Abstract
Vanishing White Matter (VWM) is a recessive neurodegenerative disease caused by mutations in translation initiation factor eIF2B and leading to progressive brain myelin deterioration, secondary axonal damage and death in early adolescence. Eif2b5R132H/R132H mice exhibit delayed developmental myelination, mild early neurodegeneration and a robust remyelination defect in response to cuprizone-induced demyelination. In the current study we used Eif2b5R132H/R132H mice for mass-spectrometry analyses, to follow the changes in brain protein abundance in normal diet- versus cuprizone-fed mice during the remyelination recovery phase. Analysis of proteome profiles suggested that dysregulation of mitochondrial functions, altered proteasomal activity and impaired balance between protein synthesis and degradation play a role in VWM pathology. Consistent with these findings, we detected elevated levels of reactive oxygen species (ROS) in mutant-derived primary fibroblasts and reduced 20S proteasome activity in mutant brain homogenates. These observations highlight the importance of tight translational control to precise coordination of processes involved in myelin formation and regeneration and point at cellular functions that may contribute to VWM pathology.
Keywords: myelination, remyelination, proteomics, cuprizone, eIF2B, VWM
Introduction
Eukaryotic translation initiation factor 2B (eIF2B) is a key constituent of the translational machinery. While eukaryotic translation initiation factor 2 (eIF2) delivers Met-tRNAMet to the 40S ribosomal subunit at each round of translation initiation, eIF2B serves as its guanine nucleotide exchange factor (GEF) and governs global protein synthesis rates under normal and stress conditions (Sonenberg & Dever 2003). Mutations in each of the five subunits of eIF2B affect the central nervous system (CNS) white matter and cause a leukodystrophy called Vanishing White Matter (VWM) disease, also known as Childhood Ataxia with CNS Hypomyelination (CACH) (OMIM#306896) (Schiffmann et al. 1994, Hanefeld et al. 1993, Leegwater et al. 2001, van der Knaap et al. 2002). The classic form of this disease, diagnosed by magnetic resonance imaging, manifests as progressive deterioration of cerebral white matter. The associated neurological symptoms include progressive motor and cognitive deficiencies that worsen upon exposure to various physical or psychological stressors, resulting in death by adolescence (reviewed in (Bugiani et al. 2010, Fogli & Boespflug-Tanguy 2006, Pronk et al. 2006, Schiffmann & Elroy-Stein 2006, van der Knaap et al. 2006)).
We previously generated a mutant mouse model for the disease by introducing a point mutation into the mouse Eif2b5 gene locus. We chose the eIF2B5 R132H mutation because it corresponds to an R136H mutation in the human protein, which in the homozygous state leads to a classical form of VWM in human patients. The GEF activity of mutant eIF2B in mice brain lysates is reduced by approximately 20%, thus compromising the ability to synthesize appropriate levels of key proteins as needed. The effect of this limitation is most notable under conditions that require the production of large amounts of proteins within short time frames in response to various physiological signals. Eif2b5R132H/R132H mice exhibit delayed developmental myelination, abnormal abundance of oligodendrocytes and astrocytes, abnormal levels of major myelin proteins, and higher proportion of small-caliber nerve axons (Geva et al. 2010). Mutants also show impaired remyelination after damage, which manifests as reduced astrogliosis following systemic injection of lipopolysaccharide (LPS) and complete failure of mutants to recover from a diet containing cuprizone, a copper chelator that induces reversible demyelination. In line with the myelin repair defect, mutant mice suffer from early neurodegeneration (Geva et al. 2010, Cabilly et al. 2012). The phenotype of Eif2b5R132H/R132H mice is significantly milder compared to that of human patients. However, this animal model exposes myelin-related mechanisms that are dependent on tight regulation of protein synthesis and are impaired due to the mutation in eIF2B.
There is an increasing body of evidence suggesting that myelin regeneration in the adult brain recapitulates the developmental myelination program (reviewed in (Fancy et al. 2011)). Accordingly, in Eif2b5R132H/R132H mice, myelin regeneration following myelin deterioration (i.e. remyelination following demyelination) is expected to be slow and abnormal, just as it is throughout early postnatal development. To learn more about the mechanisms involved in white matter development and regeneration, we used a proteomic approach to profile changes in protein levels during developmental myelination and remyelination. For this purpose, mice were fed a diet containing 0.2% cuprizone, which leads to massive demyelination due to apoptosis of mature oligodendrocytes but not oligodendroglial progenitor cells, thus allowing near-complete remyelination within weeks of cuprizone removal (Matsushima & Morell 2001). The robustness of the cuprizone model prompted us to test the effect of eIF2B5 mutation on the protein repertoire at two time points along the remyelination process. Comparison of expression pattern dynamics of hundreds of proteins in the brains of wild-type (WT) and Eif2b5R132H/R132H (Eif2b5M) mice uncovered a remarkable similarity between remyelination in WT and developmental myelination in Eif2b5M mice. However, the remyelination expression pattern was markedly different in Eif2b5M mice, consistent with their previously-identified remyelination defect. Furthermore, we detected possible involvement of specific protein complexes and pathways in myelination, suggesting a role in VWM pathology. This includes abnormal stoichiometry of translational machinery components and subunits of the 20S proteasome, highlighting the importance of accurate balance between protein synthesis and degradation, as well as abnormal stoichiometry of mitochondrial oxidative phosphorylation complexes. Indeed, we measured impaired proteasomal activity and elevated reactive oxygen species (ROS) levels in brains of Eif2b5M mice, confirming the dysregulation of proteasomal and mitochondrial functions in VWM.
Methods
Mice handling and diets
Eif2b5+/+ (wild-type, C57BL strain) and Eif2b5R132H/R132H (Eif2b5M; mutant) male siblings of heterozygote mice were used, as described previously (Geva et al. 2010). Mice were bred and housed in individually ventilated cages (Lab Products Inc., USA) in the animal facility of Tel Aviv University under a 14/10 h light/dark cycle. Animals were fed autoclavable rodent pellet (Koffolk 19–510, Koffolk Ltd, Petach Tikva, Israel) and sterile water ad libitum throughout the experiments. At the age of 6-weeks, the mice were fed with 0.2% cuprizone (Sigma) for 4 weeks (Matsushima & Morell 2001). At 10 weeks of age, the diet was replaced with a normal diet and the mice were allowed to recover for 1 or 2 weeks. Age-matched controls were fed a normal diet for the entire time (i.e., ‘normal diet-fed’). Mice were decapitated, cerebrums were collected and flash-frozen in liquid nitrogen. For each experimental setting (a certain diet, strain and time point), the cerebrums of 5 mice were pooled to yield a single proteome profile. All experimental procedures involving mice were approved by the Tel Aviv University Animal Care Committee according to national guidelines (permit #L-09-35). SILAC mice (SILAM) were fed with heavy lysine-containing diet (Lys-6) (Kruger et al. 2008). Animals were handled according to Institutional Animal Care and Use Committee guidelines at the Children's National Medical Center (Approved Protocol No. 199-07-01).
Magnetic Resonance Imaging (MRI)
Wild-type and mutant male mice were subjected to quantitative brain scans followed by voxel-based analysis to allow whole brain comparisons. Mice were scanned in a 7T/30 spectrometer (Bruker) under 10% Ketamine/6.6% Xylasene anesthesia. The diffusion tensor imaging (DTI) protocol and image analysis were previously described (Geva et al. 2010). The analysis was performed on the corpus callosum region.
Sample preparation
Mouse cerebrum half hemispheres were lysed in RIPA buffer (Sigma, St. Louis, MO) containing proteinase inhibitor cocktail (Roche, Indianapolis, IN) and phosphatase inhibitor cocktail (Roche) and homogenized by motor pestle and then sonicated on ice. Tissue lysate was centrifuged for 10 min at 13000 rpm at 4°C. Supernatant was collected and protein concentration was measured by BCA protein assay kit (ThermoScientific, Rockford, IL). The same amount of protein (50 μg) from each time point and treatment was combined with SILAM brain tissue lysate at 1:1 ratio. Total 100 μg protein was loaded and run on 4-12% Bis-Tris gel (Life Technologies, Carlsbad, CA) and gels were stained with Bio-Safe Coomassie Stain (Bio-Rad Laboratories, Hercules, CA) to visualize each band.
Mass-spectrometry
Each gel lane was cut into slices followed by in-gel trypsin-digestion as previously described (Jensen et al. 1999). Peptides from each slice were injected via an autosampler (6 uL) and loaded onto a Symmetry C18 trap column (5 μm, 300 μm i.d. × 23 mm, Waters) for 10 minutes at a flow rate of 10 μL/min, HPLC-grade water with 0.1% formic acid. Each sample was subsequently separated by a C18 reverse-phase column (3 μm, 200A, 100 μm × 15 cm, Magic C18, Michrom Bioresources) at a flow rate of 300 nL/min using an Eksigent nano-hplc system (Dublin, CA). The mobile phases consisted of HPLC-grade water with 0.1% formic acid and 90% acetonitrile. A 65 minute linear gradient from 5 to 40% acetonitrile was employed. Eluted peptides were introduced into the mass spectrometer via Michrom Bioresources CaptiveSpray. The spray voltage was set at 1.4 kV and the heated capillary at 200 °C. The LTQ-Orbitrap-XL (Thermo Fisher Scientific) was operated in data-dependent mode with dynamic exclusion in which one cycle of experiments consisted of a full-MS in the Orbitrap (300-2000 m/z) survey scan in profile mode, resolution 30,000, and five subsequent MS/MS scans in the LTQ of the most intense peaks in centroid mode using collision-induced dissociation with the collision gas (helium) and normalized collision energy value set at 35%.
For protein identification and quantification, we used MaxQuant software (Cox & Mann 2008) version 1.2.0.18 and the integrated Andromeda search engine (Cox et al. 2011)(same version). Database search was performed against forward and decoy Uniprot mouse database (UniProt release 15.4, June 2009) and the search allowed two missed cleavage, three labeled amino acids, variable modification of oxidized methionine, N-terminal acetylation, carbamidomethyl cysteine as a fixed modification and heavy lysine as label. Database search allowed a mass error window of 6 ppm and 0.5 Da for the MS and MS/MS scans, respectively. Data were filtered based on a 1% false discovery rate for proteins and peptides. A minimum of one unique or razor peptide was sufficient for protein identification.
Proteomic data analysis
A total of 3,500 proteins were detected in at least one of the experimental samples, including the SILAC sample. Among them, we focused on the 1831 proteins that were detected both in the SILAC sample and in at least one of experimental sample. For each given protein, its ‘protein level’ in a certain sample is defined as the ratio between its measured intensity in the experimental sample (unlabeled or light) compared to its measured value in the SILAC sample (heavy; Supp. Table 1). The dataset consists of eight ‘protein level profiles’, representing WT and mutant mice each of them profiled under two different feeding conditions at two different time points. The time points correspond to 11 and 12 weeks of age and are referred to as w11 and w12, respectively. The feeding refers to normal diet or a 0.2% cuprizone diet supplied between 6- and 10-weeks of age (i.e., 4-week-long), followed by normal diet for recovery. Protein levels measured at w11 and w12 in mice fed a normal diet reflect the dynamics of expression during normal brain development. Protein levels measured at w11 and w12 in cuprizone-fed mice reflect the dynamics of expression after 1-or 2-weeks recovery from cuprizone-induced demyelination.
We selected the 139 ‘eIF2B-related proteins’ (in normal diet-fed mice only) carrying the most substantial difference between protein levels in WT and protein levels in mutant mice, either in w11 or in w12. To that end, we first subtracted the level of each protein in mutant from the level of the same protein in WT, separately for each time point (referred to as ‘WT-subtracted Eif2b5M values’); proteins are then included in the ‘eIF2B-related proteins’ set if at any of the time points (w11 or w12), their absolute WT-subtracted Eif2b5M values are higher than 0.5. An enriched sub-network of the eIF2B-related proteins was constructed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity, Redwood City, CA) based on known protein-protein interactions in either human, mouse or rat. In brief, IPA constructs a sub-network so as to maximize mammalian protein-protein interactions between proteins in a given group, and then adds complexes that are enriched in this sub-network. Here, the dense network of eIF2B-related proteins (in Fig. 1b) consists of 25 proteins and two additional complexes.
Fig. 1. eIF2B mutation leads to alteration in dynamics of protein abundance and essential signaling pathways.
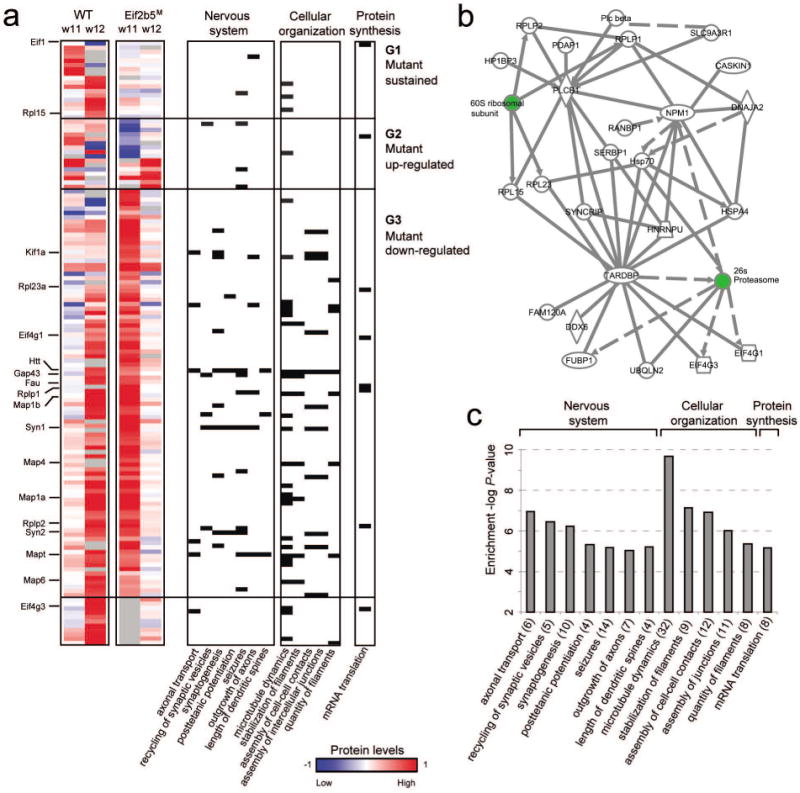
(a) The levels of the key proteins that were affected by the Eif2b5 mutation (rows, referred to as ‘eIF2B-related proteins’) in four normal-diet experiments: WT at w11, WT at w12, Eif2b5R132H/R132H mutant at w11 and Eif2b5R132H/R132H mutant at w12 (left to right columns). WT, wild-type; Eif2b5M, mutant; w11, 11-weeks of age; w12, 12-weeks of age. High (red) and low (blue) protein levels represent 2-fold change and 0.5-fold change, respectively (compared to the SILAC sample; color coding is in log2 scale). Proteins were grouped into three groups, G1–G3. The bottom group includes proteins that do not clearly belong to any of these groups. Right columns indicate protein membership (black cells) in 12 major enriched functional categories, the P-value of which is shown in c. (b) A dense mammalian protein-protein interaction sub-network that is enriched in eIF2B-related proteins. The network consists of 25 eIF2B-related proteins (white) and two additional complexes (ribosome and proteasome, green) that were automatically added due to their centrality and suggest a possible change in the balance between protein synthesis and degradation. (c) Functional categories enriched in the eIF2B-related proteins. Shown is a histogram of –log P-value of enrichment (y axis) across functional categories (x axis; also mentioned in a).
The ‘response’ is defined as the ratio between the protein levels at w12 and w11 for the same mouse strain on a similar diet. In total, four response profiles were calculated, representing WT and mutant mice fed a normal or 4-week-long cuprizone diet followed by normal diet recovery. The response under cuprizone treatment is termed ‘remyelination response’, whereas the response under normal diet is referred to as ‘developmental myelination response’. PCA was applied using the GenePattern software (Reich et al. 2006). We grouped the four response profiles using the K-means algorithm (applied on log-transformed response values). The 25 resulting groups were further grouped manually into 14 ‘response clusters’ based on a similar pattern of response dynamics (Supp. Table 2, Fig. 3a). Out of 1,831 proteins, the K-means clustering analysis was applied only on 802 proteins that were detected in all eight experimental samples and in the SILAC sample (that is, all four response values are available).
Fig. 3. Relations between cuprizone-induced remyelination in wild-type and developmental myelination in normal-diet-fed mutant mice.
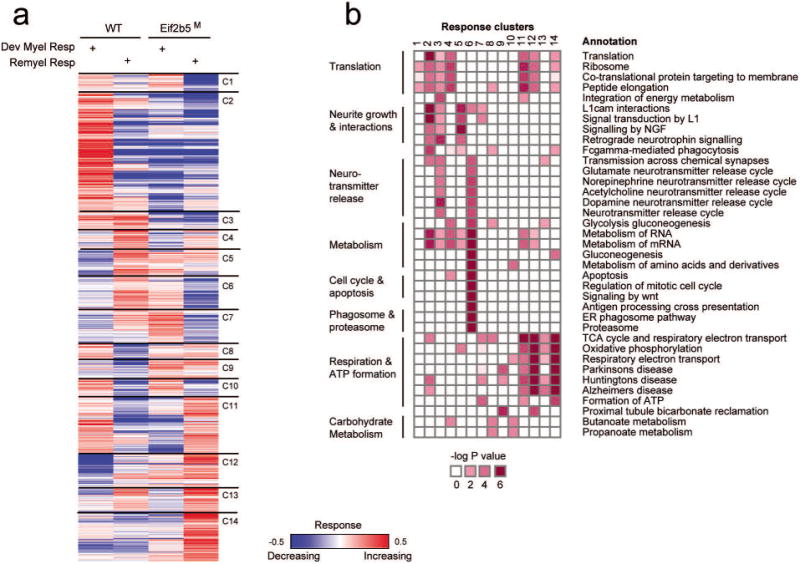
(a) Shown are the response levels for proteins (rows) of wild-type (WT) mice (columns 1,2) and mutant (Eif2b5M) mice (columns 3,4) during cuprizone-induced remyelination (Remyel Res; columns 2,4) or normal developmental myelination in the absence of cuprizone (Dev Myel Res; columns 1,3). Increasing (red) and decreasing (blue) responses indicate multiplication or division by a factor of 1.4 or higher (colors are presented in log2 scale). The proteins are partitioned into 14 clusters (C1–C14, right). (b) Pathway annotation of response clusters. Significant enrichments are shown (dark red) between response clusters (1–14, columns) and pathway annotations (rows).
Enrichment analysis was performed using the Ingenuity Pathway analysis (Ingenuity, Redwood City, CA). To test for enrichment of all proteins measured in each experiment (Supp. Table 3), the analysis was performed using all genes in the Ingenuity knowledge base as background. To test for enrichment of all specific protein groups (e.g. eIF2B-related proteins or response clusters), the analysis was performed using the 1,831 measured proteins as background. Unless otherwise stated, all reported P values were Bonferroni-corrected for multiple testing.
Transcriptomic data analysis
Transcriptome profiles of WT and Eif2b5M mice at the age of 18 and 21 days (on a normal diet) were downloaded from (Marom et al. 2011) (3 biological repeats per each experiment). Here we focused on the mRNA of all 18 gene that are part of proteasomal subunits α and β and were measured in the microarray. For each of these genes, its ‘mRNA response’ is defined as the product of subtracting the 18-day mRNA level from the 21-day mRNA level in the same mouse strain. Overall, nine mRNA response values were calculated for each gene and strain (for all pairs of 3 biological repeats at each time point), thus allowing to test the difference between the distributions of mRNA responses in the WT versus mutant strains (using a t-test).
Detection of ROS level
Primary mouse embryo fibroblasts (MEFs) were isolated from WT and mutant mice and cultured in Dulbecco modified eagle medium (DMEM) supplemented with 10% fetal calf serum, 100 un/ml penicillin, 0.1mg/ml streptomycin, 2mM L-glutamine (Biological Industries, Israel). At passage 4, cells were plated in 5 wells (per each genotype) of a 96-well plate at a density of 5,000 cells per well and incubated over-night at 37°C in a 5% CO2 incubator, followed by medium change for additional 24h incubation. Cells were then stained with Hoechst 33258 (Sigma #861405) and CellROX deep-red (Molecular Probes #C10422) at final concentrations of 2ug/ml and 5uM, respectively, for 30 minutes in 37°C. Following washing with Hank's balanced salt solution (HBSS), the cells were subjected to fluorescent acquisition by IN Cell Analyzer 2000 (GE Healthcare) followed by high content analysis using In Cell Developer Toolbox 1.9.1 software (GE Healthcare).
20S proteasome activity assay
CHEMICON®'s Proteasome Activity Assay Kit (Millipore #APT280) was used according to the manufacturer's instructions. Activity measurement is based on detection of free 7-Amino-4-methylcoumarin (AMC) fluorophore following cleavage of labeled LLVY-AMC substrate. AMC fluorescence was measured using a microplate reader (Ex= 360/40 Em= 460/40; Biotek Synergy HT). MEFs: 4.3×105 WT or mutant MEFs at passage 4 were plated in 10 cm plates for 24 h, followed by medium change and further 24h incubation. Cells were then washed twice with phosphate buffered saline (PBS), scraped into ice-cold PBS and pelleted. Cell pellets were suspended with 30 μl lysis buffer (50 mM HEPES, 5 mM EDTA, 150 nM NaCl, 1% Triton-X100) and incubated for 30 min at room temperature followed by centrifugation at 20,000 ×g for 15 min at 4°C and collection of supernatants for protein concentration measurement by Bradford protein assay. 30 μg of total protein were used per sample. Brains: Cerebrums of p21 WT or mutant mice were harvested, separated into two hemispheres, flash-frozen in liquid nitrogen, and stored in -80°C until use. Each right hemisphere was homogenized in 300 μl lysis buffer followed by sonication (3 cycles of 8sec at 0.06W (RMS)). Homogenates were centrifuged at 20,000 ×g for 20 min at 4°C, followed by collection of supernatant for protein concentration measurement by Bradford protein assay. 50 μg of total protein were used per sample.
Results
eIF2B5 mutation leads to altered expression of pathways that are essential for the nervous system
Wild-type (WT) and Eif2b5R132H/R132H (Eif2bM) mice were fed a normal diet and sacrificed at the age of 11 and 12 weeks (w11 and w12, respectively), followed by quantitative proteomic analysis of their cerebrums. To allow accurate quantification, each sample was combined with an equal amount of proteins from SILAC mice (SILAM) cerebrum. Proteins were fractionated by SDS-PAGE and subjected to in-gel digest protocol followed by LC-MS/MS analysis using LTQ-Orbitrap XL mass spectrometer. As expected, the proteome profiles demonstrated high enrichment of proteins related to brain functions (Supp. Table 3). Analysis of WT-subtracted proteome profiles reveals a strong consistency between measurements at w11 and w12 (Methods; Supp. Figure 1). For example, among 100 and 75 proteins whose absolute WT-subtracted Eif2b5M values exceeded a cutoff of 0.5 at w11 and w12, respectively, 36 proteins exceeded this cutoff at both time points (overlap P value < 1.5∙10-22; hyper-geometric test; Supp. Figure 1, top). Similar significant overlaps between w11 and w12 were also attained for other cutoffs (Supp. Figure 1, bottom), indicating the results are reproducible between the time points and suggesting that altered proteins may contribute to VWM pathogenesis. We noticed that a WT versus Eif2b5M cutoff of 0.5 yields the most significant results, and this was therefore used to construct the ‘eIF2B-related proteins’ group. This group contains the 139 proteins whose absolute WT-subtracted Eif2b5M values exceed 0.5 at w11 and/or w12 (Methods, Supp. Table 4, Fig. 1a). Figure 1b shows that 25 of the 139 eIF2B-related proteins are tightly interconnected through known protein-protein interactions. Note that both the 26S proteasome and translation machinery are major hubs in this network, highlighting the relationship between protein synthesis and degradation and suggesting a possible involvement of the ubiquitin-proteasome degradation pathway in VWM pathology.
Based on the temporal expression changes in Eif2b5M brains as the mice mature from 11 to 12 weeks of age, eIF2B-related proteins were partitioned into 3 main groups. Group 1 (18 proteins, e.g. Rpl15) consists of proteins whose expression changed in WT but remained invariant in Eif2b5M mice; groups 2 (16 proteins, e.g. Syp) consists of proteins whose expression was reduced from w11 to w12 in the WT but elevated in Eif2b5M; and group 3 (94 proteins, e.g. Kif1a, Eif4g1, Rplp1, Rplp2, Map1a, Map1b) consists of proteins whose expression increased from w11 to w12 in the WT but decreased in Eif2b5M (Fig. 1a). Remarkably, there is a strong tendency for down-regulation of proteins in the mutant compared to WT mice, as group 3 is the largest one.
Functional enrichment analysis of eIF2B-related proteins provided indications for the important contribution of eIF2B in shaping brain development (Fig. 1a,c, Supp. Table 5). For example, categories such as ‘microtubule dynamics’ (P<2.10-10, 32 proteins, e.g., Map1a,b, 4 and 6), ‘assembly of cell-cell contacts’ (P<10-7, 12 proteins, e.g., Kif1a) and ‘stabilization of filaments’ (P<7.10-8, 9 proteins, e.g., Gap43 and MAPs) are related to cytoskeletal organization, known to be essential for functional properties of cells in the nervous system. Functional categories related to nervous system development and performance were also enriched, for example ‘axonal transport’ (P< 10-7, 6 proteins, e.g., Htt, Kif1a, Mapt) and ‘synaptogenesis’ (P<6.10-7, 10 proteins, e.g., Syn1, Syn2). ‘Seizures’, which came up as a clinical symptom associated with neurological diseases involving mal-expression of certain genes (P<6.10-6, 14 proteins, e.g., Map1b, Mapt), is also one of the clinical symptom of VWM (Bugiani et al. 2010, Fogli & Boespflug-Tanguy 2006, Pronk et al. 2006, Schiffmann & Elroy-Stein 2006, van der Knaap et al. 2006). Of particular interest, as VWM results from a loss-of-function mutation in a translation initiation factor, is the enrichment of translation machinery components (P<10-5, 8 proteins), which are mostly down-regulated in the mutants as compared to the WT. These include translation initiation factors Eif1, Eif4g1 and Eif4g3 and ribosomal proteins Rplp1, Rplp2, Rpl15, Rpl23a. This interesting observation suggests the existence of co-regulatory circuits and tight feedback mechanisms within the translation machinery.
Cuprizone-induced remyelination of wild-type mice shares common expression dynamics with the delayed developmental myelination of mutant mice
Myelin formation in the brain takes place during postnatal development and upon repair from demyelination damage. To further assess the involvement of eIF2B-related proteins in myelin formation and study the proteome responses to signals related to myelination, we used the cuprizone experimental model. Supplementing the mice diet with the copper chelator cuprizone leads to massive demyelination in large areas of the mouse brain, followed by a near-complete recovery (remyelination) within a few weeks of cuprizone removal (Matsushima & Morell 2001). Thus, the cuprizone experimental model makes it possible to monitor changes along the process of remyelination. To understand the effects of the mutation on cellular functions that have a significant role in remyelination, Eif2b5M and WT mice were fed 0.2% cuprizone for 4 weeks, and protein levels in their cerebrums were analyzed after 1 or 2 weeks of recovery, i.e. at 11- and 12-weeks of age. The normal and cuprizone diet-fed mice groups, representing ‘developmental myelination’ and ‘remyelination’, respectively, were analyzed at the same ages (experimental design is detailed in Fig 2a). The effect of cuprizone was verified by time-course live magnetic resonance imaging (MRI) employing a diffusion tensor imaging (DTI) protocol and using fractional anisotrophy (FA), a marker of tissue organization, which is typically higher in myelin-rich regions. As performed and demonstrated in our previous study (Geva et al. 2010), the decrease in FA values confirmed the demyelination effect of the cuprizone diet (Fig. 2b). We next defined a ‘response’ as the fold change in the levels of each protein measured at week 12 (w12) compared to week 11 (w11). ‘Developmental myelination response’ refers to the change between w12 to w11 in the normal diet-fed mice, while ‘remyelination response’ refers to the change between w12 to w11 in the cuprizone fed-mice, i.e. between 1 and 2 weeks of recovery after cuprizone removal (see Methods and Fig. 2b). Increasing or decreasing responses (that is, >1 or <1 fold change, respectively) therefore indicate up- and down-regulation of proteins levels as the mice mature from week 11 to week 12 under the different diets. However, note that increasing or decreasing responses do not provide information about absolute protein levels.
Fig. 2. Comparison of developmental myelination and remyelination responses between different genotypes.
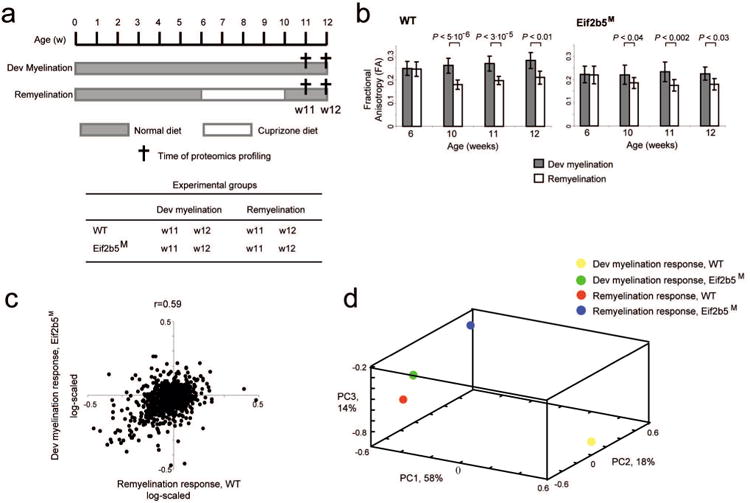
(a) Illustration of the experimental time-frame. ‘Developmental myelination’ (Dev. Myel.) refers to normal diet followed by harvesting at 11 and 12 weeks of age (w11, w12) for brain analysis. ‘Remyelination’ (Re-Myel.) refers to cuprizone-containing diet given between the age of 6-10 weeks followed by switch to normal diet and harvesting at w11 and w12 for brain analysis. Each experimental group contained 5 male mice. The brains of the 8 experimental groups, as indicated in the Table, were removed and analyzed. (b) Each mouse of the 8 experimental groups described in a was subjected to live quantitative brain scans by magnetic resonance imaging at 6, 10, 11, and 12 (when relevant) weeks of age, followed by voxel-based analysis as described in Methods. Shown is the fractional anisotropy (FA) parameter, a marker of tissue organization, of the myelin-rich corpus callosum. The plot presents the FA values (y axis) for WT (left) and Eif2b5M (right) across time points (x axis, weeks), for both developmental myelination (gray) and remyelination (white). Shown are average FA values ± SD and t-test P-values. The decreased FA value demonstrates the demyelination effect of cuprizone diet on both WT and Eif2b5M groups. (c) Shown are ‘remyelination response’ in wild-type (WT) (x axis) and normal ‘developmental myelination response’ in the mutant (Eif2b5M) strain (y axis, log scaled). Positive/negative response indicates protein level that is higher/lower at w12 versus w11 (Methods). (d) Principle component analysis (PCA) applied on the four response profiles: ‘remyelination response’ in WT (red) and mutant (blue), as well as ‘developmental myelination response’ in WT (yellow) and mutant (green). Shown is a scatter plot of each response profile for the first three principle components (PC1, PC2 and PC3). The dimension reduction by PCA indicates similarity of the ‘remyelination response’ in WT and ‘developmental myelination response’ in mutant, whereas each of the two additional response profiles differs from any other profile.
Interestingly, the WT ‘remyelination response’ and Eif2b5M ‘developmental myelination response’ were very similar but distinct from the two remaining profiles (Fig. 2c and Supp. Fig. 2). Principle component analysis of the four responses further illustrates this similarity (Fig. 2d). This observation clearly points out that the mutation is associated with impaired myelination, as manifested during development under normal conditions and upon recovery from cuprizone-induced damage. Therefore, the data provides a meaningful resource to better understand the molecular mechanisms involved in the myelination process and the role of eIF2B in the regulation of this process.
eIF2B5 mutation leads to dysregulation of the balance between protein synthesis and degradation
To further characterize the mechanisms involved and understand the abnormalities imposed by the mutation, all proteins were divided into 14 clusters according to their response pattern across the four experimental conditions (Fig. 3a and Supp. Table 2). The enrichment of specific functional categories within the different clusters (Fig. 3b) implies that the observed responses represent a real biological phenomenon. For example, proteins with functional roles in protein synthesis (8 translation initiation factors, 5 translation elongation factors, 35 ribosomal proteins; Fig. 4a) were enriched in cluster no. 2 (P<0.001, 11 proteins: 5 initiation factors, 1 elongation factor, 4 RPLs and 1 RPS) and cluster no. 11 (P<0.05, 5 RPLs and 2 RPSs). These clusters represent abnormal down-regulation of proteins in Eif2b5M relative to WT during developmental myelination (cluster no. 2) and abnormal up-regulation of proteins in Eif2b5M relative to WT during remyelination (cluster no. 11). Notably, additional components related to protein synthesis are included in other clusters (e.g., Eef1b2, Eef2, and Rps19 in cluster no. 6; Eif1, Rpl17, Rps13, Rps2, and Rps8 in cluster no. 12; and Eif4a2, Rpl18, Rpl7, Rps7 in cluster no. 14). The diversity in expression dynamics of this sub-class of proteins indicates multifaceted effects on the performance of the translation machinery, reflecting complex feedback mechanisms and downstream consequence. Downstream effects may relate to the wide-ranging changes that differentially affect the translation rate of specific mRNAs due to their unique cis-regulatory elements, which may lead to unbalanced stoichiometry within cellular complexes. Interestingly, the major myelin protein markers are found in clusters nos. 12 and 14 (e.g., MAG, cluster no. 12; MBP, CNP, MOG, PLP1, cluster no. 14; Fig. 4b), indicating abnormal up-regulation of these proteins in Eif2b5M relative to WT during remyelination. This may suggest that myelin proteins are over-produced in Eif2b5M brains but not assembled properly, further demonstrating that the myelination process is dysregulated due to the mutation.
Fig. 4. Protein synthesis and degradation.
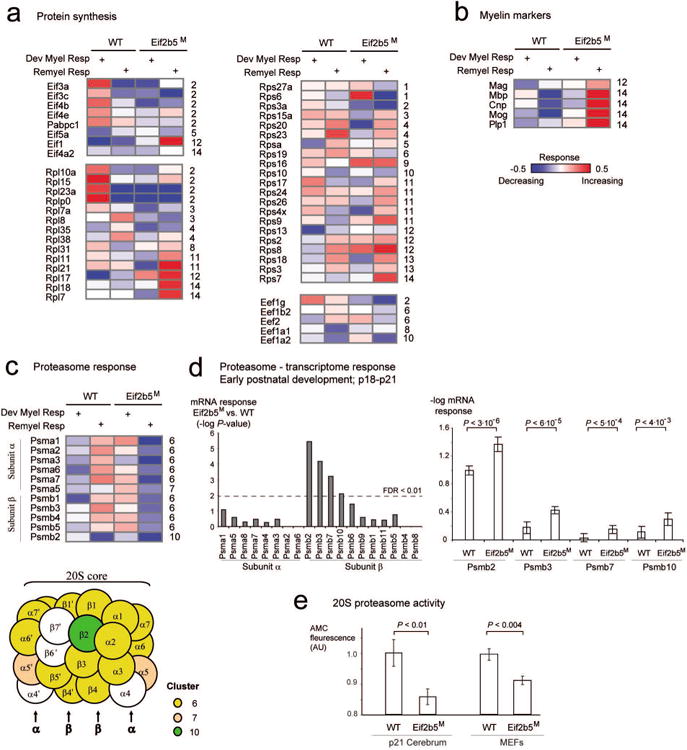
Each matrix presents response levels (rows) of proteins related to protein synthesis (a), myelin markers (b) and protein degradation (c) across four experiments (columns). The columns and color coding are shown as in Fig. 3a. For each protein, its symbol and cluster identifier are specified near the matrix (left and right, respectively; cluster identifiers are as in Fig. 3a). Notably, the proteasomal subunits α and β are specifically enriched in cluster no. 6; proteins that are part of these complexes are illustrated as circles and color coded by their response cluster identifier (c, bottom). (d) Analysis of transcriptome at p18 to p21 across proteasomal subunits α and β. Left: the histogram presents P-values for the difference between p18-p21 mRNA responses in WT versus Eif2b5M strains (y axis) across proteasomal subunits α and β (x axis). Right: Average -log mRNA responses (y axis) for WT and Eif2b5M strains across three exemplified proteasomal subunits β genes (x axis). Statistical t-test P-values are shown on top and summarized in the left panel of this figure. (e) Shown is 20S proteasome activity (y axis), which is defined as fluorescence level of free 7-Amino-4-methylcoumarin (AMC) fluorophore following cleavage of labeled LLVY-AMC substrate (Methods). Protein extracts from WT and Eif2b5M MEFs (right) or p21 brains (left) were used (x axis). Shown is average ±SD of 7 experiments (right) or 9 WT and 10 Eif2b5M brains (left).
Cluster no. 6 clearly demonstrates the global similarity between ‘remyelination response’ of WT and ‘developmental myelination response’ of Eif2b5M (Figs. 2c and 3), suggesting that the enriched functional categories within it are important for myelination during both development and repair. This cluster provides evidence for dysregulation of the balance between protein synthesis and degradation, in agreement with the involvement of the proteasome and ribosomal subunits within the eIF2B-related sub-network as depicted in Figure 1b. Notably, components of the 20S proteasome are over-represented in cluster no. 6 (Fig. 3b and 4c), which is also enriched for several other functional categories that depend heavily on protein degradation. These include metabolism, cell cycle and apoptosis (P<2.10-6, P<1.8.10-7, respectively, Fig. 3b). Intriguingly, out of the 14 subunits of the 20S proteasome chamber, 9 are found in cluster no. 6 (five alpha-type subunits, i.e., Psma1,2,3,6,7; and four beta-type subunits, i.e., Psmb1,3,4,5; see Fig. 4c). Overall, 10 out of the 11 proteasome-related core subunits detected in this study were up-regulated in WT but down-regulated in Eif2b5M mutant during remyelination.
We next reasoned that if proteasomal function is critical for remyelination, it might also be important for myelination during early postnatal brain development. To verify this, we used our previously-generated dataset of brain transcriptome profiles from WT and Eif2b5M mice at postnatal day 18 and 21 (p18 and p21), which represent the peak of myelin formation (Marom et al. 2011). Focusing on the levels of mRNAs encoding for proteasomal subunits, we tested the difference in mRNA responses (p18 to p21) between WT and Eif2b5M mice (Methods). The results indicate that the regulation of proteasomal subunit β, but not the α subunit, is dramatically altered during early postnatal development due to the mutation in Eif2b5 (Figure 4d, left). For example, the genes Psmb2, Psmb3, Psmb7 and Psmb10 achieved t-test P-value < 3.10-6, 6.10-5, 5.10-4 and 4.10-3, respectively (Figure 4d, right). Taken together, the effect of the mutation in Eif2b5 on proteasome components expression on both the transcriptome and proteome levels, although not necessarily at the same age, suggests the importance of proteasome activity to brain development, myelination and to VWM pathology.
We next checked if the upregulation of proteasome components in normal-diet-fed mutant mice results in increased proteasome activity, by measuring the release of free 7-Amino-4-methylcoumarin (AMC) fluorophore following cleavage of labeled LLVY-AMC substrate. Interestingly, the proteasome activity was 7% and 14% lower in MEFs and p21 brains, respectively, from Eif2b5M versus WT mice (Fig. 4e). This observation suggests dysregulation of appropriate 20S proteasome complex formation despite the upregulation of some of its components in Eif2b5M mice under normal conditions.
eIF2B5 mutation leads to dysregulation of mitochondrial oxidative phosphorylation
Among all the response proteins, 87 are associated with metabolism and ATP production. While 17 proteins have a role in the tricarboxylic acid cycle (TCA), 50 are part of the electron transfer chain (ETC), and 20 proteins are involved in glycolysis/gluconeogenesis (Fig. 5a,b and Supp. Fig. 3). The NADH dehydrogenase complex is particularly intriguing, because it manifests the most significant enrichment of clusters nos. 12 and 14 that are characterized by abnormal up-regulation during remyelination in Eif2b5M. In particular, cluster no. 14 contains 11 subunits of complex I (e.g., Ndufa7/a9/a13/b4/b5/b10/c2/s1/s2/s5/s7) and cluster no. 12 contains another 8 subunits of the same complex (e.g. Ndufa2/a8/a10/a12/b3/b6/s3/s6) (Fig. 5a,b). A similar significant over-representation of such up-regulation could not be observed in other (ETC and non-ETC) inner mitochondrial membrane complexes, such as the mitochondrial import translocase (Supp Fig. 3b), suggesting that the dysregulation due to the Eif2b5 R113H mutation during remyelination is mediated through complex-specific pathways. The abnormal up-regulation of NADH dehydrogenase (complex I) in the absence of similar up-regulation of F0F1-ATP synthase (complex V) (Fig. 5a,b) may lead to accumulation of protons in the mitochondrial inter-membrane space and increased generation of reactive oxygen species (ROS). To confirm that the mutation in Eif2b5 affects cellular ROS levels, MEFs isolated from WT and Eif2b5M mice were stained with CellROX, an oxidative stress fluorescent detector, followed by classification of sub-populations based on their cellular CellROX integrated intensity. Whereas only 50% of the WT cells were above the set threshold, 64% of the Eif2b5M cells were identified as ROS-positives (P<10-94, χ 2-test of independence; Fig. 5c), providing a clear indication that Eif2b5 R113H mutation leads to increased oxidative stress, even under normal condition.
Fig. 5. Mitochondrial electron transport chain (ETC) and ROS levels.
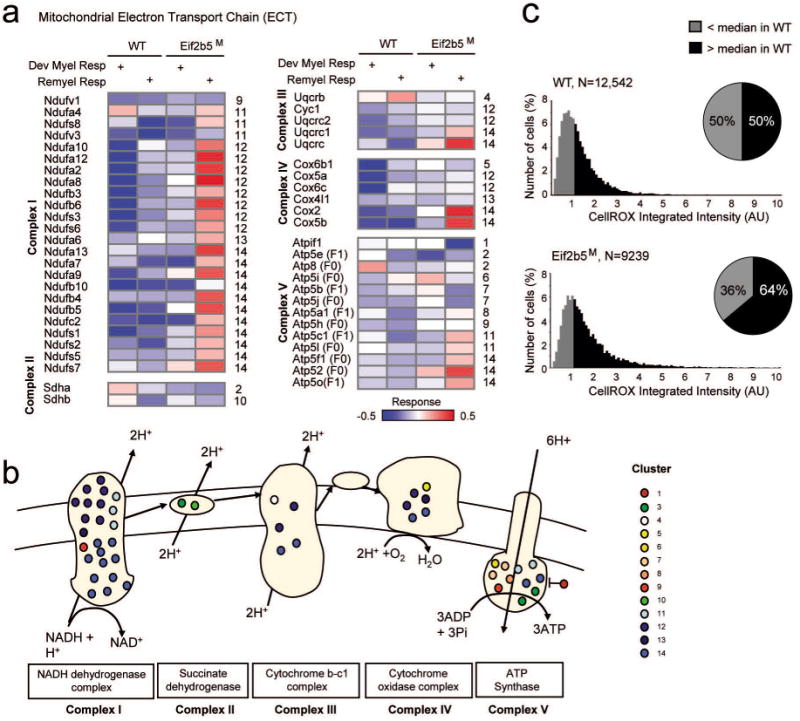
(a) Each matrix presents all proteins related to an ETC complex (rows) across experiments (columns). The columns and color coding are shown as in Fig. 3a. (b) Shown are complexes I-V. Proteins that are part of these complexes are illustrated as circles and color coded by their response cluster identifier. (c) WT and Eif2b5M MEFs were stained with Hoechst 33258 and CellROX as detailed in Methods. N= number of cells. Shown is the cellular CellROX integrated intensity. The median of CellROX integrated intensity in WT cells was set as a threshold and used to classify sub-populations of cells. ROS-positive (black) or negative (gray) cells are above or below this threshold, respectively. χ2-test of independence P-value <10-94 for the difference between the two pie charts, indicating higher proportion of Eif2b5M cells are ROS-positives compared to WT cells.
Discussion
Our previous studies have shown that Eif2b5M mice demonstrate delayed brain development, mild early neurodegeneration and a robust remyelination defect, using multiple methodologies including time course MRI, electron microscopy, immuno-histochemistry, and mRNA expression profiling (Geva et al. 2010, Marom et al. 2011, Cabilly et al. 2012). The current study shows that many ‘eIF2B-related proteins’ (60 of 94 proteins; group 3, Fig. 1a) up-regulated in WT brains during myelination are down-regulated in Eif2b5M brains, demonstrating that multiple proteins fail to reach normal levels in the mutants due to a disruption or delay in their expression program. The data reveals a remarkable similarity between the dynamics of specific complexes and functions during brain development in Eif2b5M and upon remyelination in WT mice. This finding is of particular importance because it provides an additional support for the ‘Recapitulation Hypothesis’ (Franklin & Hinks 1999), which suggested that developmental myelination and remyelination following damage share the same key stages; secondly, it further strengthens the hypothesis that hypomorphic mutations in eIF2B lead to delayed developmental myelination. The current study highlights the important role of specific protein complexes and pathways in myelination/remyelination and points at their possible role in VWM pathology. The functional enrichments in the ‘eIF2B-related proteins’ group provide an important resource to better understand the role of eIF2B as a major regulator in the myelination process. Of particular interest is the altered balance between protein production and degradation, the dysregulated mitochondrial function, and the tight relationship of these processes with cell-cycle progression. The expression dynamics of cell-cycle proteins is expected to reflect the proliferation rate of glial precursor cells during brain repair, as it does during early postnatal brain development (Van den Hove et al. 2006). The up-regulation of cell-cycle proteins in WT brains during remyelination is in agreement with this notion. The down-regulation of these proteins in Eif2b5M brains further emphasizes the negative effects of Eif2b5 mutation on the cell cycle, an important key for myelin regeneration. Importantly, our previous microarrays expression study indicated abnormal down-regulation of steady-state mRNA levels of cell-cycle genes in Eif2b5M brains during early postnatal brain development (p18 and p21, the peak of myelin formation). The prolonged cell cycle of primary astrocytes isolated from Eif2b5M newborn mice validated the negative effect of Eif2b5 mutation on cell cycle progression (Marom et al. 2011).
eIF2B5 mutation leads to dysregulation of the balance between protein synthesis and degradation
mTOR, a serine/threonine protein kinase, is a major regulator of cell growth, proliferation and survival. It senses cellular nutrient, oxygen, and energy levels and by integrating inputs from upstream pathways administers effective and precise downstream responses (Laplante & Sabatini 2012). Interestingly, mTOR-complex1 (mTORC1) signaling coordinates both protein synthesis and degradation, by promoting both functions together in response to translation inhibition. mTORC1-mediated proteasome activation is viewed as an adaptive response to support increased protein synthesis by increasing the size of the intracellular amino acid pool via NRF1-mediated transcriptional activation of genes encoding proteasomal subunits (Kimball et al. 2008, Zhang et al. 2014). The up-regulation of 20S proteasomal subunits in Eif2b5M mice during developmental myelination (Figs. 4cd) suggests a possible effect of the mutation in Eif2b5 on mTORC1 activity. This may be associated with increased translation rates to compensate for the inhibitory effect of the mutation on global translation initiation. Indeed, the rate of global protein synthesis is similar in Eif2b5M and WT mice under normal conditions (Cabilly et al. 2012), possibly reflecting the net outcome of such compensation. In contrast, although some proteasome subunits are upregulated in Eif2b5M (Fig. 4cd), the proteasome enzymatic activity is lower in Eif2b5M compared to WT (Fig. 4e), reflecting lack of complete compensation and suggesting involvement in VWM pathology. Furthermore, the up-regulated expression of 20S proteasomal subunits during remyelination in WT mice is in line with a requirement for larger amounts of amino acids to facilitate enhanced protein synthesis during myelin repair. However, in contrast to WT mice, 20S proteasome subunits are down-regulated during remyelination in Eif2b5M mice (Fig. 4c). Given that increased protein synthesis is accompanied by increased proteasomal activity, this result may highlight the inability of the mutant brain to cope with the acute demand for rapid increase in translation rate for the purpose of brain repair. The decreased proteasomal activity in the brains of Eif2b5M mice at p21, i.e. the peak of myelin synthesis (Fig. 4e), strongly supports this notion. Moreover, the data are in agreement with our previous study that demonstrated a failure of primary astrocytes and microglia isolated from Eif2b5M brains to increase the synthesis rate of multiple proteins in response to acute cellular demand following exposure to LPS (Cabilly et al. 2012).
Aberrant proteasomal function leads to pleiotropic defects in many cellular processes including cell-cycle, differentiation and apoptosis and has been also implicated in the pathogenesis of neurodegenerative diseases (Ciechanover & Brundin 2003). Proteasomal inhibition in oligodendrocytes induces mitochondrial dysfunction, oxidative stress, disruption of mitochondrial membrane potential and the accumulation of oxidized proteins (Goldbaum et al. 2006). Based on the current study, we propose that the proteasome is one of the major mediators of VWM pathology.
eIF2B5 mutation leads to dysregulation of mitochondrial oxidative phosphorylation
Mitochondrial biogenesis during the cell cycle is regulated at the protein synthesis level. Whereas subunits of ETC complex IV accumulate during S phase, the catalytic subunit of complex V, β-F1-ATP-synthase, and other critical mitochondrial components accumulate specifically at G2/M. The efficient translation of β-F1-ATP-synthase mRNA during G2/M depends on an internal ribosome entry site (IRES)-like element within its 3′ non-translated region (3′UTR) (Izquierdo & Cuezva 2000, Martinez-Diez et al. 2006). Other transcripts encoding for proteins involved in oxidative phosphorylation also contain similar translation activation cis-elements in their 3′UTRs (Di Liegro et al. 2000). Therefore, full mitochondrial membrane potential is reached at G2/M. Thus, it is not surprising that hypomorphic mutation in a translational initiation factor interferes with the accurate balance of translation required for proper mitochondrial biogenesis and activity. Moreover, the abnormal function of the translation and proteasome machineries, which are associated with abnormal cell-cycle progression, further lead to dysregulation of mitochondrial function. The current study provides a strong indication for lack of appropriate stoichiometry between subunits composing the ETC complexes. The differential responses of WT and Eif2b5M brains highlight the fundamental regulatory problem elicited by the mutation. Notably, most of ETC complexes I subunits (NADH dehydrogenase) are abnormally up-regulated in Eif2b5M brains during remyelination. The lack of a corresponding up-regulation of F0/F1 ATP-synthase may lead to accumulation of protons in the mitochondrial intermembrane space, resulting in overproduction of ROS as observed in Eif2b5M MEFs (Fig. 5c). Atpif1/IF1, an inhibitor of the F1 moiety, is induced when protons accumulate inside the mitochondrial matrix in order to prevent the reverse function of ATP-synthase (i.e. ATP hydrolysis)(Cabezon et al. 2003). It is tempting to speculate that down-regulation of Atpif1/IF1 during remyelination in Eif2b5M (Fig. 5a) is a compensatory response, indicative of F0/F1-synthase malfunction. The extreme sensitivity of the brain to oxidative stress due to high abundance of polyunsaturated fatty acids (PUFAs) suggests that Eif2b5M brains, as well as the brains of human VWM patients, suffer from increased oxidative damage (Maruyama et al. 2014).
Interestingly, several proteomic analyses identified ectopic expression of mitochondrial ETC complexes I-V in myelin, implying that oxidative phosphorylation may occur in myelin membranes in the absence of mitochondria (Taylor et al. 2004, Ishii et al. 2009, Panfoli et al. 2011, de Monasterio-Schrader et al. 2012). Consequent experiments demonstrated the activity of glycolytic and TCA cycle enzymes, ETC complexes and Fo/F1-ATP synthase in isolated myelin vesicles. Moreover, the presence of the Atpif1/IF1 inhibitor in isolated myelin was suggested to have an important role during brain ischemia (Ravera et al. 2009, Ravera et al. 2011, Ravera et al. 2013). The absence of other mitochondrial proteins in myelin vesicles ruled out mitochondrial contamination and led to the intriguing hypothesis that myelin sheaths may be able to perform aerobic metabolism by extra-mitochondrial oxidative phosphorylation, to produce ATP and provide it to axons via gap junctions (Morelli et al. 2011). The possible energetic role of myelin sheaths may explain the degeneration of chronically demyelinated axons and suggest that VWM disease could result from malfunction of energy supply by myelin. The relative contribution of defective mitochondrial function versus possible faulty extra-mitochondrial oxidative phosphorylation to VWM pathogenesis will be addressed in future studies.
The current study provides a clear demonstration of the robust impact of the mutated (hence partially functional) eIF2B on multiple molecular pathways. It assigns an important role for eIF2B as a major upstream regulator of numerous cellular circuits, leading to VWM disease. The emerging insights related to the identity and nature of these molecular circuits may enhance the development of therapeutic strategies for the treatment of VWM and other leukodystrophies.
Supplementary Material
Acknowledgments
We thank Drs. Kristy J. Brown, Adeline Vanderver and Asako Takanohashi (Children's National Medical Center, Washington DC) for their kind assistance in collection and processing of LTQ-OrbitrapXL mass-spectrometry samples. We thank Dr. Ranen Aviner for critical reading of the manuscript. This study was supported by grants from the Legacy Heritage Biomedical Science Partnership Program of the Israel Science Foundation (grant no. 1629/13, OES), the Israeli Centers of Research Excellence (I-CORE) Gene Regulation in Complex Human Disease, Center No 41/11 (IGV), and an Israeli Science Foundation fund (grant no. 1643/13, IGV). IGV is a Faculty Fellow of the Edmond J. Safra Center for Bioinformatics at Tel Aviv University and an Alon Fellow
References
- Bugiani M, Boor I, Powers JM, Scheper GC, van der Knaap MS. Leukoencephalopathy with vanishing white matter: a review. J Neuropathol Exp Neurol. 2010;69:987–996. doi: 10.1097/NEN.0b013e3181f2eafa. [DOI] [PubMed] [Google Scholar]
- Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nature structural biology. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- Cabilly Y, Barbi M, Geva M, Marom L, Chetrit D, Ehrlich M, Elroy-Stein O. Poor cerebral inflammatory response in eIF2B knock-in mice: implications for the aetiology of vanishing white matter disease. PLoS One. 2012;7:e46715. doi: 10.1371/journal.pone.0046715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- de Monasterio-Schrader P, Jahn O, Tenzer S, Wichert SP, Patzig J, Werner HB. Systematic approaches to central nervous system myelin. Cellular and molecular life sciences : CMLS. 2012;69:2879–2894. doi: 10.1007/s00018-012-0958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Liegro CM, Bellafiore M, Izquierdo JM, Rantanen A, Cuezva JM. 3′-untranslated regions of oxidative phosphorylation mRNAs function in vivo as enhancers of translation. The Biochemical journal. 2000;352 Pt 1:109–115. [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Chan JR, Baranzini SE, Franklin RJ, Rowitch DH. Myelin regeneration: a recapitulation of development? Annual review of neuroscience. 2011;34:21–43. doi: 10.1146/annurev-neuro-061010-113629. [DOI] [PubMed] [Google Scholar]
- Fogli A, Boespflug-Tanguy O. The large spectrum of eIF2B-related diseases. Biochem Soc Trans. 2006;34:22–29. doi: 10.1042/BST20060022. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Hinks GL. Understanding CNS remyelination: clues from developmental and regeneration biology. J Neurosci Res. 1999;58:207–213. [PubMed] [Google Scholar]
- Geva M, Cabilly Y, Assaf Y, Mindroul N, Marom L, Raini G, Pinchasi D, Elroy-Stein O. A mouse model for eukaryotic translation initiation factor 2B-leucodystrophy reveals abnormal development of brain white matter. Brain. 2010;133:2448–2461. doi: 10.1093/brain/awq180. [DOI] [PubMed] [Google Scholar]
- Goldbaum O, Vollmer G, Richter-Landsberg C. Proteasome inhibition by MG-132 induces apoptotic cell death and mitochondrial dysfunction in cultured rat brain oligodendrocytes but not in astrocytes. Glia. 2006;53:891–901. doi: 10.1002/glia.20348. [DOI] [PubMed] [Google Scholar]
- Hanefeld F, Holzbach U, Kruse B, Wilichowski E, Christen HJ, Frahm J. Diffuse white matter disease in three children: an encephalopathy with unique features on magnetic resonance imaging and proton magnetic resonance spectroscopy. Neuropediatrics. 1993;24:244–248. doi: 10.1055/s-2008-1071551. [DOI] [PubMed] [Google Scholar]
- Ishii A, Dutta R, Wark GM, Hwang SI, Han DK, Trapp BD, Pfeiffer SE, Bansal R. Human myelin proteome and comparative analysis with mouse myelin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14605–14610. doi: 10.1073/pnas.0905936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Cuezva JM. Internal-ribosome-entry-site functional activity of the 3′-untranslated region of the mRNA for the beta subunit of mitochondrial H+-ATP synthase. The Biochemical journal. 2000;346 Pt 3:849–855. [PMC free article] [PubMed] [Google Scholar]
- Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Do AN, Kutzler L, Cavener DR, Jefferson LS. Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. The Journal of biological chemistry. 2008;283:3465–3475. doi: 10.1074/jbc.M706643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Moser M, Ussar S, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater PA, Vermeulen G, Konst AA, et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nature genetics. 2001;29:383–388. doi: 10.1038/ng764. [DOI] [PubMed] [Google Scholar]
- Marom L, Ulitsky I, Cabilly Y, Shamir R, Elroy-Stein O. A point mutation in translation initiation factor eIF2B leads to function--and time-specific changes in brain gene expression. PLoS One. 2011;6:e26992. doi: 10.1371/journal.pone.0026992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Diez M, Santamaria G, Ortega AD, Cuezva JM. Biogenesis and dynamics of mitochondria during the cell cycle: significance of 3′UTRs. PLoS One. 2006;1:e107. doi: 10.1371/journal.pone.0000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama W, Shaomoto-Nagai M, Kato Y, Hisaka S, Osawa T, Naoi M. Role of lipid peroxide in the neurodegenerative disorders. Sub-cellular biochemistry. 2014;77:127–136. doi: 10.1007/978-94-007-7920-4_11. [DOI] [PubMed] [Google Scholar]
- Matsushima GK, Morell P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001;11:107–116. doi: 10.1111/j.1750-3639.2001.tb00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli A, Ravera S, Panfoli I. Hypothesis of an energetic function for myelin. Cell biochemistry and biophysics. 2011;61:179–187. doi: 10.1007/s12013-011-9174-8. [DOI] [PubMed] [Google Scholar]
- Panfoli I, Ravera S, Bruschi M, Candiano G, Morelli A. Proteomics unravels the exportability of mitochondrial respiratory chains. Expert review of proteomics. 2011;8:231–239. doi: 10.1586/epr.11.1. [DOI] [PubMed] [Google Scholar]
- Pronk JC, van Kollenburg B, Scheper GC, van der Knaap MS. Vanishing white matter disease: a review with focus on its genetics. Ment Retard Dev Disabil Res Rev. 2006;12:123–128. doi: 10.1002/mrdd.20104. [DOI] [PubMed] [Google Scholar]
- Ravera S, Bartolucci M, Calzia D, Aluigi MG, Ramoino P, Morelli A, Panfoli I. Tricarboxylic acid cycle-sustained oxidative phosphorylation in isolated myelin vesicles. Biochimie. 2013;95:1991–1998. doi: 10.1016/j.biochi.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Ravera S, Panfoli I, Aluigi MG, Calzia D, Morelli A. Characterization of Myelin Sheath F(o)F(1)-ATP synthase and its regulation by IF(1) Cell biochemistry and biophysics. 2011;59:63–70. doi: 10.1007/s12013-010-9112-1. [DOI] [PubMed] [Google Scholar]
- Ravera S, Panfoli I, Calzia D, Aluigi MG, Bianchini P, Diaspro A, Mancardi G, Morelli A. Evidence for aerobic ATP synthesis in isolated myelin vesicles. The international journal of biochemistry & cell biology. 2009;41:1581–1591. doi: 10.1016/j.biocel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nature genetics. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Schiffmann R, Elroy-Stein O. Childhood ataxia with CNS hypomyelination/vanishing white matter disease--a common leukodystrophy caused by abnormal control of protein synthesis. Mol Genet Metab. 2006;88:7–15. doi: 10.1016/j.ymgme.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Schiffmann R, Moller JR, Trapp BD, et al. Childhood ataxia with diffuse central nervous system hypomyelination. Ann Neurol. 1994;35:331–340. doi: 10.1002/ana.410350314. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, Pfeiffer SE. Proteomic mapping provides powerful insights into functional myelin biology. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4643–4648. doi: 10.1073/pnas.0400922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hove DL, Steinbusch HW, Scheepens A, Van de Berg WD, Kooiman LA, Boosten BJ, Prickaerts J, Blanco CE. Prenatal stress and neonatal rat brain development. Neuroscience. 2006;137:145–155. doi: 10.1016/j.neuroscience.2005.08.060. [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Leegwater PA, Konst AA, Visser A, Naidu S, Oudejans CB, Schutgens RB, Pronk JC. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol. 2002;51:264–270. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Pronk JC, Scheper GC. Vanishing white matter disease. Lancet Neurol. 2006;5:413–423. doi: 10.1016/S1474-4422(06)70440-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


