore et al., 2012), as well as 76 histone modification and 38 chromatin occupancy datasets (Shen et al., 2012b). These data from diverse primary cells/tissues and biologically relevant cell lines included GATA-2 and Scl/TAL1, among others. We derived a “chromatin occupancy signature” of the +9.5 site and compared factor occupancy and histone modification patterns at each element to the +9.5 site. This resulted in a +9.5-dissimilarity metric for each of the 797 +9.5-like elements (Table S1). Scoring was based on a 0–5 scale, in which 0 represents the +9.5 chromatin signature and 5 is entirely dissimilar.
Four of the top 20 ranked +9.5-like elements resided at loci with established developmental and/or homeostatic functions in the hematopoietic system [Bcl2l1 (Chao and Korsmeyer, 1998) Dapp1 (Bam32) (Han et al., 2003), Inpp5d (Helgason et al., 1998), and Pstpip1 (Shoham et al., 2003)]. Among the top 300 elements, 68 were GATA-2-occupied, 49 were Scl/TAL1-occupied, and 34 were GATA-2-Scl/TAL1-co-occupied (Table S2). Bcl2l1 +12.2 and Samd14 +2.5 candidate HSPC enhancer elements were conserved between human, mouse, and rat (Figure 1D). Based on conservation, we annotated human GATA-2 occupancy in 52 of the conserved elements (12.3%), including Samd14 +2.5 and Bcl2l1 +12.2, which correlate with putative enhancers, inferred from chromatin accessibility/attributes in diverse tissues (Cheng et al., 2014) (Table S2).
The highly ranked Samd14 +2.5 element resided in the first intron of Samd14, which encodes a sterile alpha motif domain protein of unknown function. GATA-2 occupied Samd14 +2.5 in human bone marrow-derived CD34+ HSPCs (Beck et al., 2013), K562 erythroleukemia (Fujiwara et al., 2009), and HUVEC (Linnemann et al., 2011) cells. In addition, Scl/TAL1 occupied Samd14 +2.5 in K562 cells, resembling the +9.5 (Figure 1E).
+9.5-like elements were assayed for enhancer activity in a transient transfection assay in G1E cells, in which +9.5 is active (Wozniak et al., 2007). While Samd14 +2.5 and Akap13 −65 had strong activity, Dapp1 +23.5 and Pstpip1 +0.7 had modest activity, and Bcl2l1+12.2 was inactive (Figure S1A). Replacing the core composite element from the inactive Bcl2l1 element with the active +9.5 did not alter +9.5 activity (Figure S1B). A Bcl2l1 +12.2 reporter containing the 3’ region of the +9.5 was active, indicating that additional 3’ determinants of activity flank the +9.5 site (Figure S1C). We tested whether Samd14 +2.5 functions as an enhancer when integrated as a LacZ fusion in E12.5 transgenic mouse embryos. This analysis revealed +9.5-like activity in hematopoietic tissues [descending aorta (DA) and fetal liver (FL)], in 4 of 6 Samd14 +2.5-LacZ embryos (Figure S1D).
To test whether GATA-2 regulates Samd14 expression, we quantitated Samd14 expression in Gata2 +9.5−/− mouse embryos in which Gata2 expression is downregulated in the E11.5 AGM and E12.5 fetal liver 4- and 20-fold, respectively (Figure 1F). Samd14 mRNA was 2- and 8-fold lower in the AGM and fetal liver, respectively. GATA-2 overexpression in Lin− fetal liver erythroid precursors upregulated Samd14 expression 7.5-fold (Figure 1G). GATA-2 regulation of Samd14 expression and Samd14 +2.5 enhancer activity suggest that Samd14 +2.5 function resembles +9.5, and GATA-2 and Samd14 may function in a common pathway.
Cistrome Constituent Function at Endogenous Loci
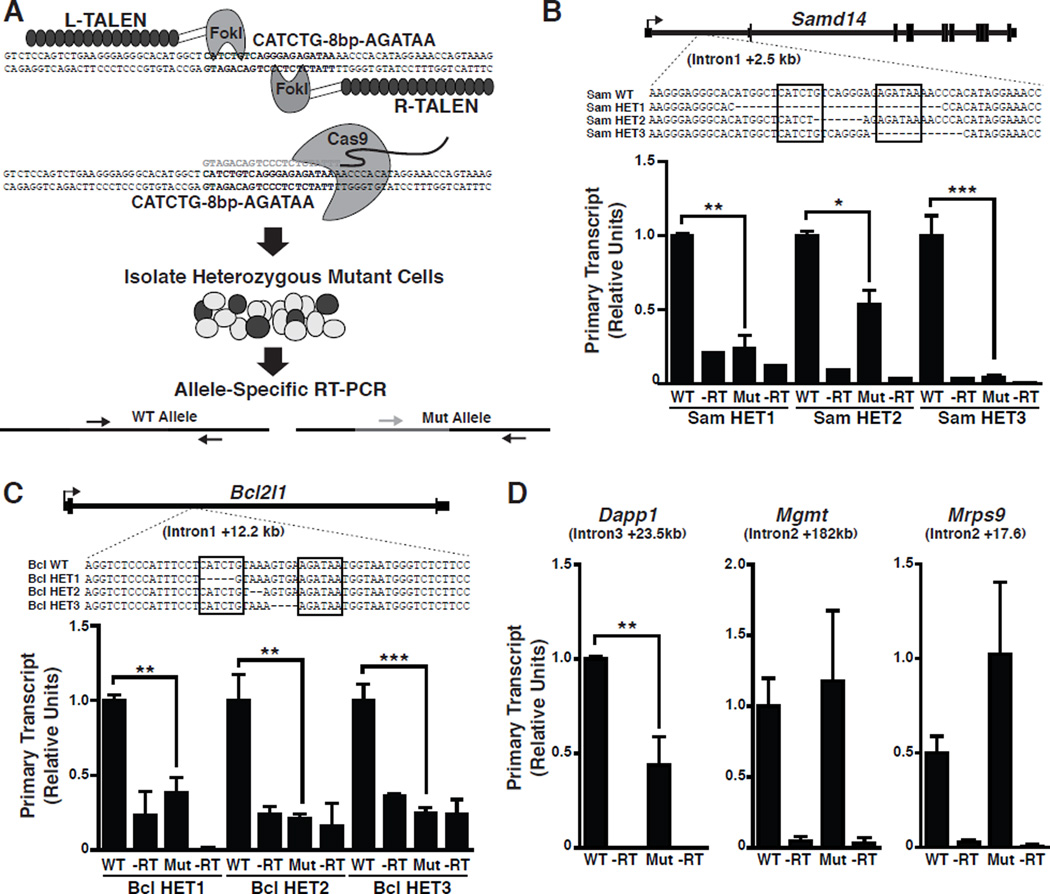
To determine if +9.5-like elements function at endogenous loci, we designed transcription activator-like effector nucleases (TALENs) (Kim et al., 2013) or clustered, regularly interspaced, short palindromic repeats (CRISPR)/Cas9 targeted endonucleases (Cho et al., 2013) to delete the respective sequences. We selected high-ranked (Samd14 +2.5, Bcl2l1 +12.2 and Dapp1 +23.5) and low-ranked +9.5-like elements (Mrps9 +17.6 and Mgmt +182) for deletion in murine G1E cells. These Gata1-null, embryonic stem cell-derived cells resemble a normal erythroid precursor (proerythroblast) and express endogenous GATA-2 and Scl/TAL1 (Weiss et al., 1997). Heterozygous clonal lines were isolated after transfection with vectors encoding TALEN pairs or a targeting sgRNA and Cas9-expression vector. Allele-specific primers were used to quantitate primary transcripts from wild type and mutant alleles (Figure 2A). Using 9.5+/− fetal liver cells, the wild type Gata2 allele is considerably more active than the mutant allele (Sanalkumar et al., 2014). Samd14 +2.5 mutations generated with TALENs revealed that deleting the entire cis-element (Sam HET1), E-box (Sam HET2), or GATA motif (Sam HET3) decreased transcription 80, 50, and >90%, respectively (Figure 2B). Thus, Samd14 +2.5 regulates endogenous Samd14 expression.
Figure 2. +9.5-like Elements Function at Endogenous Loci.
(A) TALEN or CRISPR/Cas9 strategy to delete +9.5-like elements in G1E cells. Heterozygous clonal lines were analyzed for primary transcript expression using allele-specific primers. (B) TALEN-generated heterozygous deletions in G1E clones, and allele-specific expression at Samd14 +2.5. (C) TALEN-generated heterozygous deletions in G1E clones, and allele-specific expression at Bcl2l1 +12.2. (D) Allele-specific expression in CRISPR/Cas9-generated G1E clones at the high-ranked Dapp1 +23.5 and low-ranked Mrps9 +17.6 and Mgmt +182. Statistical significance: mean +/− SEM.; *, p<0.05; **, p<0.01; ***, p<0.001. See also Figure S1.
Although Bcl2l1 +12.2 was inactive in the transfection assay (Figure S1), not all enhancers function in plasmids, and plasmid activity does not invariably predict endogenous activity. TALEN-mediated deletion of the E-box or spacer sequence from endogenous Bcl2l1 +12.2 markedly reduced transcription of the mutant vs. wild type allele in multiple clonal lines (Figure 2C). As Bcl2l1 +12.2 functions at its endogenous locus, this highlights the limitations of enhancer screening by transient transfection. CRISPR/Cas9-mediated deletion of a 22 bp sequence including the E-box and spacer of the highly ranked intronic Dapp1 +23.5 reduced mutant allele transcription 2.3-fold (Figure 2D). Gene editing was used to generate heterozygous deletions at low-ranked elements. The Mrps9 +17.6 GATA motif and spacer were deleted, while the Mgmt +182 GATA motif was deleted. Wild type and mutant primary transcripts were not significantly different (Figure 2D).
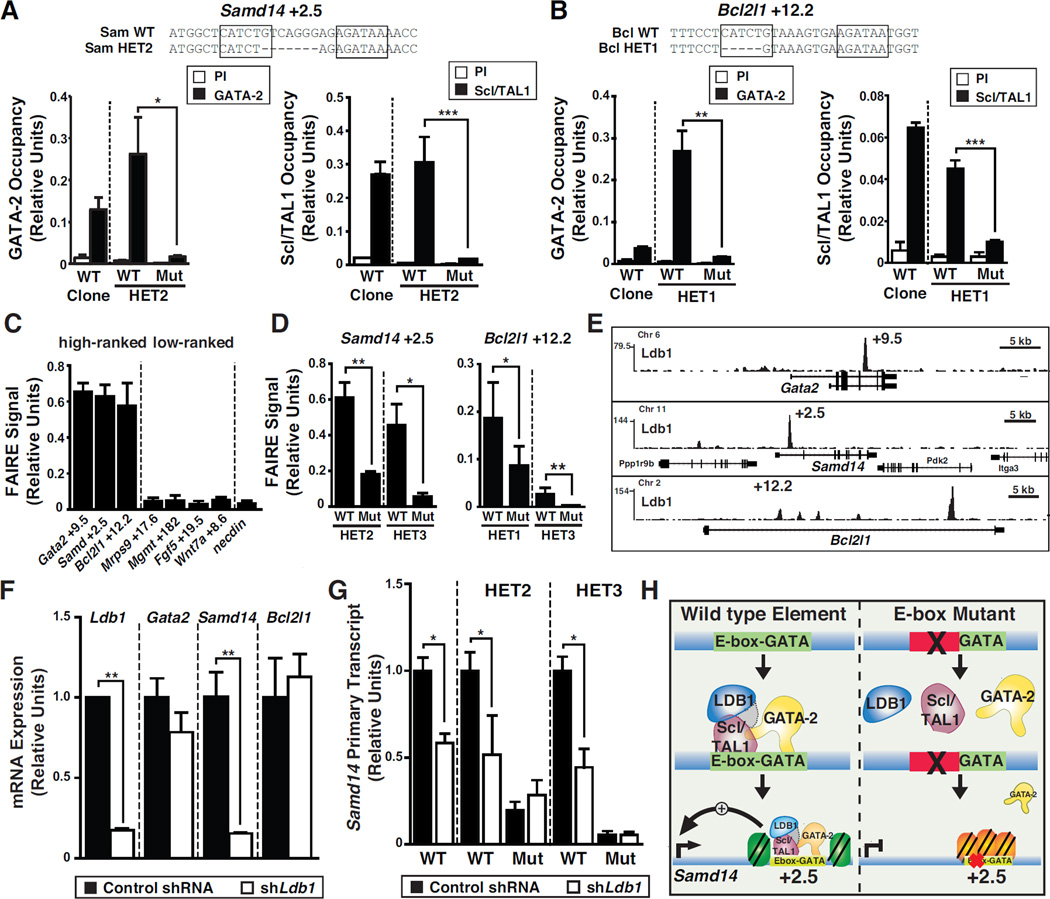
Since GATA-2 and Scl/TAL1 occupy the +9.5 (Wozniak et al., 2008), +9.5 confers open chromatin (Sanalkumar et al., 2014), and we discovered +9.5-like elements, we used allele-specific ChIP and chromatin accessibility assays to analyze function of these candidate enhancers. We compared occupancy at wild type and mutant Bcl2l1 +12.2 and Samd14 +2.5 alleles. Resembling the +9.5 deletion (Johnson et al., 2012; Sanalkumar et al., 2014), deletion of Samd14 +2.5 and Bcl2l1 +12.2 E-boxes nearly abolished GATA-2 and Scl/TAL1 occupancy (Figure 3A and 3B). Though an E-box mediates Scl/TAL1 binding to naked DNA, Scl/TAL1 co-localizes with GATA-1 at certain GATA motif-containing chromatin sites lacking an E-box (Tripic et al., 2008), and Scl/TAL1 function does not always require DNA binding activity (Kassouf et al., 2008). Scl/TAL1 and GATA-2 occupancy at endogenous +9.5-like sites required the E-box.
Figure 3. Molecular Mechanisms Underlying +9.5-like Element Function.
(A) Allele-specific GATA-2 and Scl/TAL1 occupancy at Samd14 +2.5 in G1E clones. (B) Allele-specific GATA-2 and Scl/TAL1 occupancy at Bcl2l1 +12.2 in G1E clones. (C) Chromatin accessibility in G1E cells, measured by FAIRE, of high-ranked and low-ranked +9.5-like elements vs. the negative control necdin. (D) Quantitation of allele-specific chromatin accessibility in heterozygous clonal lines of Gata2 +9.5, Samd14 +2.5 and Bcl2l1 +12.2 deletions using FAIRE. (E) ChIP-seq of Ldb1 occupancy at Gata2 +9.5, Samd14 +2.5 and Bcl2l1 +12.2 in G1E cells. (Sequence Read Archive Accession: ERA000161). (F) Gata2, Samd14, and Bcl2l1 mRNA expression following Ldb1 knockdown by retroviral shRNA infection of G1E cells. (G) shRNA knockdown of Ldb1 in heterozygous clonal G1E cells containing Samd14 +2.5 mutations. (H) Model illustrating GATA-2, Ldb1, and Scl/TAL1 function at Samd14 +2.5. Without the E-box, GATA-2, Scl/TAL1, and Ldb1 cannot occupy/regulate Samd14. Statistical significance: mean +/− SEM.; *, p<0.05; **, p<0.01; ***, p<0.001.
To test whether +9.5-like elements promote chromatin accessibility, formaldehyde-assisted isolation of regulatory elements (FAIRE) (Giresi et al., 2007) was used to quantitate open chromatin. Accessible chromatin was detected at +9.5, Samd14 +2.5, and Bcl2l1 +12.2 in G1E cells, while the low ranked Mrps9 +17.6, Mgmt +182, Wnt7a +8.6, and Fgf5 +19.5 exhibited low accessibility, indistinguishable from the negative control necdin (Figure 3C). Allele-specific FAIRE with heterozygous clones revealed markedly reduced accessibility at Samd14 +2.5 and Bcl2l1 +12.2 mutant alleles lacking the E-box (Sam HET2 and Bcl HET1), GATA motif (Sam HET3), or spacer region (Bcl HET3) (Figure 3D). Thus, prioritized +9.5-like elements confer occupancy, accessibility, and transcriptional activation.
A defining feature of E-box-GATA composite elements is the ability to assemble a complex containing GATA-1 or GATA-2 and Scl/TAL1, Ldb1, and Lmo2 (Love et al., 2014). Since +9.5-like element deletions reduced factor occupancy and accessibility, we asked whether Ldb1 functions through Samd14 +2.5 and Bcl2l1 +12.2. Ldb1 occupied +9.5 and +9.5-like sites in G1E cells (Figure 3E). shRNA-mediated Ldb1 knockdown (83% reduction) decreased Samd14 expression (6.5-fold) without affecting Gata2 or Bcl2l1 expression (Figure 3F). At Gata2, Ldb1 and BRG1 reductions are required to decrease transcription (Sanalkumar et al., 2014). shRNA-mediated Ldb1 knockdown in heterozygous G1E cells containing E-box or GATA motif deletions of Samd14 +2.5 only influenced Samd14 expression from the wild type allele (Figure 3G). Thus, Ldb1-mediated regulation of Samd14 requires Samd14 +2.5, and GATA-2-Scl/TAL1-Ldb1 occupy Samd14 +2.5, thereby activating Samd14 transcription. Deleting Samd14 +2.5 reduces chromatin accessibility, abrogates GATA-2-Scl/TAL1 occupancy, decreases transcription, and the locus is rendered Ldb1-insensitive (Figure 3H).
Samd14 Increases Hematopoietic Progenitor Levels/Activity
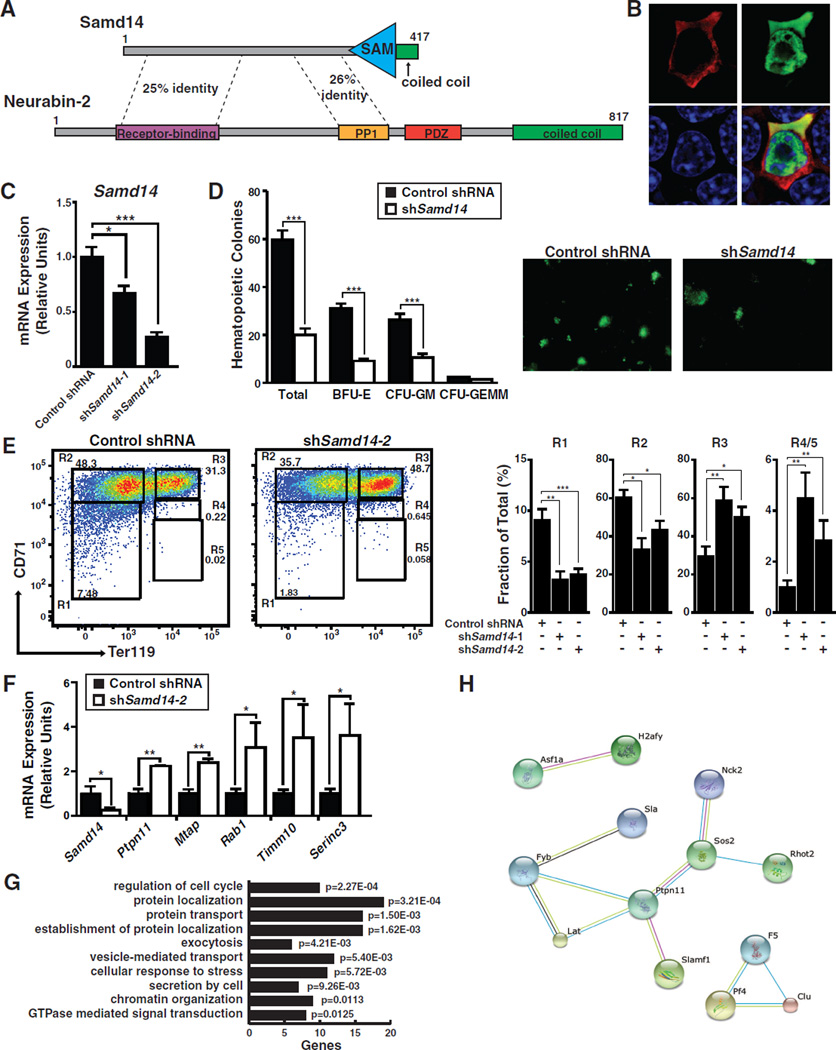
Our analysis of the +9.5-like stem/progenitor cell cistrome revealed loci with known and unknown functions. By opposing apoptosis in HSPCs, Bcl2l1 (Bcl-xL) controls hematopoiesis and is linked to myeloproliferative neoplasms and leukemia/lymphoma (Chao and Korsmeyer, 1998; Chonghaile et al., 2014). Dapp1 (Bam32) regulates B- and T-cell receptor signaling, germinal center progression, and mast cell activation (Han et al., 2003). Samd14 function in hematopoiesis or any other system is unknown. Samd14 protein contains a conserved SAM and a coiled-coil domain (Figure 4A). Mining RNA-seq data (Lara-Astiaso et al., 2014) revealed Samd14 expression in HSPCs and differentiated progeny (Figure S2A).
Figure 4. Samd14: a Regulator of Hematopoietic Progenitor Cells.
(A) Samd14 contains a C-terminal sterile-alpha motif domain, a C-terminal α-helix, and is homologous to Neurabin-2. (B) Immunofluorescence staining of G1E cells nucleofected with pMSCV-HA-Samd14-IRES-GFP expression vector (100× magnification). (C) Retroviral-mediated shRNA knockdown of Samd14 in E14.5 fetal liver cells. (D) Quantitation of GFP+ colonies, BFU-E, CFU-GM, and CFU-GEMM and representative fluorescent images at 4× magnification. (E) Flow cytometric staining of fetal liver cells for CD71 and Ter119 retrovirally-infected with control or Samd14 shRNA expanded for 3 days. Quantitation was conducted with two different Samd14 shRNAs. (F) Real-time RT-PCR validation of RNA-seq data showing genes significantly down- and up-regulated upon Samd14 knockdown. (G) DAVID analysis of genes with significantly altered expression based on RNA-seq of FACS-sorted fetal liver R1 cells using control or shSamd14 knockdown (n=3). (H) STRING analysis of genes interacting with Ptpn11 with significant expression changes. Statistical significance: mean +/− SEM.; *, p<0.05; **, p<0.01; ***, p<0.001. See also Figure S2 and S3 and Table S3.
To gain insight into Samd14 function, we compared its sequence, expression, and subcellular localization to two potential Samd14 paralogs, Ppp1r9a and Ppp1r9b. Ppp1r9a and Ppp1r9b encode regulatory subunits of the protein phosphatase I complex, neurabin-1 and neurabin-2, respectively (Terry-Lorenzo et al., 2002). Samd14 and Ppp1r9b have similar expression patterns during hematopoiesis (Lara-Astiaso et al., 2014) (Figure S2A) and in fetal erythroid precursors (Chen and Lodish, 2014) (Figure S2B). Immunostaining of expressed HA-Samd14 in G1E cells revealed a cytoplasmic localization (Figure 4B), resembling neurabin-2 in HeLa (Sagara et al., 2009) and immature dendritic cells (Bloom et al., 2008). ER-GATA-1 uniquely upregulated Samd14 expression in G1E-ER-GATA-1 cells (Figure S2C) (DeVilbiss et al., 2013). Though Ppp1r9b and Samd14 are neighboring genes (Figure 1E), heterozygous Samd14 +2.5 deletion did not affect Ppp1r9b expression (Figure S2D). Sequence alignments of intron-1 from Samd14, Ppp1r9a, and Ppp1r9b revealed a common GATA motif and partial E-box, albeit with a unique spacer length (Figure S2E). Neurabin-2 counteracts β-arrestin function to regulate GPCR signaling (Wang et al., 2004). Samd14 and Neurabin-2 share homology in GPCR- and PP1-interaction domains and additional regions (Figure 4A and S2F).
We conducted loss-of-function analysis to elucidate Samd14 function in GATA-2-expressing hematopoietic progenitors. Lineage-depleted (Lin−) E14.5 fetal liver cells were infected with control or Samd14 shRNA retrovirus. Three days post-expansion, two distinct shRNAs significantly reduced Samd14 expression (Figure 4C). In a colony-forming unit assay, Samd14 knockdown reduced BFU-E and CFU-GM colonies 3.4- and 2.5-fold, respectively (Figure 4D). To determine changes in fetal liver cellularity following Samd14 knockdown, we utilized CD71 and Ter119 markers to delineate R1–R5 populations. Early erythroid precursors reside in R1/R2 compartments, with differentiating erythroblasts in R3–R5. R1 (CD71low, Ter119−) and R2 (CD71high, Ter119−) immature erythroid precursors decreased ~2-fold, concomitant with increased mature R3 and R4/5 populations (Figure 4E).
To determine whether Samd14 knockdown altered the erythroid precursor transcriptome, RNA-seq was conducted using early erythroid precursor cells (R1) isolated from E14.5 fetal liver Lin− cells cultured for 3 days. This analysis identified 576 differentially expressed genes (q-value ≤ 0.05) upon Samd14 knockdown (254 upregulated; 322 downregulated) (Table S3). The magnitude of expression changes was low; only 1 gene was downregulated >2-fold (Samd14), and 1 was upregulated >2-fold (Rab1). RT-PCR established that Ptpn11 (Shp2), Mtap, Rab1, Timm10 and Serinc3 were upregulated 2–3-fold in R1 cells, validating the RNA-seq (Figure 4F). Gene Ontology revealed links to cell-cycle regulation and protein localization/transport (Figure 4G). Flow cytometric analysis indicated little to no change in fetal liver cell DNA content (Figure S3A), the proliferation marker Ki67 (Figure S3B), or the apoptotic marker AnnexinV (Figure S3C). The bioinformatic tool STRING revealed Ptpn11 interactions with genes upregulated by the knockdown (Figure 4H). Samd14 activity to suppress Ptpn11 expression has important implications, as Ptpn11 encodes a phosphatase that controls SCF/c-Kit signaling and regulates normal and malignant hematopoiesis (Mali et al., 2012).
Integrating Samd14 Into a Critical Hematopoietic Signaling Pathway
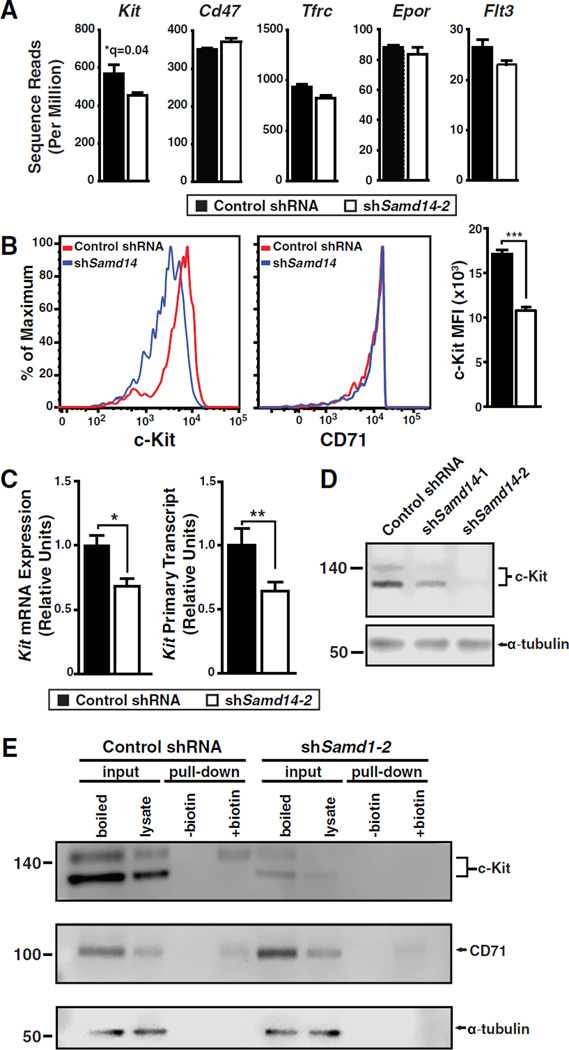
The RNA-seq analysis indicated that Samd14 knockdown decreased c-Kit expression in erythroid precursor cells (R1) (~1.4 fold, q = 0.04), and c-Kit promotes HSPC self-renewal (Deshpande et al., 2013). Transferrin receptor (Tfrc), Cd47, Epo receptor (Epor) and Flt3 were unchanged (Figure 5A). We used flow cytometry to test whether Samd14 regulates c-Kit surface expression in R1 cells. Median fluorescence intensity (MFI) of c-Kit-stained cells was reduced 40% in Samd14-knockdown vs. control (Figure 5B); CD71 surface expression was unaffected (Figure 5B). In Samd14-knockdown cells, Kit mRNA and primary transcripts (Figure 5C) were significantly reduced 1.5- and 1.6-fold, respectively. Two distinct Samd14 shRNAs downregulated c-Kit protein (Figure 5D). Cell surface c-Kit expression, assayed by cell biotinylation was detected in control cells, but not after Samd14 knockdown (Figure 5E), contrasting with membrane-bound CD71. Thus, Samd14 increased c-Kit expression, total c-Kit, and cell surface c-Kit in erythroid precursors.
Figure 5. Samd14 Upregulates c-Kit Expression.
(A) RNA-seq of c-Kit, cd47, Tfrc, Epor, and Flt3 mRNA in R1 fetal liver cells. (B) CD71low, Ter119− cells (R1 cells) were sorted from fetal liver cells 72 h post-expansion, stained with anti-c-Kit and anti-CD71 antibodies, and MFI quantitated. (C) Kit mRNA and primary transcript analysis in R1 cells. (D) Western blot analysis of c-Kit in control and Samd14-knockdown R1 cells. (E) c-Kit and CD71 surface protein analyzed by Sulfo-NHS-Biotin conjugation of surface proteins in live cells, followed by streptavidin pulldown/Western blotting. Statistical significance: mean +/− SEM.; *, p<0.05; **, p<0.01; ***, p<0.001.
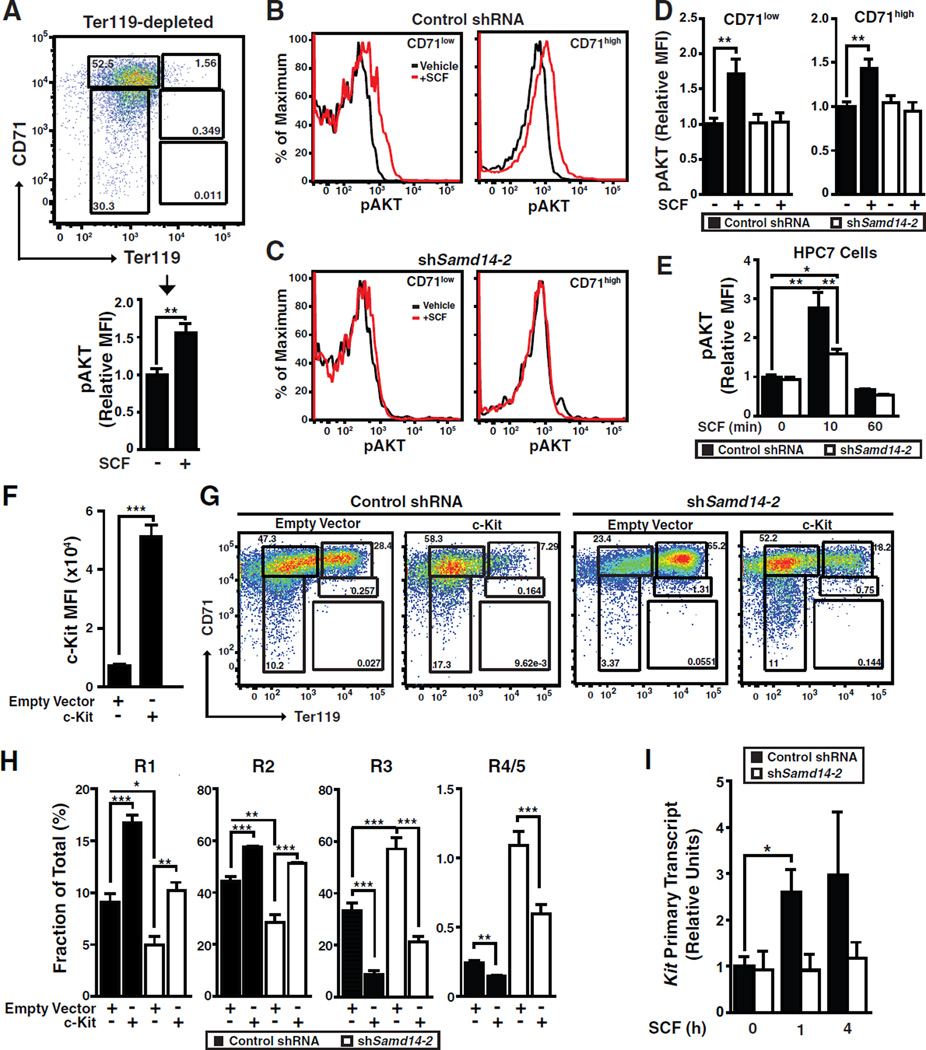
To assess consequences of Samd14 knockdown for c-Kit signaling, E14.5 fetal liver Lin− cells were expanded for 3 days, and Ter119− cells were isolated. Cells were cultured without serum and the c-Kit ligand SCF for 1 h, and then treated with SCF or vehicle. Phospho (S473)-AKT (p-AKT), a mediator of c-Kit signaling, was quantitated by phospho-flow cytometry in control and Samd14 knockdown cells. SCF induced p-AKT in control cells (Figure 6A). Cells were segregated into CD71low and CD71high populations, and SCF induced p-AKT in both control populations (Figure 6B and 6D). Samd14 knockdown rendered both populations insensitive to SCF (Figure 6C and 6D). We tested the impact of Samd14 knockdown on p-AKT induction in HPC-7 cells, an immortalized embryonic stem cell-derived multipotent hematopoietic precursor (Pinto do et al., 1998). Whereas SCF induced a transient 2.7-fold increase in p-AKT after 10 min in control shRNA-infected cells, the SCF response was reduced significantly in Samd14 knockdown cells (Figure 6E).
Figure 6. Samd14 Requirement for SCF/c-Kit Signaling.
(A) CD71 and Ter119 flow analysis of ex vivo expanded fetal liver cells following bead sorting for Ter119− cells. SCF-treated (10 ng/mL, 10 min) Ter119− cells analyzed for p-AKT MFI by flow cytometry. (B) p-AKT staining with control shRNA and CD71-low and CD-71-high fetal liver cells. (C) Phospho-flow with Samd14-knockdown CD71-low and CD-71-high fetal liver cells. (D) p-AKT MFI in control shRNA and shSamd14 fetal liver cells treated with 10 ng/mL SCF for 10 min (E) p-AKT MFI in control shRNA and shSamd14 HPC-7 cells treated with 50 ng/mL SCF for 0, 10 or 60 minutes. (F) c-Kit MFI in control and c-Kit overexpressed fetal liver cells. (G) Flow cytometry of enforced c-Kit expression, upon Samd14 knockdown, in fetal liver cells. (H) Percentage of cells in R1-R5 populations. (I) SCF treatment (50 ng/ml) of control or shSamd14-infected fetal liver cells 0, 1 and 4 h post-stimulation. Statistical significance: mean +/− SEM.; *p<0.05. **p<0.01, ***p<0.001.
Since Samd14 induced c-Kit expression on the cell surface and promoted SCF/c-Kit signaling, we reasoned that cellular deficits resulting from lowering Samd14 may be caused by insufficient c-Kit. We infected cells with c-Kit-expressing retrovirus, which increased c-Kit MFI 7.1-fold (Figure 6F). Enforced c-Kit expression in knockdown cells rescued erythroid precursors (Figure 6G). Quantitation confirmed that cells infected with both shSamd14 and c-Kit-expressing retroviruses had erythroid precursor levels resembling control cells (Figure 6H). Consistent with SCF/c-Kit signaling induction of c-Kit mRNA (Zhu et al., 2011), SCF treatment of control-infected fetal liver cells upregulated c-Kit primary transcripts and mRNA after 1 hour. As Samd14 knockdown abrogated this response (Figure 6I), Samd14 promotes SCF/c-Kit signaling as a mechanism important for hematopoiesis.
DISCUSSION
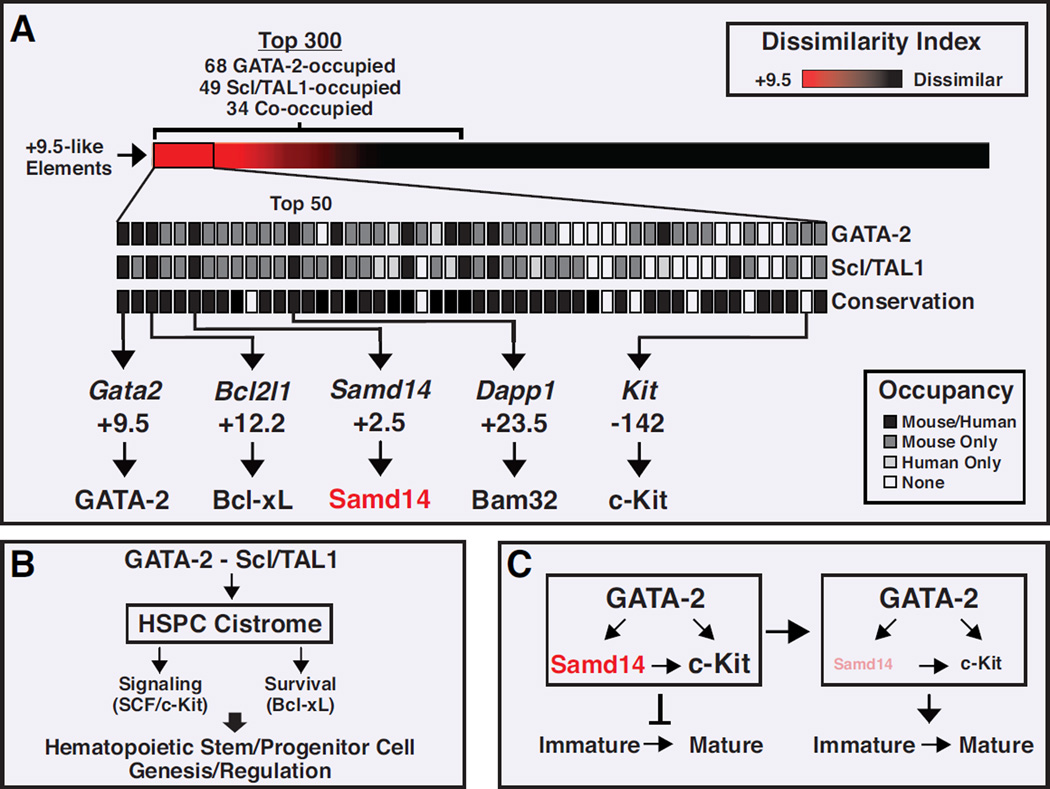
Advances in genome editing (Kim and Kim, 2014) have transformed strategies to ascribe cis-element function and transition beyond correlation-based functional inferences. Our establishment of the +9.5 as an HSC-generating cis-element (Gao et al., 2013; Johnson et al., 2012) provided a unique opportunity to discover an ensemble of HSPC-regulatory cis-elements. Based on +9.5-similarity, we stratified 797 elements and used genome editing for validation. GATA-2 occupied 68 elements (8.5%), representing a ~60-fold enrichment in GATA-2 occupancy vs. occupancy at GATA motifs genome-wide (Fujiwara et al., 2009). Eight sites were characterized by conserved GATA-2-Scl/TAL1 co-occupancy in humans (Figure 7A). Although Scl/TAL1 can co-occupy chromatin with GATA-1 and GATA-2 (Tripic et al., 2008; Wozniak et al., 2008), Scl/TAL1 dissociation from GATA factor-chromatin complexes correlates with repression in certain contexts (Tripic et al., 2008; Yu et al., 2009). Thus, GATA-2 and Scl/TAL1 are not expected to co-occupy all functional sites.
Figure 7. GATA-2/Samd14/c-Kit Feedforward Loop Revealed from HSPC Cistrome.
(A) Heat map depiction of 797 +9.5-like elements based on a dissimilarity index to the +9.5 stem cell-generating cis-element. The top 50 elements are expanded to show GATA-2- and Scl/TAL1-occupancy in mouse/human, and human/mouse conservation. Samd14 +2.5, Bcl2l1 +12.2, and Dapp1 +23.5 function was established by endogenous deletion. (B) Diagram illustrating GATA-2-regulated HSPC cistrome function to control diverse processes, including survival and SCF/c-Kit signaling. (C) Feedforward loop in which GATA-2 regulates Samd14 and c-Kit expression, and Samd14 promotes c-Kit signaling. See also Table S1 and S2
Our results provide evidence for a GATA-2-regulated HSPC cistrome with constituents residing at a panoply of genes encoding regulators of hematopoiesis, proteins with functions not linked to hematopoiesis or of unknown function (Figure 7A). The use of CRISPR/Cas9 to generate heterozygous cell lines harboring unique sequences for annealing primers specific for a wild type or a mutant allele represents a powerful approach for delineating cis-element requirements for gene regulation. Deletions of highly ranked +9.5-like sequences had large influences on endogenous gene function and cis-element occupancy by cognate factors. We defined a requirement for E-box and spacer sequences for GATA-2 occupancy at these sites.
Bcl2l1, encoding Bcl-xL, exemplifies a HSPC cistrome constituent known to control hematopoiesis. Bcl-xL confers HSPC survival (Chao and Korsmeyer, 1998). GATA-1 directly upregulates BclxL expression upon erythropoiesis (Gregory et al., 1999). GATA-2 functions through the +9.5-like element to confer Bcl2l1 expression and therefore HSPC survival (Figure 7B). After GATA switching, in which GATA-1 replaces GATA-2 at an ensemble of chromatin sites (Bresnick et al., 2010), GATA-1 usurps this function to confer survival to the developing erythroblast lacking GATA-2. This mechanism illustrates a link between +9.5-dependent control of genes important for HSC genesis/function and +9.5-like element-regulated HSPC survival. Integrating functions of other cistrome constituents will reveal additional links that constitute a systems-level developmental program, with a GATA-2 molecular switch as the common denominator (Figure 7B).
We focused on elucidating function of Samd14, a cistrome constituent of unknown function. SAMD14 SNPs are associated with blood platelet volume (Fehrmann et al., 2011), and SAMD14 is downregulated and differentially methylated in cancers (Shen et al., 2012a; Sun et al., 2008). GATA-2 function through Samd14 +2.5 controls myelo-erythroid progenitors and erythroid precursor cell maturation/function (Figure 7C). Samd14 promoted SCF/c-Kit signaling, and downregulating Samd14 abrogated SCF-mediated p-AKT, a key step in SCF/c-Kit signaling. As SCF elevates c-Kit mRNA through p-AKT (Zhu et al., 2011), and Samd14 promotes this mechanism (Figure 7C), lowering Samd14 levels reduced c-Kit mRNA and protein expression.
Frequent c-Kit mutations in malignant and non-malignant hematopoietic disorders yield constitutively active receptors (Lennartsson and Ronnstrand, 2012). AKT activation can mediate c-Kit signaling (Blume-Jensen et al., 1998; Ma et al., 2012b) and SCF/c-Kit signaling stimulates HSPC self-renewal (Deshpande et al., 2013). GATA-1 repression and GATA-2 activation of Kit correlates with occupancy of an intron 1 and an upstream cis-element, respectively (Jing et al., 2008; Munugalavadla et al., 2005). Our analysis identified a candidate +9.5-like element 141 kb upstream of the c-Kit start site. While the function of this site has not been established, it lies adjacent to a c-Kit enhancer (−147 to −154 kb) (Jing et al., 2008). GATA-2 regulation of Samd14 and Kit and Samd14 regulation of c-Kit expression/signaling conform to a Type I coherent feedforward loop (Figure 7C) (Shoval and Alon, 2010).
Samd14 and Ppp1r9b are chromosomal neighbors (Figure 1E), and Ppp1r9b encodes neurabin-2, which opposes β-arrestin-mediated suppression of GPCR function (Wang et al., 2004). Neurabin-2 controls multiple GPCRs, including α1b adrenergic (Liu et al., 2006) and thrombin (Ma et al., 2012a) receptors. Other SAM domain proteins include ephrin receptors, the p73 transcription factors, and the signaling adapter Slp76, and since SAM domains mediate diverse macromolecular interactions (Kim and Bowie, 2003), it will be instructive to determine if the SAM domain controls SCF/c-Kit signaling. As our strategy revealed a new mediator of a quintessential signaling pathway that regulates HSPC development/function, our resource is expected to reveal functions of other HSPC cistrome constituents.
EXPERIMENTAL PROCEDURES
Bioinformatics
We ranked 797 loci (mm9), matching the +9.5 sequence CATCTG-N[8]-AGATAA, based on their similarity to the +9.5 using multiple sources of genomic data (76 mouse ENCODE histone modifications and 125 transcription factor ChIP-seq datasets (Dore et al., 2012; Mouse et al., 2012; Tijssen et al., 2011; Wu et al., 2011)). We generated binary feature vectors for each locus based on individual data sources by overlapping the loci coordinates with the peak coordinates from the ChIP-seq datasets. We evaluated binary Jaccard distances of each locus to the +9.5 based on each data resource and aggregated the distances to generate a dissimilarity metric. The 797 +9.5-like loci were ranked based on this metric and annotated based on proximity to mm9 Refseq genes (Table S1).
Generation of TALEN and CRISPR/Cas9-deleted cells
TALENs were generated as described (Kim et al., 2013), and sgRNAs were generated by hemi-nested PCR-amplified construction of a U6 promoter-driven sgRNA, which was blunt-end cloned into SmaI-cut pBluescript (Addgene). 10 µg sgRNA-containing plasmids were co-nucleofected into 3 × 106 G1E cells with Cas9-expressing plasmid using Amaxa Kit R (Lonza). 72 h post-transfection, cells were cloned at limiting dilution in a 48 well plate. Cells were screened after 1 week to detect mutations using T7 endonuclease test (Cho et al., 2013; Kim et al., 2009). DNA was amplified by PCR, denatured, reannealed to facilitate heteroduplex formation, incubated with T7 Endonuclease I (New England Biosystems) for 15 min. Clones containing target mutations were sequence-validated. Validation of allele-specific primers was conducted using template or mutant cell cDNA.
Fetal liver culture
E14.5 fetal livers were disaggregated by pipetting in PBS containing 2% FBS, 2.5 mM EDTA and 10 mM glucose, and filtered (3 livers/biological replicate). Cells were lineage-depleted to enrich for progenitors using EasySep negative selection Mouse Hematopoietic Progenitor Enrichment Kit (Stem Cell Tech.). Cells were expanded in StemPro-34 media containing 2 mM L-glutamine, Pen-Strep, 0.1 mM MTG, 1 µM dexamethasone, 0.5 U/ml erythropoietin, and 1% mSCF Chinese Hamster Ovary cell conditioned medium, and maintained at 2.5 × 105 − 1 × 106/ml.
Supplementary Material
ACKNOWLEDGEMENTS
EHB is supported by NIH grants DK50107 and DK68634. SK, CND, and EHB were supported by NIH HG0070019. Cancer Center Support Grant P30CA014520 provided access to shared services. JSK is supported by Institute for Basic Science (IBS-R021-D1). KJH is supported by American Heart Association Fellowship. We thank Mitchell Weiss and Reuben Kapur for pMSCV viral vector and c-Kit vector, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number
GEO: GSE68602
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures, 3 figures, and 3 tables.
REFERENCES
- Allen PB, Ouimet CC, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci U S A. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K, Kinston SJ, Wilson NK, O'Brien TA, Gottgens B, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–e22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M. Beta-globin gene switching and DNaseI sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Bloom O, Unternaehrer JJ, Jiang A, Shin JS, Delamarre L, Allen P, Mellman I. Spinophilin participates in information transfer at immunological synapses. The Journal of cell biology. 2008;181:203–211. doi: 10.1083/jcb.200711149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P, Janknecht R, Hunter T. The kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annual review of immunology. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- Chen C, Lodish HF. Global analysis of induced transcription factors and cofactors identifies Tfdp2 as an essential coregulator during terminal erythropoiesis. Exp Hematol. 2014 doi: 10.1016/j.exphem.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, et al. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Chonghaile TN, Roderick JE, Glenfield C, Ryan J, Sallan SE, Silverman LB, Loh ML, Hunger SP, Wood B, DeAngelo DJ, et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer discovery. 2014;4:1074–1087. doi: 10.1158/2159-8290.CD-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E, Kaimakis P, Vink CS, Yokomizo T, Yamada-Inagawa T, van der Linden R, Kartalaei PS, Camper SA, Speck N, Dzierzak E. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210:2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S, Bosbach B, Yozgat Y, Park CY, Moore MA, Besmer P. KIT receptor gain-of-function in hematopoiesis enhances stem cell self-renewal and promotes progenitor cell expansion. Stem Cells. 2013;31:1683–1695. doi: 10.1002/stem.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVilbiss AW, Boyer ME, Bresnick EH. Establishing a hematopoietic genetic network through locus-specific integration of chromatin regulators. Proc Natl Acad Sci U S A. 2013;110:E3398–E3407. doi: 10.1073/pnas.1302771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Milne P, Jardine L, Zandi S, Swierczek SI, McGovern N, Cookson S, Ferozepurwalla Z, Langridge A, Pagan S, et al. The evolution of cellular deficiency in GATA2 mutation. Blood. 2014;123:863–874. doi: 10.1182/blood-2013-07-517151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS genetics. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Johnson KD, Chang YI, Boyer ME, Dewey CN, Zhang J, Bresnick EH. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210:2833–2842. doi: 10.1084/jem.20130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim S-I, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promoter erythroid cell survival by regulating bcl-xl expression. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- Han A, Saijo K, Mecklenbrauker I, Tarakhovsky A, Nussenzweig MC. Bam32 links the B cell receptor to ERK and JNK and mediates B cell proliferation but not survival. Immunity. 2003;19:621–632. doi: 10.1016/s1074-7613(03)00275-9. [DOI] [PubMed] [Google Scholar]
- Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A, Jirik F, Krystal G, Humphries RK. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Hsu AP, Ryu MJ, Wang J, Gao X, Boyer ME, Liu Y, Lee Y, Calvo KR, Keles S, et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest. 2012;122:3692–3704. doi: 10.1172/JCI61623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, Chagraoui H, Vyas P, Porcher C. Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood. 2008;112:1056–1067. doi: 10.1182/blood-2007-12-128900. [DOI] [PubMed] [Google Scholar]
- Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends in biochemical sciences. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. A library of TAL effector nucleases spanning the human genome. Nature biotechnology. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- Linnemann AK, O'Geen H, Keles S, Farnham PJ, Bresnick EH. Genetic framework for GATA factor function in vascular biology. Proc Natl Acad Sci U S A. 2011;108:13641–13646. doi: 10.1073/pnas.1108440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Yuen EY, Allen PB, Feng J, Greengard P, Yan Z. Adrenergic modulation of NMDA receptors in prefrontal cortex is differentially regulated by RGS proteins and spinophilin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18338–18343. doi: 10.1073/pnas.0604560103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love PE, Warzecha C, Li L. Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends in genetics : TIG. 2014;30:1–9. doi: 10.1016/j.tig.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Cierniewska A, Signarvic R, Cieslak M, Kong H, Sinnamon AJ, Neubig RR, Newman DK, Stalker TJ, Brass LF. A newly identified complex of spinophilin and the tyrosine phosphatase, SHP-1, modulates platelet activation by regulating G protein-dependent signaling. Blood. 2012a;119:1935–1945. doi: 10.1182/blood-2011-10-387910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Mali RS, Martin H, Ramdas B, Sims E, Kapur R. Role of intracellular tyrosines in activating KIT-induced myeloproliferative disease. Leukemia. 2012b;26:1499–1506. doi: 10.1038/leu.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali RS, Ma P, Zeng LF, Martin H, Ramdas B, He Y, Sims E, Nabinger S, Ghosh J, Sharma N, et al. Role of SHP2 phosphatase in KIT-induced transformation: identification of SHP2 as a druggable target in diseases involving oncogenic KIT. Blood. 2012;120:2669–2678. doi: 10.1182/blood-2011-08-375873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto do OP, Kolterud A, Carlsson L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 1998;17:5744–5756. doi: 10.1093/emboj/17.19.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger MA, Schroeder T. Hematopoiesis. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a008250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara M, Kawasaki Y, Iemura SI, Natsume T, Takai Y, Akiyama T. Asef2 and Neurabin2 cooperatively regulate actin cytoskeletal organization and are involved in HGF-induced cell migration. Oncogene. 2009;28:1357–1365. doi: 10.1038/onc.2008.478. [DOI] [PubMed] [Google Scholar]
- Sanalkumar R, Johnson KD, Gao X, Boyer ME, Chang YI, Hewitt KJ, Zhang J, Bresnick EH. Mechanism governing a stem cell-generating cis-regulatory element. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1400065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Takahashi M, Byun HM, Link A, Sharma N, Balaguer F, Leung HC, Boland CR, Goel A. Boswellic acid induces epigenetic alterations by modulating DNA methylation in colorectal cancer cells. Cancer Biol Ther. 2012a;13:542–552. doi: 10.4161/cbt.19604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012b;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, Kastner DL. Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci U S A. 2003;100:13501–13506. doi: 10.1073/pnas.2135380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval O, Alon U. SnapShot: network motifs. Cell. 2010;143:326-e321. doi: 10.1016/j.cell.2010.09.050. [DOI] [PubMed] [Google Scholar]
- Snow JW, Trowbridge JJ, Johnson KD, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. Context-dependent function of "GATA switch" sites in vivo. Blood. 2011;117:4769–4772. doi: 10.1182/blood-2010-10-313031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR, Arthur DC, Gu W, Gould CM, Brewer CC, et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics and immunity. Blood. 2013 doi: 10.1182/blood-2013-07-515528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Iijima T, Kano J, Kobayashi H, Li D, Morishita Y, Okubo C, Anami Y, Noguchi M. Frequent aberrant methylation of the promoter region of sterile alpha motif domain 14 in pulmonary adenocarcinoma. Cancer science. 2008;99:2177–2184. doi: 10.1111/j.1349-7006.2008.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry-Lorenzo RT, Carmody LC, Voltz JW, Connor JH, Li S, Smith FD, Milgram SL, Colbran RJ, Shenolikar S. The neuronal actin-binding proteins, neurabin I and neurabin II, recruit specific isoforms of protein phosphatase-1 catalytic subunits. J Biol Chem. 2002;277:27716–27724. doi: 10.1074/jbc.M203365200. [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated protein distinguish active from repressive GATA transcription factor complexes. Blood. 2008;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F-Y, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhao J, Brady AE, Feng J, Allen PB, Lefkowitz RJ, Greengard P, Limbird LE. Spinophilin blocks arrestin actions in vitro and in vivo at G protein-coupled receptors. Science. 2004;304:1940–1944. doi: 10.1126/science.1098274. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Boyer ME, Grass JA, Lee Y, Bresnick EH. Context-dependent GATA factor function: combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J Biol Chem. 2007;282:14665–14674. doi: 10.1074/jbc.M700792200. [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Keles S, Lugus JJ, Young K, Boyer ME, Tran TT, Choi K, Bresnick EH. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol. Cell. Biol. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HH, Ji K, Alderson N, He Z, Li S, Liu W, Zhang DE, Li L, Feng GS. Kit-Shp2-Kit signaling acts to maintain a functional hematopoietic stem and progenitor cell pool. Blood. 2011;117:5350–5361. doi: 10.1182/blood-2011-01-333476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.