Abstract
MicroRNAs are noncoding RNAs of approximately 22–24 nucleotides which are capable of interacting with the 3′ untranslated region of coding RNAs (mRNAs), leading to mRNA degradation and/or protein translation blockage. In recent years, differential microRNA expression in distinct cardiac development and disease contexts has been widely reported, yet the role of individual microRNAs in these settings remains largely unknown. We provide herein evidence of the role of miR-27 and miR-125 regulating distinct muscle-enriched transcription factors. Overexpression of miR-27 leads to impair expression of Mstn and Myocd in HL1 atrial cardiomyocytes but not in Sol8 skeletal muscle myoblasts, while overexpression of miR-125 resulted in selective upregulation of Mef2d in HL1 atrial cardiomyocytes and downregulation in Sol8 cells. Taken together our data demonstrate that a single microRNA, that is, miR-27 or miR-125, can selectively upregulate and downregulate discrete number of target mRNAs in a cell-type specific manner.
1. Introduction
MicroRNAs are noncoding RNAs of approximately 22–24 nucleotides which are capable of interacting with the 3′ untranslated region of coding RNAs (mRNAs), leading to mRNA degradation and/or protein translation blockage [1]. Understanding microRNA biogenesis has been greatly achieved; however, knowledge about the tissue distribution and functional consequences remains more elusive. In recent years, an increasing body of evidence has demonstrated a highly relevant role of microRNAs in multiple aspects of cardiac development and diseases [2, 3].
Functional evidence of the role of microRNAs in developing heart was demonstrated by selective inhibition of Dicer in tissue-restricted manner. Conditional ablation of Dicer using Nkx2.5Cre driver mice resulted in embryonic lethality with pericardial oedema and cardiac hypoplasia [4, 5]. Furthermore, Dicer inhibition using alpha-MHC-Cre mice also resulted in cardiac developmental impairment [6]. Thus, these studies highlight the importance of microRNA biogenesis for heart development. In addition, diverse studies have provided evidences of differential expression of microRNAs during heart development, both during embryogenesis [7–9] and at postnatal stages [10, 11] supporting a pivotal role of microRNAs during heart development. Moreover, recent studies reported microarray analyses which determine whether miRNAs are deregulated in common cardiovascular physiopathological conditions, such as hypertrophic and/or dilated cardiomyopathy, heart failure, or atrial fibrillation [12–17]. Taken together these data demonstrate a key role for microRNAs in cardiac development and disease. However, the role of individual microRNAs in these settings remains largely unknown.
We have previously reported a discrete number of differentially expressed microRNAs during cardiac development and we further elaborated on the functional role of miR-27 as regulator of the transcription factor Mef2c [7]. Furthermore, we recently reported that a large number of these microRNAs also display differential expression during iPS-derived cardiomyogenesis [18]. Among those, miR-125 displayed increased expression during both cardiac development and iPS-derived cardiomyogenesis, suggesting that it might play a pivotal role during muscle development. We therefore went on into this study dissecting the discrete role of miR-27 and miR-125, respectively, regulating muscle-enriched transcription factors.
2. Material and Methods
2.1. Cell Culture and MicroRNA Transfection Assays
HL1 cells (6∗105 cells per well) [19] and Sol8 (ATCC, USA) cells were transfected with corresponding pre-miR (Ambion, USA), respectively, at 50 nM using lipofectamine 2000 (Invitrogen, USA) according to manufacturer's guidelines. Negative controls included nontransfected cells as well as FAM-labeled pre-miR negative control transfected cells, which also allowed transfection efficiency evaluation. In all cases, transfection efficiencies were greater than 50%, as revealed by observation of FAM-labeled pre-miR transfection. After 4 hours after transfection, HL1 cells were cultured in appropriate cell culture media and collected after 48 hours as previously reported [7, 20].
2.2. qRT-PCR Analyses
mRNA qRT-PCR was performed in Mx3005Tm QPCR System with an MxPro QPCR Software 3.00 (Stratagene, USA) and SSOFast EvaGreen detection system (BioRad, USA). Two internal controls, mouseβ-actin and Gapdh, were used in parallel for each run. Each PCR reaction was performed at least three times to obtain representative averages. Primers sequences are provided in Table 1.
Table 1.
List of the oligonucleotides sequences used in the qPCR assays. Note that all primers were designed using the Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) online tool, fixing the primer length to 100–200 nucleotides and an annealing temperature of 60°C. MgCl2 concentration was always the same since SSOFast EvaGreen Master mix was used in all qPCR experiments.
| Gapdh | Fw: 5′-TCTTGCTCAGTGTCCTTGCTGG-3′ | 180 pb |
| Rv: 5′-TCCTGGTATGACAATGAATACGC-3′ | ||
|
| ||
| β-Actin | Fw: 5′-CCAGAGGCATACAGGGAC-3′ | 144 pb |
| Rv: 5′-TGAGGAGCACCCTGTGCT-3′ | ||
|
| ||
| Myocd | Fw: 5′-TTTTCAATTCCATCCCCAAC-3′ | 210 pb |
| Rv: 5′-CCCAGGGATCTTTGGAATTT-3′ | ||
|
| ||
| Mdfi | Fw: 5′-CAGGCTCTGAACAGCATTGA-3′ | 125 pb |
| Rv: 5′-GGTTCTGAGAGGTGGTCGTG-3′ | ||
|
| ||
| Mstn | Fw: 5′-GGCTCTTTGGAAGATGACGA-3′ | 188 pb |
| Rv: 5′-GGAGTCTTGACGGGTCTGAG-3′ | ||
|
| ||
| Runx1 | Fw: 5′-TACCTGGGATCCATCACCTC-3′ | 164 pb |
| Rv: 5′-GACGGCAGAGTAGGGAACTG-3′ | ||
|
| ||
| Mef2c | Fw: 5′-GGGGTGAGTGCATAAGAGGAC-3′ | 288 pb |
| Rv: 5′-AGAAGAAACACGGGGACTATGGG-3′ | ||
|
| ||
| Mef2d | Fw: 5′-TCTCCCAGTCTACCCACTCG-3′ | 162 pb |
| Rv: 5′-CAGGTGAACTGAAGGCTGGT-3′ | ||
MicroRNA qRT-PCR was performed using Exiqon LNA microRNA qRT-PCR primers and detection kit according to manufacturer's guidelines. All reactions were always run in triplicate using 5S as normalizing control, as recommended by the manufacturer. The Livak & Schmittgen method was used to analyze the relative quantification RT-PCR data [21] and normalized in all cases taking as 100% the wild-type (control) value, as previously described [22].
3. Results
3.1. Search for Muscle-Enriched Transcription Factor Candidate Targets for miR-27 and miR-125
Using TargetScan search engine, mouse miR-27 is predicted to target over a thousand genes. Hand-curate literature search on PubMed indicating previous involvement of these predicted targets in the cardiovascular and/or skeletal muscle biology setting suggests that approximately only a third of these genes (324/1002; ~32%) might be targeted in this context. A subclassification of these genes demonstrates that approximately ~13% corresponded to transcription factors. Among them, Runx1 and Mef2c have been already validated as direct miR-27 targets [7, 23]. We focus our attention on those transcription factors playing a role in either cardiac or skeletal muscle development, such as myostatin (Mstn), myocardin (Myocd), and MyoD family inhibitor (Mdfi). Using a similar approach, we also search for putative muscle-related transcription factors that might be putatively targeted by miR-125, which resulted in the identification of myocyte enhancer factor 2D (Mef2d).
3.2. Divergent Tissue-Specific miRNA Effects in Muscle Cells
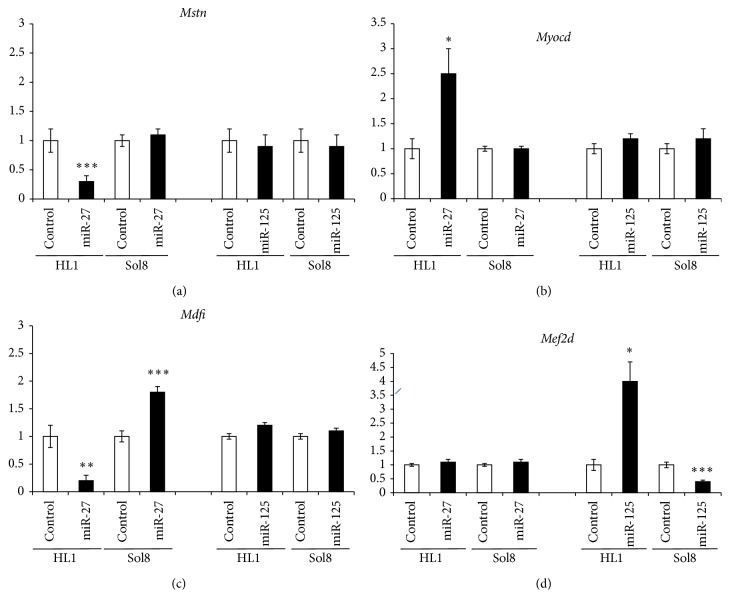
In order to dissect the functional role of miR-27 and miR-125 in muscle cells, we overexpressed these microRNAs in two distinct muscle cell types, Sol8 skeletal muscle myoblasts and HL1 atrial cardiomyocytes, respectively. After 48 hours of transfection, expression levels of these microRNAs and distinct muscle-enriched transcription factors were measured by qPCR as compared to lipofectamine nontransfected control cells. Figure 1 demonstrates that similar levels of microRNA overexpression were achieved for miR-27 and miR-125, respectively, in HL1 and Sol8 cells. Furthermore, functional assessment of microRNA overexpression was assayed by measuring Mef2c and Runx1 expression levels, since these transcription factors were previously reported as direct targets of miR-27 [7, 23]. Figure 2 shows that overexpression of miR-27 selectively results in downregulation of both Mef2c and Runx1 in cardiomyocytes (HL1) and skeletal myoblasts (Sol8), whereas no significant changes were observed upon miR-125 overexpression. Interestingly, selectively overexpression of miR-27 leads to downregulation of Mstn in HL1 atrial cardiomyocytes but not in Sol8 skeletal muscle myoblasts, as illustrated in Figure 3. On the other hand, miR-27 overexpression leads to significant upregulation of Myocd in HL1 atrial cardiomyocytes, whereas no changes are observed in Sol8 cells (Figure 3). Surprisingly, overexpression of miR-27 results in downregulation of Mdfi in HL1 cardiomyocytes and Mdfi upregulation in Sol8 cells. Importantly, expression of Mstn, Myocd, or Mdfi is not altered in Sol8 or HL1 cells after miR-125 expression, supporting the miR-27 specificity of these effects (Figure 3).
Figure 1.
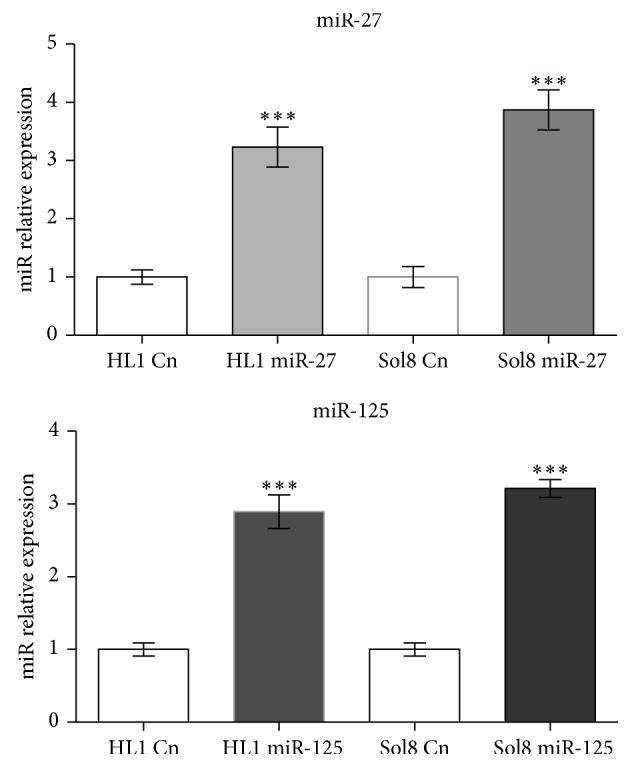
qPCR analyses of miR-27 and miR-125 expression levels in HL1 and Sol8 cells transfected with pre-miR-27 and pre-miR-125, respectively, as compared to nontransfected (lipofectamine only) control cells. Observe that a similar overexpression level is achieved for both miR-27 and miR-125 in HL1 and Sol8 cells, respectively (n = 3). *** P < 0.001, **** P < 0.0001.
Figure 2.
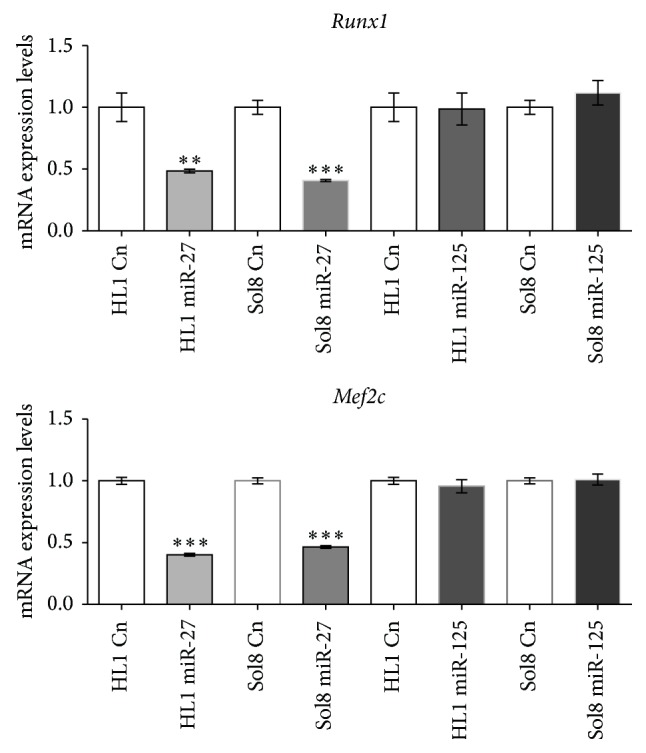
qPCR analyses of Runx1 and Mef2c expression levels in HL1 and Sol8 cells transfected with pre-miR-27 and pre-miR-125, respectively, as compared to nontransfected (lipofectamine only) control cells. Note that Runx1 and Mef2c expression levels are significantly downregulated in miR-27 but not in miR-125, overexpressing cells (HL1 and Sol8) (n = 3). ** P < 0.01, *** P < 0.001.
Figure 3.
qPCR analyses of Mstn (a), Myocd (b), Mdfi (c), and Mef2d (d) expression in HL1 and Sol8 cells, respectively, transfected with miR-27 and miR-125, as stated in the corresponding panel. Observe that overexpression of miR-27 leads to downregulation of Mstn and upregulation of Myocd in HL1 cells, but not in Sol8 cells, while miR-125 overexpression does not alter any of these genes. Importantly, miR-27 overexpression downregulates Mdfi in HL1 cells, while it is upregulated in Sol8 cells. In the case of miR-125 overexpression, a similar effect is observed for Mef2d but in reverse mode; that is, miR-125 upregulates Mef2d in HL1 cells and downregulates it in Sol8 cells (n = 3). * P < 0.05, ** P < 0.01, and *** P < 0.001.
In line with the data obtained for miR-27, overexpression of miR-125 resulted in selective upregulation of Mef2d in HL1 atrial cardiomyocytes and Mef2d downregulation in Sol8 cells. Importantly, miR-27 overexpression led to no significant changes of Mef2d expression neither in Sol8 nor in HL1 cells.
4. Discussion
MicroRNAs have been demonstrated to play essential roles in multiple biological processes, such as embryonic development, cell tissue specification, and cell proliferation, as well as in distinct pathological conditions, such as cancer and cardiovascular diseases. miR-27 has been indeed implicated in several of these contexts, such as embryonic development [7], angiogenesis [24, 25], adipogenesis [26–28], and atherosclerosis [29]. In particular, miR-27 has been reported to selectively regulate Pax3 [20, 30], Runx1 [23], and Mef2c [7]. Similarly, miR-125 has been documented to play essential roles in stem cell differentiation [31, 32] and distinct cancer types [33–35], yet its role in muscle biology is more elusive [36].
In this study, we report that miR-27 overexpression leads to selective downregulation of Mstn and Mdfi in HL1 atrial cardiomyocytes, suggesting a direct role of miR-27 regulating these genes. Surprisingly, miR-27 overexpression leads to upregulation of Myocd in HL1 atrial cardiomyocytes. Selective microRNA-mediated downregulation of target genes is widely documented [37, 38], although some reports also demonstrate upregulation of target genes [39, 40] such as for miR-373 [Place et al., 2007]. Thus, our data suggest that miR-27 can equally act upregulating or downregulating genes in the cardiac muscle context. Intriguingly, in Sol8 cells, miR-27 overexpression does not alter Mstn or Myocd, yet it upregulates Mdfi. These data suggest that miR-27 does not regulate Mstn and Myocd in the skeletal muscle context, while it upregulates Mdfi. Taken together these data demonstrate a cell-type specific role of miR-27 for the same target genes. A similar finding is also documented for miR-125, since miR-125 overexpression in HL1 atrial cardiomyocytes upregulates Mef2d, while it is downregulated in Sol8 cells.
To our knowledge this is the first report that demonstrates distinct effects for a single microRNA for the same target gene in distinct cellular contexts. In a mechanistic way, this implies that miR-27 is capable of interacting with Mstn and Mdfi 3′UTR in cardiomyocytes but not in skeletal myoblasts, while miR-125 upregulates and downregulates Mef2d depending on the cell context. Importantly, overexpression of miR-125 does not modify expression of Mstn, Myocd, or Mdfi in any cell context, while miR-27 overexpression does not alter Mef2d expression in HL1 or Sol8 cells. These findings reinforce the notion of a specific regulatory role of miR-27 in Mstn, Myocd, or Mdfi and of miR-125 in Mef2d, in line with TargetScan predictions. Furthermore, they suggest that either a selective blocking mechanism is operative in one cell type, for example, skeletal myoblasts, or a coadjuvant facilitating interactive factor is exclusively expressed in the other cell type, for example, cardiomyocytes. Further research is required to sort out these hypotheses. We are aware that our biological assay does not provide direct biochemical evidence of microRNA-mRNA interaction, yet it reveals the overall biological output of miR-27/miR-125 overexpression, respectively. However, it is important to realize that 3′UTR luciferase report assays in heterologous systems, such as 3T3 fibroblasts or HeLa cells, will be rather inappropriate to give the cell-type specific effects revealed in our assays.
In summary, we provide evidence that miR-27 and miR-125, respectively, can selectively upregulate and downregulate discrete number of target mRNAs in a cell-type specific manner.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Espinoza-Lewis R. A., Wang D.-Z. MicroRNAs in heart development. Current Topics in Developmental Biology. 2012;100:279–317. doi: 10.1016/b978-0-12-387786-4.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W.-J., Yin K., Zhao G.-J., Fu Y.-C., Tang C.-K. The magic and mystery of MicroRNA-27 in atherosclerosis. Atherosclerosis. 2012;222(2):314–323. doi: 10.1016/j.atherosclerosis.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y., Ransom J. F., Li A., et al. Dysregulation of cardiogenesis , cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 5.Saxena A., Tabin C. J. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):87–91. doi: 10.1073/pnas.0912870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J.-F., Murchison E. P., Tang R., et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(6):2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinchilla A., Lozano E., Daimi H., et al. MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovascular Research. 2011;89(1):98–108. doi: 10.1093/cvr/cvq264. [DOI] [PubMed] [Google Scholar]
- 8.Babiarz J. E., Ravon M., Sridhar S., et al. Determination of the human cardiomyocyte mRNA and miRNA differentiation network by fine-scale profiling. Stem Cells and Development. 2012;21(11):1956–1965. doi: 10.1089/scd.2011.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vacchi-Suzzi C., Hahne F., Scheubel P., et al. Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0052442.e52442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu J., Hanna P., Van Wagoner D. R., et al. Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circulation: Cardiovascular Genetics. 2012;5(3):327–335. doi: 10.1161/CIRCGENETICS.111.961631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porrello E. R., Mahmoud A. I., Simpson E., et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(1):187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rooij E., Sutherland L. B., Liu N., et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Rooij E., Sutherland L. B., Thatcher J. E., et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda S., Kong S. W., Lu J., et al. Altered microRNA expression in human heart disease. Physiological Genomics. 2007;31(3):367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 15.Sucharov C., Bristow M. R., Port J. D. miRNA expression in the failing human heart: functional correlates. Journal of Molecular and Cellular Cardiology. 2008;45(2):185–192. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo X., Zhang H., Xiao J., Wang Z. Regulation of human cardiac Ion channel genes by MicroRNAs: theoretical perspective and pathophysiological implications. Cellular Physiology and Biochemistry. 2010;25(6):571–586. doi: 10.1159/000315076. [DOI] [PubMed] [Google Scholar]
- 17.Cooley N., Cowley M. J., Lin R. C. Y., et al. Influence of atrial fibrillation on microRNA expression profiles in left and right atria from patients with valvular heart disease. Physiological Genomics. 2012;44(3):211–219. doi: 10.1152/physiolgenomics.00111.2011. [DOI] [PubMed] [Google Scholar]
- 18.Bonet F., Hernandez-Torres F., Esteban F. J., Aranega A., Franco D. Comparative analyses of microrna microarrays during cardiogenesis: functional perspectives. Microarrays. 2013;2:81–96. doi: 10.3390/microarrays2020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozano-Velasco E., Contreras A., Crist C., Hernández-Torres F., Franco D., Aránega A. E. Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Developmental Biology. 2011;357(1):165–178. doi: 10.1016/j.ydbio.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 21.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Domínguez J. N., Navarro F., Franco D., Thompson R. P., Aránega A. E. Temporal and spatial expression pattern of β1 sodium channel subunit during heart development. Cardiovascular Research. 2005;65(4):842–850. doi: 10.1016/j.cardiores.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Feng J., Iwama A., Satake M., Kohu K. MicroRNA-27 enhances differentiation of myeloblasts into granulocytes by post-transcriptionally downregulating Runx1. British Journal of Haematology. 2009;145(3):412–423. doi: 10.1111/j.1365-2141.2009.07632.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q., Gallagher R., Ufret-Vincenty R., Li X., Olson E. N., Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(20):8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbich C., Kaluza D., Frömel T., et al. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood. 2012;119(6):1607–1616. doi: 10.1182/blood-2011-08-373886. [DOI] [PubMed] [Google Scholar]
- 26.Kang T., Lu W., Xu W., et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. Journal of Biological Chemistry. 2013;288(48):34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L., Trajkovski M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism: Clinical and Experimental. 2014;63(2):272–282. doi: 10.1016/j.metabol.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y., Zhang X., Ding X., et al. miR-27 inhibits adipocyte differentiation via suppressing CREB expression. Acta Biochimica et Biophysica Sinica. 2014;46(7):590–596. doi: 10.1093/abbs/gmu036. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M., Wu J.-F., Chen W.-J., et al. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis. 2014;234(1):54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Crist C. G., Montarras D., Pallafacchina G., et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boissart C., Nissan X., Giraud-Triboult K., Peschanski M., Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139(7):1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 32.Emmrich S., Rasche M., Schöning J., et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes & Development. 2014;28(8):858–874. doi: 10.1101/gad.233791.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M., Liu Z., Zhao Y., et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. The Journal of Biological Chemistry. 2010;285(28):21496–21507. doi: 10.1074/jbc.m109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leotta M., Biamonte L., Raimondi L., et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells in vitro. Journal of Cellular Physiology. 2014;229(12):2106–2116. doi: 10.1002/jcp.24669. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J. X., Gao S., Pan Y. Z., Yu C., Sun C. Y. Overexpression of microRNA-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Molecular Medicine Reports. 2014;10(2):995–1002. doi: 10.3892/mmr.2014.2271. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Ha T., Zou J., et al. MicroRNA-125b protects against myocardial ischaemia/reperfusion injury via targeting p53-mediated apoptotic signalling and TRAF6. Cardiovascular Research. 2014;102(3):385–395. doi: 10.1093/cvr/cvu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heidersbach A., Saxby C., Carver-Moore K., et al. MicroRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. eLife. 2013;2 doi: 10.7554/eLife.01323.e01323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wystub K., Besser J., Bachmann A., Boettger T., Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genetics. 2013;9(9) doi: 10.1371/journal.pgen.1003793.e1003793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdisciplinary Reviews: RNA. 2012;3(3):311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 40.Lee S., Vasudevan S. Post-transcriptional stimulation of gene expression by MicroRNAs. Advances in Experimental Medicine and Biology. 2013;768:97–126. doi: 10.1007/978-1-4614-5107-5-76. [DOI] [PubMed] [Google Scholar]