Abstract
With different approaches to finding prognostic or diagnostic biomarkers for Alzheimer’s disease (AD), many studies pursue only brief lists of biomarkers or disease specific pathways, potentially dismissing information from groups of correlated biomarkers. Using a novel Bayesian graphical network method, with data from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging, the aim of this study was to assess the biological connectivity between AD associated blood-based proteins. Briefly, three groups of protein markers (18, 37, and 48 proteins, respectively) were assessed for the posterior probability of biological connection both within and between clinical classifications. Clinical classification was defined in four groups: high performance healthy controls (hpHC), healthy controls (HC), participants with mild cognitive impairment (MCI), and participants with AD. Using the smaller group of proteins, posterior probabilities of network similarity between clinical classifications were very high, indicating no difference in biological connections between groups. Increasing the number of proteins increased the capacity to separate both hpHC and HC apart from the AD group (0 for complete separation, 1 for complete similarity), with posterior probabilities shifting from 0.89 for the 18 protein group, through to 0.54 for the 37 protein group, and finally 0.28 for the 48 protein group. Using this approach, we identified beta-2 microglobulin (β2M) as a potential master regulator of multiple proteins across all classifications, demonstrating that this approach can be used across many data sets to identify novel insights into diseases like AD.
Keywords: Alzheimer’s disease, Bayesian, biomarkers, graphical networks, imputation
INTRODUCTION
The concept that an ideal biomarker should be directly related to disease pathophysiology and be informative of the disease process, even in the very early pre-clinical phase [1], seems unlikely for the complex and often heterogeneous Alzheimer’s disease (AD). It is also pertinent that an efficacious biomarker be non-invasive, easily translatable to routine clinical testing or eventually microfluidic high-throughput population screening and expedient serial monitoring. Despite enormous resources being poured into the search for candidate biomarkers that fit this definition, a consensus is yet to come to fruition. However, peripheral tissues, especially blood fractions have been mined for biomarkers that match at least one or more of the above characteristics.
With the decreasing cost of non-invasive blood-based biomarker screening, it is now likely that a successful biomarker for the early diagnosis of AD will consist of a panel of analytes from a range of ‘panomic’ screening techniques and sample components.
Biomarker screening for AD has elucidated a long list of candidates from various platforms, with insufficient cross-validation. However, the ‘gold standard’ peripheral biomarker for AD that will reliably identify individuals on a path toward AD, or even correlate with promising, but invasive and impractical cerebrospinal fluid (CSF) [2] and positron emission tomography (PET) biomarkers [3], is yet to emerge. Nevertheless, much hope is dedicated to the idea that such a marker does exist in the periphery. Multiple research groups have found a panoply of individual markers using assemblies of statistical methods, methods that are primarily designed to choose the best representative from groups of biomarkers.
Recently a number of approaches to screening large sample data sets have been sought to screen for biomarkers that have diagnostic and prognostic utility [4–16]. However in many cases, dependant upon the volume of data accumulated, and the ‘pan-omic’ approach to sample screening and subsequent data interrogation, biological networks have been uncovered with one or more targets that meet diagnostic or prognostic utility, but the direct relationship to pathology has been unexplained [17–26]. A single analyte (or analyte panel) may be insufficient to allow the researcher to understand how the marker fits in the cascade of disease process, which could lead to novel therapies. For this reason, others have turned to a Bayesian network classifier to integrate diverse data sets, incorporate biological information, and infer/impute missing data from well characterized networks, where all the nodes may not have been initially screened [27–31].
In this study we applied a novel approach for Bayesian inference of multiple graphical networks, using data from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. We assessed the biological networks identified using three biomarker sets, and highlight the importance of biomarker connectivity in understanding biological processes related to disease pathology.
METHODS
Population sample and biomarker selection
Of the total 1,112 participants from the AIBL study at baseline, 659 healthy control (HC), 112 mild cognitive impairment (MCI), and 186 Alzheimer’s disease (AD) subjects with complete data for each of the biomarker panels tested, were selected for analyses. Two neuropsychological composite scores (episodic memory composite score and non-memory composite score [32] as well as the Mini–Mental State Examination (MMSE) were tested as part of the demographic assessment. Biomarkers were selected in three sets; Set A) the top 18 biomarkers from [24], Set B) the top 37 biomarkers as selected using a Linear Models for Microarray Data (LIMMA) analysis with a q-value cut off of 0.0003, and Set C) the top 48 biomarkers selected using a LIMMA analysis with a q-value cut off of 0.05. Biomarker lists and accompanying Venn diagrams are shown in Supplementary Table 1 and Supplementary Figure 1. Biomarkers included both those proteins measured using the Rules Based Medicine (RBM) Human Discovery xMAP® panel [24], and those clinical pathology measures routinely tested as part of the AIBL protocol [33]. Further information regarding sample preparation and processing, including biomarker selection, can be found in [24]. Biomarker data was log transformed and qq-normalized prior to analyses. As an internal validation, we split the HC subject into two groups, high performing HC (hpHC), and normal HC (HC) via an unsupervised mixed modelling approach (using six neuropsychological test scores), resulting in a total of four groups for comparison, hpHC, HC, MCI, and AD.
Statistical methodology
Sample demographics were tabled and compared using χ2 and generalized linear modelling. We then use a graphical model approach, which describes the conditional dependence relationships among random variables, in order to make inference on the protein interaction networks. Specifically we use the approach of [34] to assess the relationships between biomarkers both within and between clinical groups. This Bayesian approach is designed to simultaneously infer multiple undirected networks in situations where some networks may be unrelated, while others may have a similar structure. The proposed approach infers a separate graphical model for each group but allows for shared structures, when supported by the data. Moreover, this approach allows obtaining a measure of relative network similarity across groups. This measure of similarity reflects how appropriate the assumption that the networks for any two groups have common edges is, based on the data for each group. This Bayesian approach was run using the default hyperparameter setting and posterior inference procedure as described in [34].
RESULTS
Population demographics
While both MCI and AD groups had significantly older participants than both HC and hpHC groups (p < 0.0001), there was no significant difference in the distribution of males and females per group (p = 0.84). Both MCI and AD groups had more participants with the variant APOEε4 allele than the HC groups, while all three neuropsychological score measures showed lower scores for participants within the MCI and AD groups as compared with the HC groups (p < 0.0001).
Biological networks
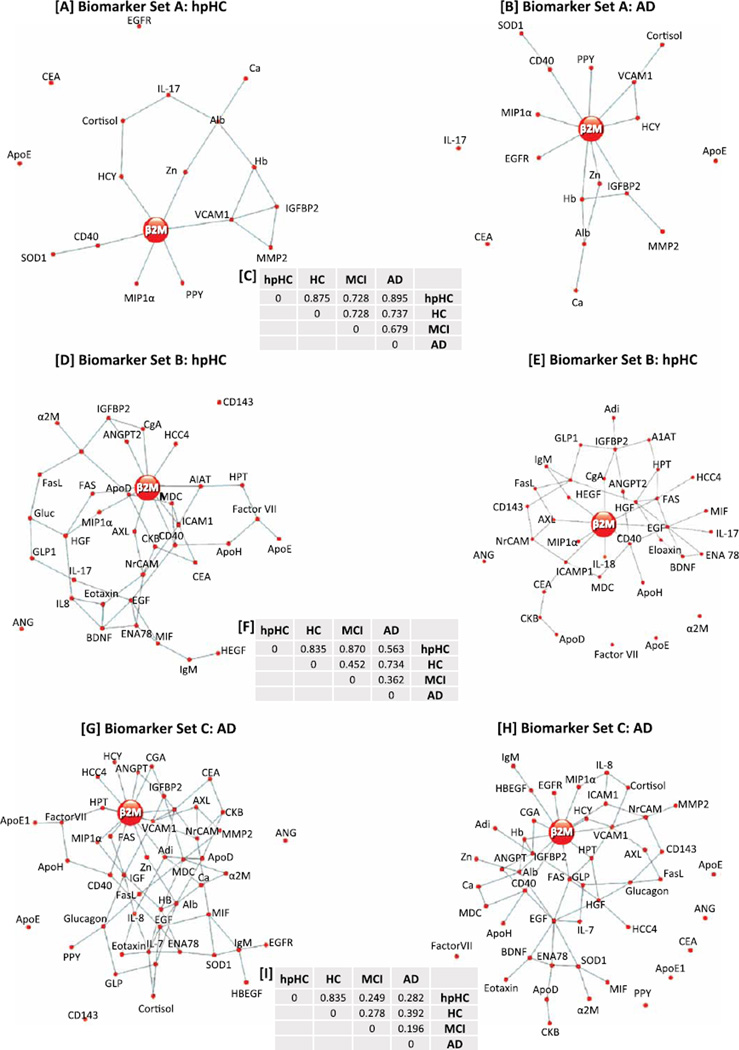
The inferred biomarker connections, along with the posterior probability of similarity between networks were plotted for each of the different clinical groups and each of the three different sets of proteins where the posterior probability of connection was greater than 0.5 (Fig. 1, Supplementary Table 2). Immediately noticeable across all the plots, was the network hub surrounding beta-2 microglobulin (β2M), with differing numbers of connections between β2M and other biomarkers dependent upon classification group and the number of biomarkers analyzed. Using biomarker set A and comparing the connections for β2M between hpHC and AD groups, we found six biomarkers common to both groups [pancreatic polypeptide (PPY), macrophage inflammatory protein 1 alpha (MIP1α), homocysteine (HCY), CD40, zinc (Zn) and vascular cell adhesion molecule 1 (VCAM1)], while the AD group had an extra three unique biomarker connections [hemoglobin (Hb), insulin growth factor binding protein 2 (IGFBP2), epidermal growth factor receptor (EGFR)] (Fig. 1A, B). Due to only a small number of differences in biomarker connections between the clinical groups, the posterior probability of network similarity between clinical groups was quite high (hpHC versus HC: 0.86, hpHC versus AD: 0.90).
Fig. 1.
A) Biomarker Set A (18 biomarkers): calculated connections between biomarkers for the hpHC group. B) Biomarker Set A (18 biomarkers): calculated connections between biomarkers for the AD group. C) Posterior probability of biomarker connection between classification groups for Biomarker Set A. D) Biomarker Set B (37 biomarkers): calculated connections between biomarkers for the hpHC group. E) Biomarker Set B (37 biomarkers): calculated connections between biomarkers for the AD group. F) Posterior probability of biomarker connection between classification groups for Biomarker Set B. G) Biomarker Set C (48 biomarkers): calculated connections between biomarkers for the hpHC group. H) Biomarker Set C (48 biomarkers): calculated connections between biomarkers for the AD group. I) Posterior probability of biomarker connection between classification groups for Biomarker Set C.
Increasing the number of biomarkers in the analyses to 37 (set B) both increased the complexity of the differences between clinical groups and decreased the posterior probability of similarity between the hpHC and AD networks (hpHC versus HC: 0.84, hpHC versus AD: 0.55). Interestingly, β2M was connected to five biomarkers in both the hpHC and AD groups [chromogranin A (CgA), tumor necrosis factor (TNF) receptor superfamily, member 6 (FAS), receptor tyrosine kinase (AXL), CD40, intercellular adhesion molecule 1 (ICAM1)]; connected to three unique biomarkers in the hpHC group [alpha 1 antitrypsin (A1AT), angiopoientin 2 (ANGPT2), human chemokine 4 (HCC4)]; and connected to further four unique biomarkers in the AD group [epidermal growth factor (EGF), MIP1α, interleukin 8 (IL8), and heparin-binding EGF- like growth factor (HBEGF)] (Fig. 1C, D).
Further increasing the number of biomarkers to 48, and including two different measures of apolipoprotein E (one commercial ELISA [35], one RBM[24]), decreased the posterior probability of similarity between the hpHC and AD networks (hpHC versus HC: 0.84, hpHC versus AD: 0.28). Again assessing the connections around β2M, we find seven biomarker connections in common between the hpHC and AD groups [HCY, CgA, FAS, MIP1α, VCAM1, CD40, haptoglobin (HAPT)], while the hpHC group had an extra four unique connections [HCC4, ANGPT2, macrophage-derived chemokine (MDC)], and the AD group had an extra five connections [Hb, albumin (Alb), EGFR, HBEGF, ICAM1] (Fig. 1E, F).
Although β2M was clearly the most frequently connected biomarker across all marker sets and clinical groups, we also sought those proteins that formed mini biomarker hubs (smaller than the β2M hub) across the clinical groups. Epidermal growth factor (EGF) emerged, with five and eight connections for the hpHC and AD groups respectively, within biomarker set B, and six and eight connections for the hpHC and AD groups, respectively, within biomarker Set C (Fig. 1D, E, G, H). Other biomarkers with greater than four connections in either biomarker set A/B included CD40, HGF, VCAM1, ICAM1, IGFBP2, BDNF, albumin, MDC, adiponectin, glucagon, and hemoglobin. Furthermore, a brief analyses of 41 of the 48 biomarkers that had information available via IPA, identified important and well known complexes such as NFκB, IL12, and P13K, and other markers including PDGF BB, TNF, and ERK1/2. A graphical representation of the IPA analyses is presented in Supplementary Figure 2.
Lastly, we assessed the differences and similarities between graphical networks for hpHC and AD groups for all three biomarker sets using the iGraph R package (http://cran.r-project.org/web/packages/igraph/igraph.pdf). Supplementary Figure 3 shows an increasing number of connections appearing in the hpHC group that were not seen in the AD group between networks with increasing numbers of biomarkers (Supplementary Figure 3A, C, E), but more importantly a greater focus around β2M post intersection of hpHC and AD groups (Supplementary Figure 3B, D, F).
DISCUSSION
The aim of this research was to assess biomarker network interaction using three sets of overlapping proteins and four clinical classifications. We used a novel Bayesian graphical network approach [34] to assess the differences between the networks, and present the identified biomarker β2M as a central network regulator. We show that by increasing the number of biomarkers in the analyses spectrum, we see stepwise increases in both the complexity of the network, and in the information provided by the interaction networks. It can clearly be seen that there is a plethora of information that can be mined from these analyses that would be otherwise missed in variable selection/dimension reduction analyses. We have, for the sake of brevity, and due to the strength of the β2M network across all clinical groups and biomarker sets, chosen to focus on the key information from the interaction network surrounding β2M.
In the initial Doecke et al. paper, β2M was shown to be significantly increased by 1.24 fold (p = 0.006) and was increased in AIBL, ADNI, and TARC datasets [18, 24]. Its relationship to other makers in the plasma proteome was a consistent feature in the Bayesian graphical network analysis. The centralized relationship of so many proteins leads us to conclude that β2M may be a master regulator of a number of downstream pathways, a significant finding that may have been over looked if not investigated using this Bayesian approach. In support of this conclusion, β2M is involved in a range of biological pathways, primarily through its activity in stabilizing class I MHC complexes.
β2M is the light chain of the MHC-class I complex [36], which is important in t-cell regulation and the immune system pathway [37]. It also has a role in iron uptake, through interactions with the hemochromatosis protein (HFE), which is a transferrin protein receptor [38]. The MHC class I complex has also been reported to affect receptor activity, in particular that of insulin receptors, albeit only in the absence of β2M [39]. The broad range of interactions of MHC I complexes illustrates that β2M is indeed a central member of a number of regulatory pathways, most likely through its chaperone-like activity in stabilizing the MHC complex.
Interestingly, β2M in certain microenvironments can form toxic fibrillar amyloid aggregates, particularly linked to dialysis related amyloidosis [36, 40–42]. Some studies have shown that the β2M fibrils (not the monomers) are the cytotoxic species of the protein and when aggregated can lead to membrane disruption and permeabilization [40, 41, 43].
β2M has also been implicated in some non-renal, cardiovascular conditions suggesting its role in physiology is still being elucidated [44]. β2M is highly expressed on motor neurons and shown to play a role in the progression in a murine model of motor neuron disease [45]. One of the major hallmarks of AD is the accumulation of amyloid fibril formation and there appears to be some commonalities between amyloid-β [46] and β2M propensity for fibril formation and membrane disruption. β2M obviously has a role to play in the sequence of events in AD and needs to be followed up with more research.
The current research has demonstrated that increasing the number of proteins in the network elucidates further biomarkers that may have important roles in the underlying biological disease mechanisms. Analyzing multiple biomarker sets with overlapping markers has had the advantage of demonstrating the robustness of biomarker relationships across biomarker sets. We see multiple markers besides β2M consistently acting as mini-biomarker hubs, connected to the same biomarkers across the biomarker sets (VCAM1, CD40, EGF, HGF), while others are consistently not connected (Ang, ApoE4).
A possible limitation of this study stems from using only one assay platform to conduct the analyses. Further work is underway to validate these findings using a separate protein array platform. Yeh et al. showed that increasing the number of biomarkers in analyses via the integration of biological knowledge enabled the reconstruction of gene regulatory networks [47]. Our research follows a similar premise, where increasing the number of proteins in the network identified novel interactions for β2M in AD. Similarly, Wang and colleagues recently used Bayesian network classifiers to integrate data from multiple platforms to identify biomarkers confirming previously published results [31]. Previous research using Bayesian networks to define marker connections in AD has primarily been performed using imaging data [28, 29], however these methods have not defined the posterior probability of both within and between group connections.
The strength of the methodology used in the current study is demonstrated by the posterior probabilities shown in Fig. 1. We show that increasing the number of biomarkers increases the network differences, with posterior probabilities of network similarity between hpHC and AD groups decreasing from 0.895 using 18 markers to 0.282 using 48 markers. Assessing the hpHC and HC groups, we saw only a very minor decrease in posterior probability; 0.875 using 18 markers to 0.835 using 48 markers, demonstrating that increasing the complexity of the model did not decrease the sensitivity of the inter-group comparisons.
Our novel methodology to interrogate the biomarker interaction networks both within and between groups for relationships has elucidated biological pathways and identified critical targets that may be useful in future biomarker screening. With increasing interest demonstrated in using protein array technology to investigate protein-disease pathology relationships, we advocate the use of graphical network methodologies to ascertain a better understanding of the underlying biological relationships that can potentially explain disease pathology.
In summary, the current study has interrogated a small set of biomarkers from a large and well-characterized study of ageing, with the express aim of searching for changes in biomarker interaction networks. We find that by increasing the search space to include a large number of biomarkers, we gain a better understanding of biological interactions that may elucidate disease specific pathways. Since many biomarker selection studies choose only the best candidates to represent the disease classification, it is our belief that more information could be assembled from many studies that opt for that smaller set of biomarkers to functional modules that predict disease status, and we look forward to verification of our biological network results in other populations in the near future.
Supplementary Material
Table 1.
| hpHC | HC | MCI | AD | p-value* | |
|---|---|---|---|---|---|
| n | 323 | 336 | 112 | 186 | |
| Age | 69.64 (6.38) | 70.73 (6.9) | 76.18 (7.69) | 78.8 (8.47) | p < 0.0001 |
| Gender (F/M) | 188/135 | 188/148 | 63/49 | 111/75 | p = 0.843 |
| APOEε4 (−ve/+ve) | 247/76 | 239/97 | 55/57 | 71/115 | p < 0.0001 |
| MMSE | 29 (1.12) | 29 (1.25) | 26 (2.6) | 20 (5.22) | p < 0.0001 |
| Composite score 1a | 0.24 (0.54) | −0.09 (0.57) | −1.31 (0.57) | −1.82 (0.56) | p < 0.0001 |
| Composite score 2b | 0.3 (0.67) | −0.15 (0.61) | −0.92 (0.79) | −1.85 (0.69) | p < 0.0001 |
Calculated as the average of the z score for California Verbal Learning Test Second Edition long delayed recall and Rey Complex Figure Test 30 minute delayed recall.
Calculated as the average of the z scores for Rey Complex Figure Test copy, Digit Symbol Coding, Boston Naming Test, Letter Fluency, Category Fluency, Digit Span (forwards), and Digit Span (backwards).
p-values calculated using χ2 test, and generalized linear model for the marginalized means.
ACKNOWLEDGMENTS
This research was also supported by the Science Industry and Endowment Fund (sief.org.au), and the National Health and Medical Research Council (NHMRC) via the Dementia Collaborative Research Centres program (DCRC) and the Commonwealth Scientific and Industrial Research Organisation (CSIRO) (http://aibl.csiro.au). Pfizer International has contributed financial support to assist with analysis of blood samples and to further the AIBL research program. Christine Peterson’s research has been supported under the NIH/NCI T32 Pre-Doctoral Training Program in Biostatistics for Cancer Research (NIH Grant NCI T32 CA096520), and by a training fellowship from the Keck Center of the Gulf Coast Consortia, on the NLM Training Program in Biomedical Informatics, National Library of Medicine (NLM) T15LM007093. AIBL would like to thank the volunteers and their partners and carers for donating their time to this study which would not be possible without their support.
ABBREVIATIONS
- A1AT
Alpha 1 antitrypsin
- α2M
Alpha 2 macropglobulin
- β2M
Beta 2 microglobulin
- Adi
Adiponectin
- Alb
Albumin
- Ang
Angiotensinogen
- ANGPT2
Angiopointen 2
- ApoD
Apolipoprotein D
- ApoE
Apolipoprotein E
- ApoH
Apolipoprotein H
- AXL
AXL receptor tyrosine kinase
- BDNF
Brain-derived neurotrophic factor
- Ca
Calcium
- CD143
Angiotensin-converting enzyme
- CD40
TNF receptor superfamily member 5
- CEA
Carcinoembryonic antigen
- CgA
Cromogranin A
- CKB
Creatine Kinase
- EGF
Epidermal Growth Factor
- EGFR
Epidermal growth factor receptor
- ENA78
C-X-C motif chemokine 5
- FAS
TNF receptor superfamily, member 6
- FasL
TNF receptor superfamily, member 6 receptor
- GLP1
Glucagon-like peptide-1
- HEGF
Human Epidermal Growth Factor
- Hb
Hemoglobin
- HBEGF
Human Epidermal Growth Factor
- HCC4
Human CC chemokine-4
- HCY
Homocysteine
- HGF
Hepatocyte Growth Factor Level
- HPT
Hygromycin phosphotransferase
- ICAM1
Inter-Cellular Adhesion Molecule 1 Level
- IGFBP2
Insulin-like growth factor-binding protein 2
- IgM
Immunoglobulin M
- IL-17
Interluekin-17
- IL-8
Interleukin-8
- MDC
Macrophage-derived Chemokine Level
- MIF
Macrophage Migration Inhibiting Factor Level
- MIP1α
Macrophage Inflammatory Protein alpha
- MMP2
matrix metalloproteinase-2
- NrCAM
Plasminogen Activator Inhibitor-1 Level
- PPY
Pancreatic Polypeptide
- SOD1
Superoxide dismutase 1
- VCAM1
Vascular Cell Adhesion Molecule 1
- Zn
Zinc
Footnotes
Authors’ disclosures available online (http://www.j-alz.com/disclosures/view.php?id=2579).
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-141497.
REFERENCES
- 1.Klunk WE. Biological markers of Alzheimer’s disease. Neurobiol Aging. 1998;19:145–147. doi: 10.1016/s0197-4580(98)00013-x. [DOI] [PubMed] [Google Scholar]
- 2.Sui X, Liu J, Yang X. Cerebrospinal fluid biomarkers of Alzheimer’s disease. Neurosci Bull. 2014;30:233–242. doi: 10.1007/s12264-013-1412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villemagne VL, Burnham S, Bourgeat P, Brown B, Ellis KA, Salvado O, Szoeke C, Macaulay SL, Martins R, Maruff P, Ames D, Rowe CC, Masters CL Australian Imaging B, Lifestyle Research G. Amyloid beta deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: A prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 4.Rembach A, Ryan TM, Roberts BR, Doecke JD, Wilson WJ, Watt AD, Barnham KJ, Masters CL. Progress towards a consensus on biomarkers for Alzheimer’s disease: A review of peripheral analytes. Biomark Med. 2013;7:641–662. doi: 10.2217/bmm.13.59. [DOI] [PubMed] [Google Scholar]
- 5.Zetterberg H. Unresolved questions in Alzheimer’s research: Will biomarkers help? Biomark Med. 2014;8:61–63. doi: 10.2217/bmm.13.120. [DOI] [PubMed] [Google Scholar]
- 6.Yang H, Lyutvinskiy Y, Herukka SK, Soininen H, Rutishauser D, Zubarev RA. Prognostic polypeptide blood plasma biomarkers of Alzheimer’s disease progression. J Alzheimers Dis. 2014;40:659–666. doi: 10.3233/JAD-132102. [DOI] [PubMed] [Google Scholar]
- 7.Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, Jiang T, Tan L. Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J Alzheimers Dis. 2014;40:1017–1027. doi: 10.3233/JAD-132144. [DOI] [PubMed] [Google Scholar]
- 8.Sutphen CL, Fagan AM, Holtzman DM. Progress update: Fluid and imaging biomarkers in Alzheimer’s disease. Biol Psychiatry. 2014;75:520–526. doi: 10.1016/j.biopsych.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes ME, Barmada MM, Kamboh MI, Visweswaran S. The application of network label propagation to rank biomarkers in genome-wide Alzheimer’s data. BMC Genomic. 2014;15:282. doi: 10.1186/1471-2164-15-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder HM, Carrillo MC, Grodstein F, Henriksen K, Jeromin A, Lovestone S, Mielke MM, O’Bryant S, Sarasa M, Sjogren M, Soares H, Teeling J, Trushina E, Ward M, West T, Bain LJ, Shineman DW, Weiner M, Fillit HM. Developing novel blood-based biomarkers for Alzheimer’s disease. Alzheimers Dement. 2014;10:109–114. doi: 10.1016/j.jalz.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGhee DJ, Ritchie CW, Thompson PA, Wright DE, Zajicek JP, Counsell CE. A systematic review of biomarkers for disease progression in Alzheimer’s disease. PLoS One. 2014;9:e88854. doi: 10.1371/journal.pone.0088854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriksen K, O’Bryant SE, Hampel H, Trojanowski JQ, Montine TJ, Jeromin A, Blennow K, Lonneborg A, Wyss-Coray T, Soares H, Bazenet C, Sjogren M, Hu W, Lovestone S, Karsdal MA, Weiner MW Blood-Based Biomarker Interest G. The future of blood-based biomarkers for Alzheimer’s disease. Alzheimers Dement. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Ramirez J, Wu J. Network-based biomarkers in Alzheimer’s disease: Review and future directions. Front Aging Neurosci. 2014;6:12. doi: 10.3389/fnagi.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Xiao S, Liu Y, Lin Z, Su N, Li X, Li G, Zhang M, Fang Y. The efficacy of plasma biomarkers in early diagnosis of Alzheimer’s disease. Int J Geriatr Psychiatry. 2014;29:713–719. doi: 10.1002/gps.4053. [DOI] [PubMed] [Google Scholar]
- 15.Rosen C, Hansson O, Blennow K, Zetterberg H. Fluid biomarkers in Alzheimer’s disease - current concepts. Mol Neurodegener. 2013;8:20. doi: 10.1186/1750-1326-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann S, Delaby C, Touchon J, Hirtz C, Gabelle A. Biomarkers of Alzheimer’s disease: The present and the future. Rev Neurol (Paris) 2013;169:719–723. doi: 10.1016/j.neurol.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A, Kaye JA, Leszek J, Miller BL, Minthon L, Quinn JF, Rabinovici GD, Robinson WH, Sabbagh MN, So YT, Sparks DL, Tabaton M, Tinklenberg J, Yesavage JA, Tibshirani R, Wyss-Coray T. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 18.O’Bryant SE, Xiao G, Barber R, Huebinger R, Wilhelmsen K, Edwards M, Graff-Radford N, Doody R, Diaz-Arrastia R Texas Alzheimer’s R, Care C, Alzheimer’s Disease Neuroimaging I. A blood-based screening tool for Alzheimer’s disease that spans serum and plasma: Findings from TARC and ADNI. PLoS One. 2011;6:e28092. doi: 10.1371/journal.pone.0028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soares HD, Potter WZ, Pickering E, Kuhn M, Immermann FW, Shera DM, Ferm M, Dean RA, Simon AJ, Swenson F, Siuciak JA, Kaplow J, Thambisetty M, Zagouras P, Koroshetz WJ, Wan HI, Trojanowski JQ, Shaw LM Biomarkers Consortium Alzheimer’s Disease Plasma Proteomics P. Plasma biomarkers associated with the apolipoprotein E genotype and Alzheimer disease. Arch Neurol. 2012;69:1310–1317. doi: 10.1001/archneurol.2012.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faux NG, Rembach A, Wiley J, Ellis KA, Ames D, Fowler CJ, Martins RN, Pertile KK, Rumble RL, Trounson B, Masters CL, The ARG. Bush AI. An anemia of Alzheimer’s disease. Mol Psychiatry. 2014;19:1227–1234. doi: 10.1038/mp.2013.178. [DOI] [PubMed] [Google Scholar]
- 21.Burnham SC, Faux NG, Wilson W, Laws SM, Ames D, Bedo J, Bush AI, Doecke JD, Ellis KA, Head R, Jones G, Kiiveri H, Martins RN, Rembach A, Rowe CC, Salvado O, Macaulay SL, Masters CL, Villemagne VL Alzheimer’s Disease Neuroimaging I, Australian Imaging B, Lifestyle Study Research G. A blood-based predictor for neocortical Abeta burden in Alzheimer’s disease: Results from the AIBL study. Mol Psychiatry. 2014;19:519–526. doi: 10.1038/mp.2013.40. [DOI] [PubMed] [Google Scholar]
- 22.Rembach A, Faux NG, Watt AD, Pertile KK, Rumble RL, Trounson BO, Fowler CJ, Roberts BR, Perez KA, Li QX, Laws SM, Taddei K, Rainey-Smith S, Robertson JS, Vandijck M, Vanderstichele H, Barnham KJ, Ellis KA, Szoeke C, Macaulay L, Rowe CC, Villemagne VL, Ames D, Martins RN, Bush AI, Masters CL group Ar. Changes in plasma amyloid beta in a longitudinal study of aging and Alzheimer’s disease. Alzheimers Dement. 2014;10:53–61. doi: 10.1016/j.jalz.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Rembach A, Doecke JD, Roberts BR, Watt AD, Faux NG, Volitakis I, Pertile KK, Rumble RL, Trounson BO, Fowler CJ, Wilson W, Ellis KA, Martins RN, Rowe CC, Villemagne VL, Ames D, Masters CL, group Ar. Bush AI. Longitudinal analysis of serum copper and ceruloplasmin in Alzheimer’s disease. J Alzheimers Dis. 2013;34:171–182. doi: 10.3233/JAD-121474. [DOI] [PubMed] [Google Scholar]
- 24.Doecke JD, Laws SM, Faux NG, Wilson W, Burnham SC, Lam CP, Mondal A, Bedo J, Bush AI, Brown B, De Ruyck K, Ellis KA, Fowler C, Gupta VB, Head R, Macaulay SL, Pertile K, Rowe CC, Rembach A, Rodrigues M, Rumble R, Szoeke C, Taddei K, Taddei T, Trounson B, Ames D, Masters CL, Martins RN Alzheimer’s Disease Neuroimaging I, Australian Imaging B, Lifestyle Research G. Blood-based protein biomarkers for diagnosis of Alzheimer disease. Arch Neurol. 2012;69:1318–1325. doi: 10.1001/archneurol.2012.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watt AD, Perez KA, Rembach AR, Masters CL, Villemagne VL, Barnham KJ. Variability in blood-based amyloid-beta assays: The need for consensus on pre-analytical processing. J Alzheimers Dis. 2012;30:323–336. doi: 10.3233/JAD-2012-120058. [DOI] [PubMed] [Google Scholar]
- 26.Lui JK, Laws SM, Li QX, Villemagne VL, Ames D, Brown B, Bush AI, De Ruyck K, Dromey J, Ellis KA, Faux NG, Foster J, Fowler C, Gupta V, Hudson P, Laughton K, Masters CL, Pertile K, Rembach A, Rimajova M, Rodrigues M, Rowe CC, Rumble R, Szoeke C, Taddei K, Taddei T, Trounson B, Ward V, Martins RN Group AR. Plasma amyloid-beta as a biomarker in Alzheimer’s disease: The AIBL study of aging. J Alzheimers Dis. 2010;20:1233–1242. doi: 10.3233/JAD-2010-090249. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Yu J, Zhang S, Bao F, Wang P, Huang X, Li J. Bayesian network analysis reveals alterations to default mode network connectivity in individuals at risk for Alzheimer’s disease. PLoS One. 2013;8:e82104. doi: 10.1371/journal.pone.0082104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Li R, Fleisher AS, Reiman EM, Guan X, Zhang Y, Chen K, Yao L. Altered default mode network connectivity in Alzheimer’s disease–a resting functional MRI and Bayesian network study. Hum Brain Mapp. 2011;32:1868–1881. doi: 10.1002/hbm.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Chen K, Yao L, Jin Z, Guo X Alzheimer’s Disease Neuroimaging I. Structural interactions within the default mode network identified by Bayesian network analysis in Alzheimer’s disease. PLoS One. 2013;8:e74070. doi: 10.1371/journal.pone.0074070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartolucci A, Bae S, Singh K, Griffith HR. An examination of Bayesian statistical approaches to modeling change in cognitive decline in an Alzheimer’s disease population. Math Comput Simul. 2009;80:561–571. doi: 10.1016/j.matcom.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Zuo Y, Liu L, Man Y, Tadesse MG, Ressom HW. Identification of functional modules by integration of multiple data sources using a bayesian network classifier. Circ Cardiovasc Genet. 2014;7:206–217. doi: 10.1161/CIRCGENETICS.113.000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rembach A, Watt AD, Wilson WJ, Villemagne VL, Burnham SC, Ellis KA, Maruff P, Ames D, Rowe CC, Macaulay SL, Bush AI, Martins RN, Masters CL, Doecke JD. Plasma amyloid-beta levels are significantly associated with a transition toward Alzheimer’s disease as measured by cognitive decline and change in neocortical amyloid burden. J Alzheimers Dis. 2014;40:95–104. doi: 10.3233/JAD-131802. [DOI] [PubMed] [Google Scholar]
- 33.Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, Lautenschlager NT, Lenzo N, Martins RN, Maruff P, Masters C, Milner A, Pike K, Rowe C, Savage G, Szoeke C, Taddei K, Villemagne V, Woodward M, Ames D Group AR. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int Psychogeriatr. 2009;21:672–687. doi: 10.1017/S1041610209009405. [DOI] [PubMed] [Google Scholar]
- 34.Peterson C, Vannucci M, Karakas C, Choi W, Ma L, Maletic-Savatic M. Inferring metabolic networks using the Bayesian adaptive graphical lasso with informative priors. Stat Interface. 2013;6:547–558. doi: 10.4310/SII.2013.v6.n4.a12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta VB, Laws SM, Villemagne VL, Ames D, Bush AI, Ellis KA, Lui JK, Masters C, Rowe CC, Szoeke C, Taddei K, Martins RN Group AR. Plasma apolipoprotein E and Alzheimer disease risk: The AIBL study of aging. Neurology. 2011;76:1091–1098. doi: 10.1212/WNL.0b013e318211c352. [DOI] [PubMed] [Google Scholar]
- 36.Burmeister WP, Gastinel LN, Simister NE, Blum ML, Bjorkman PJ. Crystal structure at 2.2 A resolution of the MHC-related neonatal Fc receptor. Nature. 1994;372:336–343. doi: 10.1038/372336a0. [DOI] [PubMed] [Google Scholar]
- 37.Halabelian L, Ricagno S, Giorgetti S, Santambrogio C, Barbiroli A, Pellegrino S, Achour A, Grandori R, Marchese L, Raimondi S, Mangione PP, Esposito G, Al-Shawi R, Simons JP, Speck I, Stoppini M, Bolognesi M, Bellotti V. Class I major histocompatibility complex, the trojan horse for secretion of amyloidogenic beta2-microglobulin. J Biol Chem. 2014;289:3318–3327. doi: 10.1074/jbc.M113.524157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardoso CS, de Sousa M. HFE, the MHC and hemochromatosis: Paradigm for an extended function for MHC class I. Tissue Antigens. 2003;61:263–275. doi: 10.1034/j.1399-0039.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 39.Ramalingam TS, Chakrabarti A, Edidin M. Interaction of class I human leukocyte antigen (HLA-I) molecules with insulin receptors and its effect on the insulin-signaling cascade. Mol Biol Cell. 1997;8:2463–2474. doi: 10.1091/mbc.8.12.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue WF, Hellewell AL, Gosal WS, Homans SW, Hewitt EW, Radford SE. Fibril fragmentation enhances amyloid cytotoxicity. J Biol Chem. 2009;284:34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue WF, Hellewell AL, Hewitt EW, Radford SE. Fibril fragmentation in amyloid assembly and cytotoxicity: When size matters. Prion. 2010;4:20–25. doi: 10.4161/pri.4.1.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson DE, Hurst RD. Polyphenolic phytochemicals–just antioxidants or much more? Cell Mol Life Sci. 2007;64:2900–2916. doi: 10.1007/s00018-007-7237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milanesi L, Sheynis T, Xue WF, Orlova EV, Hellewell AL, Jelinek R, Hewitt EW, Radford SE, Saibil HR. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc Natl Acad Sci U S A. 2012;109:20455–20460. doi: 10.1073/pnas.1206325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanga Z, Nock S, Medina-Escobar P, Nydegger UE, Risch M, Risch L. Factors other than the glomerular filtration rate that determine the serum beta-2-microglobulin level. PLoS One. 2013;8:e72073. doi: 10.1371/journal.pone.0072073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staats KA, Schonefeldt S, Van Rillaer M, Van Hoecke A, Van Damme P, Robberecht W, Liston A, Van Den Bosch L. Beta-2 microglobulin is important for disease progression in a murine model for amyotrophic lateral sclerosis. Front Cell Neurosci. 2013;7:249. doi: 10.3389/fncel.2013.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams TL, Serpell LC. Membrane and surface interactions of Alzheimer’s Abeta peptide–insights into the mechanism of cytotoxicity. FEBS J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 47.Yeh HY, Cheng SW, Lin YC, Yeh CY, Lin SF, Soo VW. Identifying significant genetic regulatory networks in the prostate cancer from microarray data based on transcription factor analysis and conditional independency. BMC Med Genomic. 2009;2:70. doi: 10.1186/1755-8794-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.