Abstract
Purpose
Golden retriever muscular dystrophy (GRMD) is a widely used canine model of Duchenne muscular dystrophy (DMD). Recent studies have shown that magnetic resonance imaging (MRI) can be used to non-invasively detect consistent changes in both DMD and GRMD. In this paper, we propose a semi-automated system to quantify MRI biomarkers of GRMD.
Methods
Our system was applied to a database of 45 MRI scans from 8 normal and 10 GRMD dogs in a longitudinal natural history study. We first segmented six proximal pelvic limb muscles using two competing schemes: 1) standard, limited muscle range segmentation and 2) semi-automatic full muscle segmentation. We then performed pre-processing, including: intensity inhomogeneity correction, spatial registration of different image sequences, intensity calibration of T2-weighted (T2w) and T2-weighted fat suppressed (T2fs) images, and calculation of MRI biomarker maps. Finally, for each of the segmented muscles, we automatically measured MRI biomarkers of muscle volume and intensity statistics over MRI biomarker maps, and statistical image texture features.
Results
The muscle volume and the mean intensities in T2 value, fat, and water maps showed group differences between normal and GRMD dogs. For the statistical texture biomarkers, both the histogram and run-length matrix features showed obvious group differences between normal and GRMD dogs. The full muscle segmentation shows significantly less error and variability in the proposed biomarkers when compared to the standard, limited muscle range segmentation.
Conclusion
The experimental results demonstrated that this quantification tool can reliably quantify MRI biomarkers in GRMD dogs, suggesting that it would also be useful for quantifying disease progression and measuring therapeutic effect in DMD patients.
Keywords: Duchenne muscular dystrophy, golden retriever muscular dystrophy, MRI, canine model, segmentation, statistical texture analysis
I.INTRODUCTION
Duchenne muscular dystrophy (DMD) is a fatal, X-linked, muscle disorder characterized by progressive degeneration of skeletal and cardiac muscle [1]. It is the most common form of muscular dystrophy, affecting approximately 1 in 3,500 live born males [2, 3]. DMD is caused by mutations in the DMD gene, which encodes the cytoskeletal protein dystrophin. Proximal leg muscle weakness is generally the first observed clinical manifestation of DMD. The ensuing progressive muscle weakness usually leaves affected boys severely disabled by their early teenage years, and results in death due to respiratory and cardiac failure [2]. Currently, no therapy halts or reverses progression of DMD. Although cellular and gene therapies are promising, key questions must first be addressed in relevant animal models. Spontaneous forms of X-linked muscular dystrophy due to dystrophin deficiency have been characterized in mice, [4] cats, [5] and dogs. [6, 7] Because golden retriever muscular dystrophy (GRMD) dogs develop progressive and fatal disease strikingly similar to the human condition, this model has increasingly been used in preclinical trials [8].
Although muscle biopsy has been widely used in the diagnosis of muscle diseases including DMD, the invasiveness limits its use and now most patients are diagnosed using molecular methods. As a result, non-invasive outcome measures, especially quantitative methods, are needed for studies in animal models and in DMD patients. Magnetic resonance imaging (MRI) has been used as to provide data on disease progression in both natural history and treatment trials for GRMD [9, 10, 11] and DMD [12, 13, 14]. Observed MRI changes in DMD include an increase in T2 and decrease in T1 relaxation times due to accumulation of fat in affected muscles. There is an associated increase in whole body fat and decrease in muscle mass [12, 15]. These MRI imaging features have been successfully correlated with functional measurements [16, 17]. In fact, one study suggested that MRI is more sensitive than functional tests in predicting disease progression [15]. The potential role of MRI as a biomarker in the GRMD model has also been reported [9, 10, 11]. In the Thibaud et al. study [10], 2-month-old GRMD and carrier dogs were assessed. GRMD dogs had an abnormally high T2-/T1-weighted signal ratio, greater T2-weighted image heterogeneity, and more enhanced muscle signal on T1-weighted images after Gd-DTPA injection compared to heath controls. A subsequent study of 3-month-old to 7-year-old muscular dystrophy dogs by Kobayashi et al. [9] demonstrated increased T2 values and fat suppressed T2-weighted intensity, as well as greater enhancement with gadolinium, consistent with inflammation and necrosis. Yokota et al.’s study [11] contrasted muscular dystrophic dogs treated with a morpholino cocktail with both untreated muscular dystrophic dogs and wild-type littermates from 2 months to 5 years of age. Treated muscular dystrophic dogs had consistently decreased T2 values compared to those that were untreated.
In existing DMD and GRMD studies, muscle segmentation and MRI-based biomarker quantification are manually driven. Because full manual assessment is cumbersome, time consuming and subject to rater errors, computerized quantification methods are needed for advancing MRI biomarkers as objective surrogate outcome measures for pre-clinical and clinical trials. In this paper, we present a novel semi-automatic MRI quantification tool that provides potential imaging biomarkers for monitoring disease progression in pre-clinical and clinical trials of GRMD and DMD. In addition, we propose new run-length matrix based MRI statistical texture analysis technique to create statistical texture biomarkers, which discriminate between normal and GRMD dogs.
II.METHOD
II.1. Study Subjects
The database used in this study included 10 GRMD and 8 normal dogs. Data from MRI were included in a range of phenotypic tests performed over their first year of life. The proximal pelvic limbs of all dogs were scanned at approximately 3 and 6 months of age. Additional studies were completed at 9 months in five GRMD and four normal dogs. Totally 45 MRI scans were acquired.
All the dogs were produced through a GRMD colony maintained at the University of North Carolina at Chapel Hill (UNC-CH) (PI, Kornegay). Dogs were scanned on a Siemens 3T Allegra Head-Only MRI scanner with a circular polarization (CP) head coil or Siemens 3T Tim Trio Whole-Body MRI scanner with a 32-channel body coil at the UNC Biomedical Research Imaging Center (BRIC). Dogs were anesthetized, placed on an MRI gantry in the sternal (prone) position with the pelvic limbs extended and positioned in the coil centered at the midpoint of the femur.
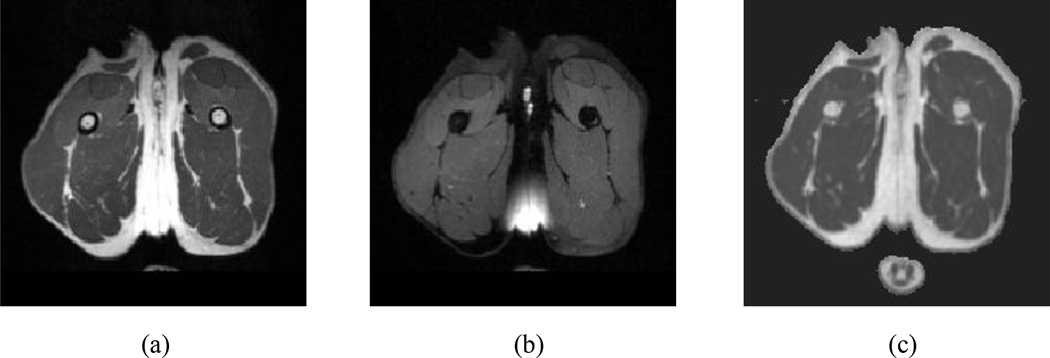
The image protocol of the MRI scans is listed in Table 1. T2-weighted (T2w) image sequence [Fig. 1 (a)] was acquired using a variable-flip-angle turbo spin echo (TSE) sequence. No fat saturation magnetization preparation was applied. T2-weighted fat suppressed (T2fs) images sequence were then acquired using the same variable-flip-angle TSE sequence [Fig. 1 (b)] with the same scanning parameters except that a fat saturation preparation was applied. During the fat saturation procedure, a frequency-selection pulse was applied to excite the fat; dephasing gradient pulses were applied immediately after the frequency-selection pulse to spoil the fat signal. Thus, the remaining signal contained close to zero contribution from fat
Table 1.
Image protocol for MRI scans of canine model. A MRI scan includes a T2-weighted (T2w) image sequence, T2-weighted fat suppressed (T2fs) image sequence, and multi-slice, ten-echo Carr-Purcell-Meiboom-Gill sequence (T2FIT).
| TR (ms) |
TE (ms) |
FOV (mm) |
Slice Thickness (mm) |
Number of slices |
Matrix | Orientation | Scan Time (min) |
Image resolution (mm) |
|
|---|---|---|---|---|---|---|---|---|---|
| T2w | 3000 | 406–409 | 256 | 1 | 160 | 256 × 256 | transverse | 27 | 1×1×1 |
| T2fs | 3000 | 406–409 | 256 | 1 | 160 | 256 × 256 | transverse | 27 | 1×1×1 |
| T2FIT | 3000 | 7–70;Δ7 | 256 | 2 | 200 | 224 × 256 | transverse | 37 | 1×1×3 |
Fig. 1.
MRI scan of the proximal pelvic limb at mid-thigh of a normal control dog in transverse view. (a) T2-weighted (T2W) image and. (b) T2-weighted fat suppressed (T2fs) image, and (c) the transverse view of a T2 value map derived from T2FIT.
To obtain T2 value maps, a multi-slice, ten-echo Carr-Purcell-Meiboom-Gill (CPMG) sequence [T2FIT, Fig. 1 (c)] was applied with the parameters listed in Table 1. To make the acquisition time manageable, the T2FIT only covered the mid-femur section of the proximal pelvic limbs. The computation of T2 value maps is described in detail in section II.2.2.2.
II.2. Image Analysis Method
Our biomarker quantification scheme is composed of three modules: muscle segmentation, pre-processing, and biomarker analysis. As a prerequisite, we first segmented six major proximal pelvic limb muscles of dogs in the MRI images. Then, the process continued with several preprocessing steps and the calculation of the MRI biomarker including texture-based biomarkers. Finally, regional statistics of the MRI biomarkers were calculated for each segmented muscle and these biomarkers were assessed in the context of GRMD disease progression.
II.2.1. Muscle segmentation
In this study, we quantified MRI-based biomarkers of GRMD dogs and normal littermates in six proximal pelvic limb muscles: cranial sartorius (CS), rectus femoris (RF), semitendinosus (ST), biceps femoris (BF), gracilis (GR), and adductor magnus (AD). These muscles were selected to have a balanced representation of flexors and extensors, variable sizes and variable histopathological progression, based on our prior pathologic studies [6, 18]. For instance, histopathologic changes in the CS occur earlier than those in the vastus lateralis [19]. As the anatomical structures of the left and right limbs have a high degree of bilateral symmetry, i.e. differences between the left and a mirrored right leg of the same subject are expected to be small when compared to inter-subject differences, we focused on the left limb in this research for the purpose of tool development and feasibility/validation analysis. We expect that muscle segmentation of the right limb and the subsequent biomarker quantification can be obtained by mirroring the MR images and by applying the processing framework for the left limb.
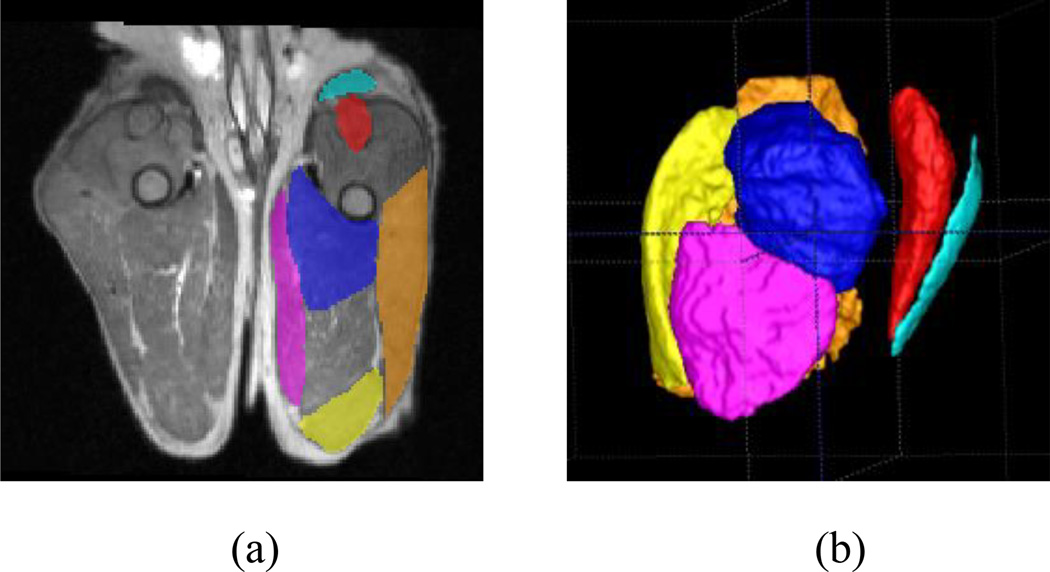
In existing DMD and GRMD MRI studies, muscles were usually segmented manually. To reduce the workload of muscle segmentation, the muscles are generally segmented in a limited range, typically mid-section in the transverse view. For instance, in our previous work [20], we manually segmented the muscles in five slices close to the femoral mid-point. The midpoint of the femur was determined by first identifying the femoral head and condyle as the proximal and distal anatomical limits, respectively, and then dividing the distance between these landmarks in half. As the large majority of a muscle is excluded using such a limited segmentation approach, measurements of muscle volume as well as other biomarkers are quite likely inaccurate (see section III for our evaluation on this topic). To address this issue, we performed an interpolation-based semi-automatic whole muscle segmentation. First, prior to the segmentation, the MRI data were rigidly realigned to a standardized coordinate space, where transverse slices were perpendicular to the femur axis and the sagittal and dorsal slices were parallel to the lateral and cranial axes, respectively. An initial segmentation was then performed by three of the authors (ZF, YS, and JW), each manually delineating the outline of every muscle in every fifth slice only (per muscle, an average of 24 manually-segmented slices) using the ITK-SNAP software (www.itksnap.org) [21]. The manual segmentation was performed independently, i.e., the raters were blinded to the diagnostic information of the dogs and blinded to segmentations from other raters. Thus, the manual segmentation of the muscles was unbiased. Figure 3 (a) shows the manual segmentation of the six muscles of a normal dog in a transverse slice. The segmentations of the remaining slices (4 slices between each 2 manually-segmented slices) were then interpolated via a straightforward linear interpolation scheme from the neighboring manually-segmented slices. Figure 3 (b) shows the interpolated muscle volumes in three dimensional (3D) views. Given the non-folded appearance of proximal pelvic limb muscles, we did not expect a large error for such an interpolated segmentation approach.
Fig. 3.
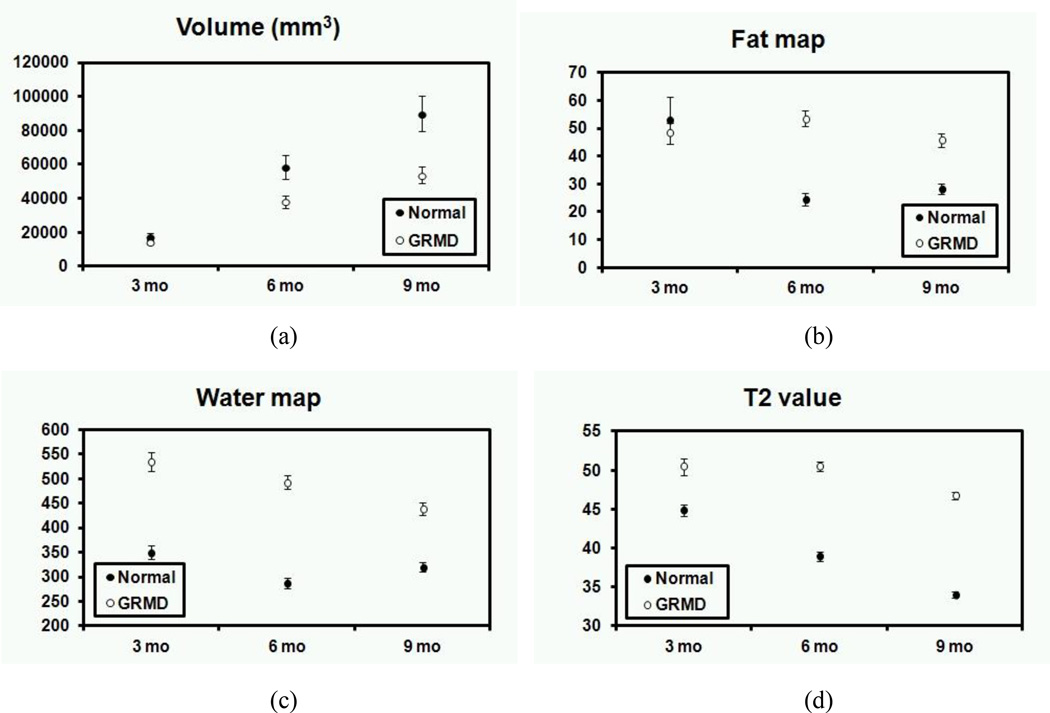
The relationship between age and the average value of aveage value of (a) muscle volume (two tailed t-test assuming unequal variance for all the data of normal and GRMD dogs, p = 0.002), (b) fat map (p = 0.002), (c) water map (p < 0.001), and (d) T2 value (p < 0.001) for normal and GRMD dogs with standard errors.
Unlike the limited mid-femur range segmentations, this straightforward interpolation scheme allowed us to compute a set of MRI biomarkers, incorporating intensity information across most of the entire muscle’s length.
We created a reference dataset to assess the semi-automated segmentation method. ZF manually delineated the full muscles of CS, RF, and ST in the MRI scans of two normal dogs and two GRMD dogs. We used the biomarkers (Section II.2.3.) calculated in the manually segmented muscles as the reference standard. We employed the percent difference ratio D [Eq. (2)] of all biomarkers calculated in the evaluated muscle segmentation (Be) and reference muscle segmentation (Br, manual muscle segmentation) as the performance metric to measure agreement between our biomarker quantification tool and the reference standard.
| 2 |
II.2.2. Pre-processing
The pre-processing was divided into five steps: (1) intensity inhomogeneity correction, (2) calculation of T2 value map, (3) registration of T2w, T2fs, and T2 value maps, (4) intensity calibration of T2w and T2fs, and (5) calculation of MRI biomarker maps.
II.2.2.1. Intensity inhomogeneity correction
Intensity inhomogeneity refers to the slow, non-anatomic intensity variations of the same tissue in a MRI image, typically caused by B1 inhomogeneity, receiver coil non-uniformity or static field inhomogeneity [22]. We used the N4 algorithm [23] [24] to correct for such intensity inhomogeneities. N4 aims at correcting intensity inhomogeneity by seeking the multiplicative bias field that maximizes the high frequency content in the intensity histogram.
II.2.2.2. Calculation of T2 value map
We calculated T2 value map on a voxel-by-voxel basis for the proximal pelvic limbs of dogs in the transverse plane of T2FIT by fitting an exponential decay curve to the signal intensity of the corresponding voxels using a linear-least-squares curve-fitting algorithm [25]. In T2FIT, the intensity of a voxel as a function of echo time is fit to a monoexponential function [Eq. (1)].
| 1 |
where i is an index corresponding to the ith TE of the T2FIT and Ii(x,y,z) is the image intensity for the voxel (x,y,z) in the ith TE of the T2FIT. The monomial coefficient from the linear-least-square curve fit was weighted inversely and assigned to each voxel of T2 value map as the final T2 value map value. The dynamic range of the T2 value map was 16 bits.
The central slice of a T2 value map obtained from the T2FIT is shown in Fig. 1 (c). T2 value maps are sensitive to signal from both water and fat tissue. Thus, T2 value maps capture changes in both fat and water content within muscles, such as those occurring due to disease progression. Furthermore, the T2 value maps are quantitative measurements (in ms) and not affected by the MR signal inhomogeneity. Thus, they can be directly employed for intra and inter-subject comparisons.
II.2.2.3. Image registration
Due to the possible presence of motion during the MRI acquisition, we co-registered the T2fs and T2 value maps to the T2w via standard rigid transformation using three translation parameters and three rotation parameters.
In the study presented in this paper, we used T2w as the fixed image as it provides better signal than the T2fs and higher resolution than the T2 value maps. We employed a mutual information metric [26] based implementation available within the Insight Segmentation and Registration Toolkit (ITK, www.itk.org) to register T2fs and T2 value maps to T2w for each MRI scan.
II.2.2.4. Intensity calibration
In contrast to the T2 value maps, T2w and T2fs are not inherently calibrated, so their intensity values cannot be directly compared across MRI scans from different subjects or even when acquired from the same subject at different times. To overcome this limitation, we calibrated the T2w and T2fs via an intensity-rescaling technique based on the mean intensity observed within subcutaneous fat regions. These fat regions were assumed to be composed of nearly pure fat in all dogs (independent of diagnosis). Because the fat signal is pre-saturated in T2fs, fat regions show very low intensity in T2fs but exhibit high intensity in T2w. For this calibration procedure, we first manually identified a subcutaneous fat region in three contiguous transverse slices at mid-thigh of the proximal pelvic limb in the registered T2w and T2fs. The mean intensity of the subcutaneous fat region in calibrated T2w was then (arbitrarily) set to 1000 and the one in calibrated T2fs was set to 100. Thus, the intensities Ic of the calibrated T2w or T2fs were defined as:
Ic(x, y, z) = 900 / (m1 – m2) × I (x, y, z) + (100 × m1 – 1000 × m2) / (m1 – m2)
where I (x, y, z) is the intensity at voxel (x, y, z) in the non-calibrated T2w or T2fs and m1 and m2 are the mean intensities of the subcutaneous fat regions in non-calibrated T2w and T2fs, respectively.
II.2.2.5. Calculation of MRI biomarker maps
To evaluate the importance of the fat and water signals in the MRI images, we first quantified the loss in intensity between intensity calibrated T2w and T2fs to compute an estimate of the fat signal in each voxel and thus create a fat map (FM):
FM (x, y, z) = IT2w (x, y, z) – IT2fs (x, y, z)
Because the fat signals were suppressed in the T2fs, the water signal in each voxel was simply estimated from the intensity of the voxel in the intensity calibrated T2fs image, allowing us to create a water map (WM) by
WM (x, y, z) = IT2FS (x, y, z)
IT2w (x, y, z) and IT2fs(x, y, z) are the intensities of voxel (x, y, z) in the denoised and registered T2w and T2fs. Denoising was established via standard anisotropic diffusion smoothing [27]. Anisotropic diffusion is a technique to reduce image noise without removing significant edges, lines or other details that are important for the interpretation of the images. The performance of anisotropic diffusion smoothing is controlled by three parameters, the number of iterations, time step, and conductance, where the number of iterations and the time step are parameters used for controlling the smoothness of the image and conductance is used for controlling the edge contrast in the smoothed image. Without noise reduction, significant image noise may considerably impact the WM and FM map computation and could lead to incorrect water and fat signal related biomarkers as the FM is calculated via direct differences on the T2w and the T2fs images. However, the smoothing process may have side effects, such as the fusion of fat and muscle signals at the boundary of the subcutaneous fat and muscle regions. This problem can be reduced by using a relatively high conductance to enhance the contrast of different tissues in the smoothed image. In this study, we empirically selected the parameters for anisotropic diffusion smoothing by visually observing the smoothing effect in a subset of MRI scans (3 normal and 3 GRMD scans). The number of iterations was set to 5, time step was set to 0.625 s, and conductance was set to 1.
II.2.2.6. Statistical texture biomarker
Statistical texture analysis has been widely used in medical imaging applications to characterize texture patterns within the image [28, 29, 30, 31]. In muscular dystrophy MRI studies, dystrophic muscles usually present a unique texture pattern [32]. Thus, the statistical texture analysis technique has great potential to discriminate normal from dystrophic muscle on MRI [32,33].
In this study, we automatically generated contiguous quadrilateral volumes of interest (VOIs) with a fixed size (empirically chosen at 31 × 31 × 31 mm, as the average diameter of the proximal limb of golden retriever dogs is approximately 30 mm. Furthermore, we intentionally chose an odd number so that we could unambiguously determine the center voxel of each VOI. The distance between adjacent VOIs is 50% of the VOI size, resulting in 50% overlap between adjacent VOIs. Areas of the VOI outside the segmented muscle were ignored when calculating texture features.
II.2.2.6.1. Histogram texture features
We employed standard intensity histogram-based first-order texture features and run-length matrix-based high-order texture features to analyze the muscle MR images [34]. We calculated the histogram texture features on the (intensity calibrated) T2w image as it provides better signal than the T2fs and higher resolution than the T2 value maps. Given the expected difference in MR appearance, we calculated standard deviation (SD) of the T2w intensities [Eq. (3)] for each muscle in each VOI. We then calculated the average of the SD in all VOIs for each muscle. We hypothesized that T2w standard deviation would be higher in GRMD versus normal dogs and that it would change significantly across time in GRMD dogs but not in normal dogs.
| 3 |
where I(x) is the intensity of voxel x, Ī is the mean intensity of a muscle in a VOI, and n is the number of voxels of a muscle in a VOI.
II.2.2.6.2. Three-dimensional run-length matrix features
Compared to the histogram texture features, the run-length matrix features take into account both the intensity information and spatial distribution of voxels, and thus nicely complement the histogram texture features.
Run-length Matrix
A gray-level ‘run’ is defined as a set of consecutive voxels of similar intensity level in a given direction within a predefined similarity range, commonly chosen at 1.5% percent of the full intensity range (as determined by 64 run-length matrix bins). An element of a standard run-length matrix measures the number of occurrences of a run with a specific length and specific intensity level in a given direction. However, such direction-specific run-lengths lack the power to characterize the 3D extension of the texture features in 3D MRI scans, because they only assess the intensity distribution in a single direction. In this study, we developed an extended method to directly measure 3D run-length matrix-based texture features.
First, to reduce the influence of voxels with very bright intensities caused by image noise, the top 5 % of the intensities were set to the value of the 95th percentile. We then employed a grayscale 3D connected-component labeling technique to identify 3D run-length components with similar intensity for each muscle in each VOI. We chose a similarity range of 1.5 percent of the cropped intensity range. This allowed us to establish 3D run-length matrices for each VOI in each muscle, which assess run-lengths via 3D volumetric runs rather than the conventional, single-direction length measurements.
Finally, we calculated five commonly used run-length matrix features, including short run emphasis (SRE), long run emphasis (LRE), gray-level non-uniformity (GLN), run-length non-uniformity (RLN), and run percentage (RP) [34], for each muscle in each VOI. We then calculated the average of each feature in all VOIs for each muscle.
II.2.3. Biomarker analysis
Our quantification tool was designed to measure a set of MR parameters and their changes across time. Because muscle atrophy/hypertrophy, inflammation, and fat infiltration are characteristic histopathological features of DMD / GRMD, MRI derived biomarkers of interest within muscle regions included three categories of biomarkers: (1) muscle volume, (2) mean intensities in fat percentage (FP), water percentage (WP), and T2 maps, and (3) statistical texture features. These biomarkers were used to longitudinally and cross-sectionally assess and compare the structural tissue changes of GRMD dogs and normal littermates. We performed the two tailed t-test assuming unequal variances to evaluate the group significant differences (p-value < 0.05 was considered statistically significant) between normal and GRMD dogs.
II.2.3.1. Muscle volume
Muscle volume was calculated straightforwardly from the muscle segmentation via voxel counting. Based on our previous studies [6, 18], we hypothesized that (1) most of the muscles of GRMD dogs would have smaller volumes than those of normal littermates; a notable exception is the CS, which is spared and may even undergo hypertrophy [18] followed by atrophy, with increased endomysial space due to fat and connective tissue; (2) muscle volumes of normal dogs would increase with age, whereas muscle volumes of most GRMD muscles would not increase with age or would do so at a lesser rate than normal dogs, again with the exception of the CS.
II.2.3.2. Intensity map based biomarkers
For the biomarkers derived from FM, WM, and T2 value map, we computed the arithmetic mean after the application of an erosion operation with a circular kernel (radius = 3 voxels) on the muscle segmentation to reduce the influence of partial-volume voxels. We hypothesized that (1) muscles of GRMD dogs would contain more fat than normal dogs and thus have higher mean intensities in FM and T2 value map than normal dogs; (2) the mean intensities in all the biomarker maps of normal dogs would not change significantly with age, whereas the mean intensities of GRMD dogs would vary among muscles, with those for some muscles increasing and others potentially decreasing with age.
II.2.3.3. Statistics of texture analysis based biomarkers
For the statistical texture based biomarkers, we also computed the arithmetic mean on the eroded muscle segmentations. For these biomarkers, we hypothesized that muscles of GRMD dogs would contain more fat and inflammation than normal dogs and thus (1) GRMD dogs would have higher SRE, GLN, RLN, and RP of run-length matrix than normal dogs; (2) GRMD dogs would have lower LRE of run-length matrix than normal dogs; and (3) the run-length matrix features of normal dogs would not change significantly with age, whereas those of GRMD dogs would significantly change with age.
III. RESULTS
Figure 3 shows the mean value with standard error of the biomarkers of each muscle in normal and GRMD dogs at 3-, 6-, and 9-months of age, respectively. Normal muscles had consistently larger volumes (two tailed t-test assuming unequal variance for all the data of normal and GRMD dogs, p = 0.002) and had lower T2 value map (p < 0.001), FM (p = 0.002), and WM (p < 0.001) values when compared to GRMD dogs at most of the age points, except for FM at 3-months of age. Notably, consistent with our original hypothesis, the GRMD CS had a larger volume than that of normal dogs. For normal dogs, the mean intensities in T2 value map showed a consistent decline with age, whereas the mean intensities in FM and WM did not change. For GRMD dogs, the mean intensities in WM showed a decline with age, whereas the mean intensities in FM did not change; the mean intensities in T2 value map also showed a significant decline between 6- and 9-months of age, whereas the mean intensities in T2 value map did not change over this period.
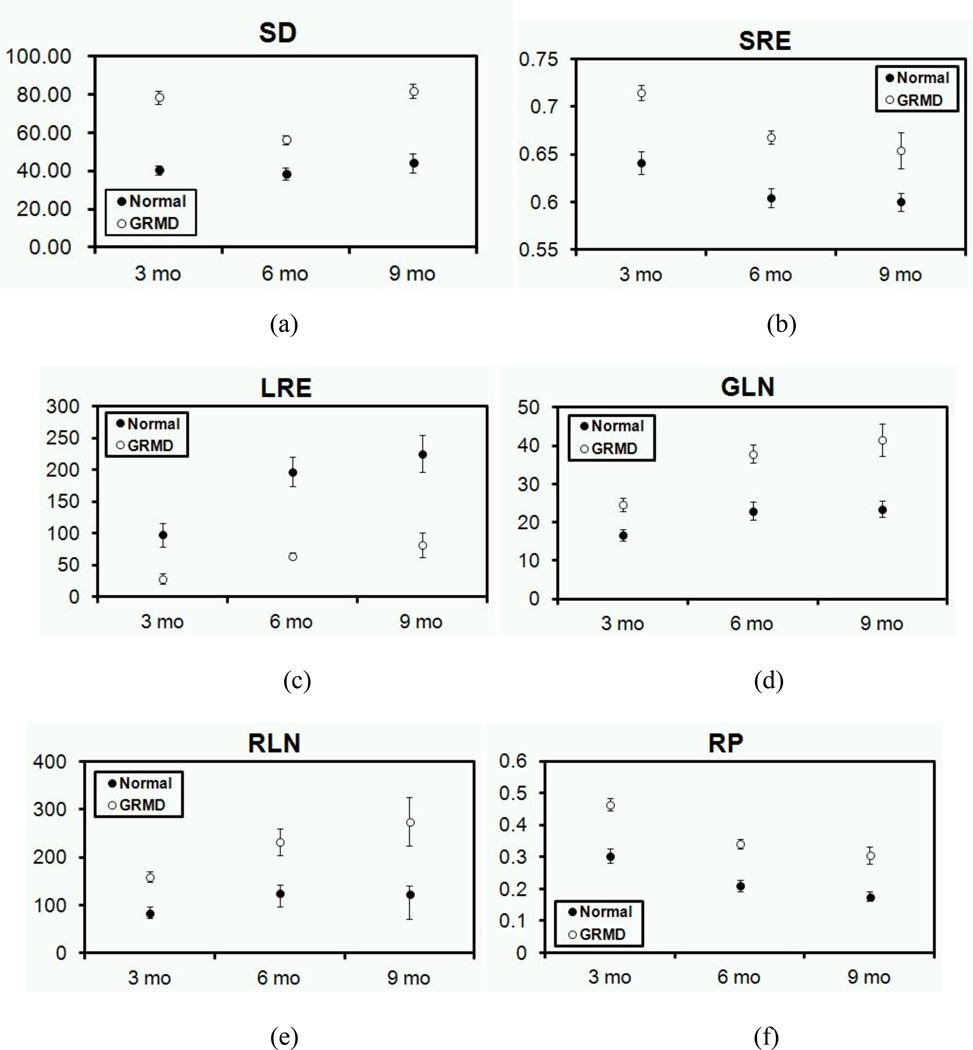
Figure 5 (a) shows the average value of SD of histogram with standard error of muscles in T2w of normal and GRMD dogs at 3-, 6-, and 9-months of age. Overall, the SD differed significantly between normal and GRMD (p < 0.001, two tailed t-test assuming unequal variance).
Figures 5 (b)-(f) show the run-length matrix texture features with standard error for each muscle in normal and GRMD dogs at 3-, 6-, and 9-months of age, respectively. Overall, the SRE (p < 0.001, two tailed t-test assuming unequal variance), GLN (p < 0.001), RLN (p < 0.001), and RP (p < 0.001) of GRMD muscles were higher while the LRE (p < 0.001) was lower, when compared to normal muscles at each age. With respect to longitudinal changes across time, the change trend for GLN and RP did not differ significantly between normal and GRMD muscles with age (Fig. 5 b-f), whereas, the SRE, LRE, and RLN differed between normal and GRMD muscles with age. In particular, the LRE of GRMD muscles showed a significant increment with age, whereas the LRE of normal muscles did not change.
Muscle segmentation is an essential step for the quantification tool. Table 2 shows the imaging-map-based biomarkers and volume performance calculated from the interpolation-based muscle segmentation and reference muscle volume of two normal dogs and two GRMD dogs in CS, RF, and ST. The average percent error ratios over the mean intensities of FM, WM, and T2 value map were quite low at 1.28%, 0.73%, and 0.40%, respectively. We also calculated the biomarker performance from the limited-range muscle segmentation (only five consecutive mid-femur slices) (Table 2). The average error ratios over the mean intensities of FM, WM, and T2 value map were considerably higher at 28.12%, 2.70%, and 2.95%, respectively. Overall, the errors were an order of magnitude larger in the limited-range muscle segmentations than in the interpolated full muscle segmentations. By use of the paired two tailed t-test, we found that the errors were significantly higher in the limited-range muscle segmentation than in the interpolated full muscle segmentation (p = 0.001, p = 0.04, p = 0.04 for fat map, water map, and T2 value map, respectively).
Table 2.
The average difference of the biomarkers calculated from the interpolation-based muscle segmentation and the reference manual full muscle segmentation and the average difference of the biomarekers from the manual muscle segmentation in a limited range (five slices) and reference muscle segmentation of two normal dogs and two GRMD dogs in cranial sartorius (CS), femoris (RF), and semitendinosus (ST).
| Interpolation based segmentation |
Manual segmentation in limited range |
|||||
|---|---|---|---|---|---|---|
| Fat Map | Water Map | T2 Value | Fat Map | Water Map | T2 Value | |
| CS | 1.14% | 1.41% | 0.98% | 35.74% | 4.08% | 6.14% |
| RF | 1.84% | 0.61% | 0.05% | 29.24% | 1.10% | 1.65% |
| ST | 0.84% | 0.16% | 0.18% | 19.38% | 2.91% | 1.06% |
| Average | 1.28% | 0.73% | 0.40% | 28.12% | 2.70% | 2.95% |
In order to test whether the above results may be true in GRMD only, we investigated the relative error-rates of biomarkers in GRMD (RGRMD) versus normal (Rnormal) dogs by calculating the ratio between RGRMD and Rnormal, i.e., RGRMD / Rnormal. The ratios for FM, WM and T2 value of the segmentation in mid-femur slices were 0.72, 5.85, and 1.10, respectively; the ratios for fat map, water map and T2 value of the interpolation-based whole muscle segmentation were 1.06, 4.10, and 0.98, respectively. These results do not show obvious evidence to support that the elevated error rates are present only in GRMD. Furthermore, we also calculated the difference of biomarkers derived from interpolation-based full muscle segmentation and the limited range muscle segmentation over all normal dog scans in our study for cranial sartorius (CS), femoris (RF), and semitendinosus (ST). The result shows a comparable error levels (21.27%, 4.80%, and 2.94% for fat map, water map, and T2 value, respectively) to the ones shown in Table 2 (28.12%, 2.70%, and 2.95% for fat map, water map, and T2 value, respectively). While there is no ground truth full muscle segmentation available for all these scans, it can safely be said that the interpolation-based full muscle and mid-femur limited muscle biomarkers show large differences. Given that the interpolation-based full muscle segmentation showed clearly better performance when compared to cases with ground truth, this indicates that limiting the segmentation to a small range introduces significant variation/errors in MR biomarker measurements. Thus, the interpolation-based whole muscle segmentation scheme allowed us to compute fairly stable and accurate values for the biomarkers, whereas the limited range segmentation based measures show higher variance and errors. .
We also investigated the segmentation inter-rater variability between ZF and JW on the semi-automatic segmentation scheme (Table 3). The comparison results showed that the average performance across raters over mean intensities in FM, WM, and T2 value map were appropriately low at 6.36%, 5.28%, and 1.21%, respectively. The inter-rater variability for normal and GRMD dogs showed similar performance (p > 0.99, paired two tailed t-test). This relatively small inter-rater difference indicates that our biomarker quantification tool is reliable and stable.
Table 3.
Inter-rater variability: biomarker differences of cranial sartorius (CS), rectus femoris (RF), semitendinosus (ST), biceps femoris (BF), gracilis (GR), and adductor magnus (AD) between ZF and JW’s segmentation.
| Fat Map | Water Map | T2 Value | |
|---|---|---|---|
| CS | 7.63% | 5.05% | 4.22% |
| RF | 8.47% | 4.11% | 0.78% |
| ST | 5.56% | 0.69% | 0.66% |
| BF | 2.48% | 0.42% | 0.41% |
| GR | 9.74% | 1.03% | 1.04% |
| AD | 4.26% | 0.28% | 0.14% |
| Average | 6.36% | 1.93% | 1.21% |
IV.DISCUSSION
Muscle segmentation plays an important role in DMD MRI biomarker quantification. The results from our study demonstrated that MRI biomarkers computed from the muscle regions determined by semi-automatic full muscle segmentation have a lower error rate than biomakers from regions determined by the standard limited range manual muscle segmentations (5 mid-femur slices) [9, 10, 35]. For the muscle volume, one could expect that full muscle segmentation would provide more accurate measurement results than in limited range segmentation. The magnitude of the error observed in the other biomarkers was more surprising. This result is likely due to the inhomogeneous nature of muscle lesions in DMD / GRMD subjects. Furthermore, it strongly highlights that the whole or at least a large segment of the muscle should ideally be segmented to miminize sampling error, while excluding irrelevant surrounding tissues. Our experimental results showed that a straightforward interpolation scheme allowed computation of a fairly stable and accurate set of MRI biomarkers. Although the muscles of normal dogs may contain less heterogeneity, the quantification at mid-femur of normal dogs still may not be optimal, because not all the muscles are growing along the entire femur bone. For instance, the mid-femoral point is generally the distal of gracilis. Because the sectional area of the distal is small, the quantification would be easily affected by the human errors (muscle segmentation) and image noises. Therefore, whole muscle volume based biomarker quantification is highly recommended for the muscular dystrophic studies.
The overall computational scheme of this tool is composed of “automatic” and “interactive” processing steps. The automatic processing part consists of the pre-processing and biomarker calculation. The average automatic processing time is about 15 minutes / scan (CentOS 5.6 Linux, Intel 64 bit 1.66GHz quad-core, 128GB RAM). The interactive processing part consists of the muscle segmentation and identification of subcutaneous fat regions, in which muscle segmentation is the most labor-intensive task. Manual full muscle segmentation for each muscle would require an average of 60 minutes, whereas the semi-automatic muscle segmentation for each muscle, including the manual delineation of muscle regions in every 5th slice and the automatic segmentation interpolation, averaged less than 15 minutes. Our experimental results indicate that the target segmentation accuracy requirements of the automatic muscle segmentation are relatively relaxed, as the GRMD biomarkers could be estimated fairly stably in the presence of smaller segmentation errors (as were likely present in these semi-automatic segmentations). In our ongoing research, we are investigating the use of fully automatic multi-atlas-based muscle segmentation schemes.
While fat ultimately increases histologically in both DMD and GRMD, the relatively young age of these dogs and short 6–9 month time course of the study may not have been sufficient to capture more chronic fatty infiltration. Another potential reason is the inherently large variability of the fat measures in images acquired with the selected fat suppression-based MR protocols. Ultimately spectroscopic methods such as proton MR spectroscopy may be more sensitive to these early, small changes in lipid levels at low fat fractions.
The T2 value maps covered the mid-femur section of the proximal pelvic limbs (Section II. 1). As described above, MRI biomarkers calculated from full muscle segmentation show considerably less error than from limited range muscle segmentations. Thus, a T2 value map should ideally cover the entire length of a muscle or at least a large segment of the muscle to maximize the number of pixels included in the calculation. As a result of experiments presented here, we modified our imaging protocol in ongoing studies such that the T2FIT sequence (used for calculating T2 value map) covers the full proximal pelvic limb, while adjusting the slice thickness and number of echoes to keep a comparable scan time (TR 3000 ms / TE 10, 20, 30, 40, 50 ms; slice thickness 7 mm; 30 slices per echo). We have tested the modified imaging protocol on two GRMD dogs (see Table 4 for T2 value based biomarker results). The average biomarker difference between limited and full muscle segmentations for T2 value map was 15.14% (p < 0.0001, paired two tailed t-test). The smallest muscles had the greatest difference between the two methods. This suggests that the T2-value-based biomarker should also be assessed within the full muscle or include enough pixels within a given muscle to minimize the effects of sampling error.
Table 4.
The average T2 value map-based biomarkers of cranial sartorius (CS), rectus femoris (RF), semitendinosus (ST), biceps femoris (BF), gracilis (GR), and adductor magnus (AD) calculated in the entire proximal pelvic limb T2 value map and mid-femur T2 value map.
| T2 Value |
||
|---|---|---|
| Entire proximal limb T2 value map |
Mid-femur T2 value map |
|
| CS | 60.62 | 45.66 |
| RF | 55.94 | 46.73 |
| ST | 53.90 | 46.48 |
| BP | 55.00 | 48.14 |
| GR | 53.38 | 46.72 |
| AD | 53.08 | 47.26 |
| Average | 55.32 | 46.83 |
| Difference | 15.14% | |
We observed that T2w and T2fs images generally contain higher intensity inhomogeneities than the T2 value maps. Although we applied the N4 algorithm to correct intensities, the strong intensity inhomogeneity in T2w and T2fs could not be completely eliminated. Furthermore, the T2w and T2fs images were calibrated using manually- identified subcutaneous fat. Because the manual identification of subcutaneous fat is subject to human rater error, the intensity calibration may not be fully accurate. It was, therefore, not surprising to find that the inherently intensity calibrated T2 value map based biomarkers were more reliable than those of our fat and water map based biomarkers.
Although the statistical texture analysis technique has been used in many imaging applications of muscle diseases, to our knowledge, it has not been widely used for studies of muscular dystrophy. One characteristic histopathologic feature of DMD and GRMD is the patchy, inhomogeneous and differential involvement of lesions within even the same muscle. The commonly used mean intensity marker will not be sufficiently sensitive to capture this characteristic inhomogeneity. In this study, we employed histogram features and adapted 3D run-length matrix features to investigate the difference in MR appearance of normal and GRMD muscles cross-sectionally and longitudinally across time. Because of fatty change , one would expect GRMD muscles to have coarser texture than normal muscles. Therefore, the affected muslces would have more short runs, less long runs, and a higher total number of runs than normal muscles. Furthermore, because muscle lesions are irregular, the uniformity in the distribution of runs over grey levels and over the lengths of runs would generally be higher in affected versus normal muscles. For these reasons, the run-length matrix features distinguished GRMD from normal muscles. Overall, the statistical texture biomarkers had advantages in quantifying group differences for the GRMD and normal dogs and longitudinal disease progression in GRMD dogs, suggesting that these biomarkers will be useful in DMD clinical trials.
In this study, we proposed several MRI derived biomarkers and statistical texture analysis based biomarkers. Histopathologic / functional correlation would be required to validate these markers and is ongoing, using functional indices of isometric distal limb force [37] and repeated eccentric contractions; muscle weight; and histopathological measures of myofiber and endomysial space (non-contractile tissue) per unit area [38, 39].. In this paper, we focused on developing a biomarker quantification tool. Thus, validation of the proposed biomarkers using histopathologic / functional findings is outside the paper’s scope and will be described separately..
As a final note, we plan to disseminate this biomarker quantification tool publicly as open source on NIH’s Neuro-Imaging Toolkit and Resource Clearinghouse (NITRC, http://www.nitrc.org/). We hope this quantification tool will constitute a valuable resource for the DMD research community.
V.CONCLUSIONS
In this study, we developed a semi-automatic MRI biomarker quantification tool incorporating enhanced histogram based statistical features and novel 3D run-length matrix based statistical texture biomarkers in the GRMD model of DMD. Our quantification tool showed clear group differences for the affected and normal dogs and longitudinal disease progression in affected dogs, albeit with operator input for the segmentation of muscles and subcutaneous fat regions. Our quantification tool provided useful MRI biomarkers for GRMD. However, it also had some limitations: (i) the muscle segmentation method was not fully automated and required human rater input; (ii) the subcutaneous fat regions were identified manually for the intensity calibration; and (iii) the intensity inhomogeneous correction was imperfect. In our ongoing research, we are focusing on development of fully automatic muscle segmentation techniques to promote efficiency and reliability of our scheme.
Fig. 2.
(a) Manual muscle segmentation of ST = semitendinosus (yellow), RF = rectus femoris (red), CS = cranial sartorius (light blue), AD = adductor magnus (blue), GR = gracilis (pink), and BF = biceps femoris (brown) in a transverse view and (b) linearly interpolated muscle volumes in 3D view.
Fig. 4.
The relationship between age and the average value of each histogram and run-length matrix texture feature for normal and GRMD dogs with standard errors. (a) standard deviation of histogram (two tailed t-test assuming unequal variance for all the data of normal and GRMD dogs, p < 0.001), (b) SRE (p < 0.001), (c) LRE (p < 0.001), (d) GLN (p < 0.001), (e) RLN (p < 0.001), and (f) RP (p < 0.001).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant Nos. R42 NS059095-03 (NINDS) (Styner), P30-HD003110-41 (NICHD) (Styner) and 1U24NS059696-01A1 (NINDS) (Kornegay), the Muscular Dystrophy Association (Kornegay), Wellstone center for Muscular Dystrophy Research [USPHS (NIAMS) 1U54AR056953-01], The North Carolina Translational and Clinical Sciences (NC TraCS) Institute [Tracs50K (50KR71104)] and UNC Intellectual and Developmental Disabilities Research Center. The authors thank Weili Lin, and Kathleen Wilber for their help in acquisition of MRI scans and their helpful discussions and Janet and Dan Bogan and Jennifer Dow for technical assistance in managing the dogs.
REFERENCES
- 1.Moser H. Duchenne muscular dystrophy: pathogenetic aspects and genetic prevention. Hum Genet. 1984;66:17–40. doi: 10.1007/BF00275183. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 4.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–92. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaschen FP, Hoffman EP, Gorospe JR, Uhl EW, Senior DF, Cardinet GH, 3rd, Pearce LK. Dystrophin deficiency causes lethal muscle hypertrophy in cats. J Neurol Sci. 1992;110:149–159. doi: 10.1016/0022-510x(92)90022-d. [DOI] [PubMed] [Google Scholar]
- 6.Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle Nerve. 1988;11:1056–1064. doi: 10.1002/mus.880111008. [DOI] [PubMed] [Google Scholar]
- 7.Cooper BJ, Winand NJ, Stedman H, Valentine BA, Hoffman EP, Kunkel LM, Scott M-O, Fischbeck KH, Kornegay JN, Avery RJ, Williams JR, Schmickel RD, Sylvester JE. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334:154–156. doi: 10.1038/334154a0. [DOI] [PubMed] [Google Scholar]
- 8.Kornegay JN, Bogan JR, Bogan DJ, Childers MK, Li J, Nghiem P, Detwiler DA, Larsen CA, Grange RW, Bhavaraju-Sanka RK, Tou S, Keene BP, Howard JF, Jr, Wang J, Fan Z, Schatzberg SJ, Styner MA, Flanigan KM, Xiao X, Hoffman EP. Canine models of Duchenne muscular dystrophy and their use in therapeutic strategies. Mamm Genome. 2012;23:85–108. doi: 10.1007/s00335-011-9382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi M, Nakamura A, Hasegawa D, Fujita M, Orima H, Takeda S. Evaluation of dystrophic dog pathology by fat-suppressed T2-weighted imaging. Muscle Nerve. 2009;40:815–826. doi: 10.1002/mus.21384. [DOI] [PubMed] [Google Scholar]
- 10.Thibaud JL, Monnet A, Bertoldi D, Barthelemy I, Blot S, Carlier PG. Characterization of dystrophic muscle in golden retriever muscular dystrophy dogs by nuclear magnetic resonance imaging. Neuromuscul Disord. 2007;17:575–584. doi: 10.1016/j.nmd.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S, Hoffman E. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamminen AE. Magnetic resonance imaging of primary skeletal muscle diseases: patterns of distribution and severity of involvement. Br J Radiol. 1990;63:946–950. doi: 10.1259/0007-1285-63-756-946. [DOI] [PubMed] [Google Scholar]
- 13.Lovitt S, Moore SL, Marden FA. The use of MRI in the evaluation of myopathy. Clin Neurophysiol. 2006;117:486–495. doi: 10.1016/j.clinph.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Marden FA, Connolly AM, Siegel MJ, Rubin DA. Compositional analysis of muscle in boys with Duchenne muscular dystrophy using MR imaging. Skeletal Radiol. 2005;34:140–148. doi: 10.1007/s00256-004-0825-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu GC, Jong YJ, Chiang CH, Jaw TS. Duchenne muscular dystrophy: MR grading system with functional correlation. Radiology. 1993;186:475–480. doi: 10.1148/radiology.186.2.8421754. [DOI] [PubMed] [Google Scholar]
- 16.Karpati G, Ajdukovic D, Arnold D, Gledhill RB, Guttmann R, Holland P, Koch PA, Shoubridge E, Spence D, Vanasse M, Watters GV, Abrahamowicz M, Duff C, Worton RG. Myoblast transfer in Duchenne muscular dystrophy. Ann Neurol. 1993;34:8–17. doi: 10.1002/ana.410340105. [DOI] [PubMed] [Google Scholar]
- 17.Miller RG, Sharma KR, Pavlath GK, Gussoni E, Mynhier M, Lanctot AM, Greco CM, Steinman L, Blau HM. Myoblast implantation in Duchenne muscular dystrophy: The San Francisco Study. Muscle Nerve. 1997;20:469–478. doi: 10.1002/(sici)1097-4598(199704)20:4<469::aid-mus10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 18.Kornegay JN, Cundiff DD, Bogan DJ, Bogan JR, Okamura CS. The cranial sartorius muscle undergoes true hypertrophy in dogs with golden retriever muscular dystrophy. Neuromuscul Disord. 2003;13:493–500. doi: 10.1016/s0960-8966(03)00025-7. [DOI] [PubMed] [Google Scholar]
- 19.Childers MK, Okamura CS, Bogan DJ, Bogan JR, Sullivan MJ, Kornegay JN. Myofiber injury and regeneration in a canine homologue of Duchenne muscular dystrophy. Am J Phys Med Rehab. 2001;80:175–181. doi: 10.1097/00002060-200103000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Fan Z, Kornegay J, Styner M. MRI-based quantification of Duchenne muscular dystrophy in a canine model. Proc. SPIE Med. Imaging. 2011;7965:79650G1–79650G9. [Google Scholar]
- 21.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Hou Z. A review on MR image intensity inhomogeneity correction. Proc Int J Biomed Imaging. 2006;2006 doi: 10.1155/IJBI/2006/49515. 49515-1-49515-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 25.Kim HK, Laor T, Horn PS, Racadio JM, Wong B, Dardzinski BJ. T2 mapping in Duchenne muscular dystrophy: distribution of disease activity and correlation with clinical assessments. Radiology. 2010;255:899–908. doi: 10.1148/radiol.10091547. [DOI] [PubMed] [Google Scholar]
- 26.Pluim JP, Maintz JB, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans. Med. Imaging. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 27.Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell. 1990;12:629–639. [Google Scholar]
- 28.Chuah TK, Poh CL, Sheah K. Quantitative texture analysis of MRI images for detection of cartilage-related bone marrow edema. Proc IEEE Eng Med Biol Soc. 2011;2011:5112–5115. doi: 10.1109/IEMBS.2011.6091266. [DOI] [PubMed] [Google Scholar]
- 29.Mayerhoefer ME, Schima W, Trattnig S, Pinker K, Berger-Kulemann V, Ba-Ssalamah A. Texture-based classification of focal liver lesions on MRI at 3.0 Tesla: a feasibility study in cysts and hemangiomas. J Magn Reson Imaging. 2010;32:352–359. doi: 10.1002/jmri.22268. [DOI] [PubMed] [Google Scholar]
- 30.Holli K, Lperi AL, Harrison L, Luukkaala T, Toivonen T, Ryymin P, Dastidar P, Soimakallio S, Eskola H. Characterization of breast cancer types by texture analysis of magnetic resonance images. Acad Radiol. 2010;17:135–141. doi: 10.1016/j.acra.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Yu C, Jiang G, Liu W, Tong L. 3D texture analysis on MRI images of Alzheimer’s disease. Brain Imaging Behav. 2012;6:61–69. doi: 10.1007/s11682-011-9142-3. [DOI] [PubMed] [Google Scholar]
- 32.Herlidou S, Rolland Y, Bansard JY, Le Rumeur E, De Certaines JD. Comparison of automated and visual texture analysis in MRI: characterization of normal and diseased skeletal muscle. Magn Reson Imaging. 1999;17:1393–1397. doi: 10.1016/s0730-725x(99)00066-1. [DOI] [PubMed] [Google Scholar]
- 33.Mahmoud-Ghoneim D, Cherel Y, Lemaire L, De Certaines JD, Maniere A. Texture analysis of magnetic resonance images of rat muscles during atrophy and regeneration. Magn Reson Imaging. 2006;24:167–171. doi: 10.1016/j.mri.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Galloway M. Texture analysis using gray level run lengths. Comput Graph Imaging Process. 1975;4:172–179. [Google Scholar]
- 35.Mathur S, Lott DJ, Senesac C. Age-related differences in lower-limb muscle cross-sectional area and torque production in boys with Duchenne muscular dystrophy. Arch Phys Med Rehabil. 2010;91:1051–1058. doi: 10.1016/j.apmr.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J. MICCAI Workshop. 2012 [Google Scholar]
- 37.Tegler CJ, Grange RW, Bogan DJ, Markert CD, Case D, Kornegay JN, Childers MK. Eccentric contractions induce rapid isometric torque drop in dystrophic-deficient dogs. Muscle Nerve. 2010;42:130–132. doi: 10.1002/mus.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Childers MK, Bogan JR, Bogan DJ, Greiner H, Staley J, Holder M, Grange RW, Kornegay JN. Chronic administration of a leupeptin-derived calpain inhibitor fails to ameliorate severe muscle pathology in a canine model of Duchenne muscular dystrophy. Front Pharmacol. 2011;2:89. doi: 10.3389/fphar.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophoton Internat. 2004;11:36–42. [Google Scholar]