Abstract
We hypothesized that mutations that inactivate phosphodiesterase (PDE) activity and lead to increased cyclic AMP (cAMP) and cyclic GMP (cGMP) levels may be associated with prostate cancer (PCa). We sequenced the entire PDE coding sequences in the DNA of 16 biopsy samples from PCa patients. Novel mutations were confirmed in the somatic or germline state by Sanger sequencing. Data were then compared to the 1000 Genome Project. PDE, CREB and pCREB protein expression was also studied in all samples, in both normal and abnormal tissue, by immunofluorescence. We identified 3 previously described PDE sequence variants that were significantly higher in PCa. Four novel sequence variations, one each in the PDE4B, PDE6C, PDE7B and PDE10A genes, respectively, were also found in the PCa samples. Interestingly, PDE10A and PDE4B novel variants that were present in 19% and 6% of the patients, respectively, were found in the tumor tissue only. In patients carrying PDE defects, there was pCREB accumulation (p<0.001), and an increase of the pCREB/CREB ratio (patients 0.97± 0.03; controls 0.52± 0.03; p-value < 0.001) by immunohistochemical analysis. We conclude that PDE sequence variants may play a role in the predisposition and/or progression to PCa at the germline and/or somatic state, respectively. Larger such studies are needed to confirm these findings.
Keywords: Prostate cancer, PCa, phosphodiesterases, PDE family, CREB, pCREB, cAMP, cGMP
Introduction
Phosphodiesterases (PDEs) are grouped into 21 families based on their structural homology and biochemical features (Azevedo, et al. 2014). They function as enzymes that regulate intracellular levels of cyclic AMP (cAMP) and cyclic GMP (cGMP) which in turn affect the function of a multitude of molecules (Halpin 2008). Recently, protein–protein interactions have been described for several of the PDEs (Jensen, et al. 2009). These data add to the complexity of PDEs in modulating multiple signaling pathways, such as those of the protein kinases A, B and C (PKA, PKB, PKC, respectively) and others (Baillie, et al. 2000; Hoffmann, et al. 1998; MacKenzie, et al. 2000; Pozuelo Rubio, et al. 2005; Shakur, et al. 2001; Zhao, et al. 2002). PKA, in particular, is essential for PDE function both for mediating cAMP effects, and through A-kinase anchoring proteins (AKAPs), which bind and localize PDEs in the cell (Pan, et al. 2012). PDE4 is the most thoroughly studied PDE in terms of interactions with other molecules, such as arrestins and receptors for activated C kinase 1 (RACK1) (Pan et al. 2012).
Pharmacological exploitation of PDEs has led to the discovery of drugs with selective action against specific isoforms. Selective PDE inhibitors, such as Sildenafil®, are widely used for the treatment of conditions ranging from erectile dysfunction to heart failure and pulmonary hypertension (Tsertsvadze, et al. 2009). Other PDE inhibitors are considered for therapeutic treatment in diseases such as asthma and chronic obstructive pulmonary disease (Kodimuthali, et al. 2008). PDE5A inhibitors (PDE5i) relax smooth muscle cells (SMCs) by increasing intracellular levels of Ca2+, and raise cGMP levels though activation of the K+ channels in endothelial cells (Luedders, et al. 2006). PDE5is are used to treat benign prostatic hyperplasia (BPH) (Hotston, et al. 2006; Wang 2010; Wong, et al. 2009) and it has been shown in vitro that they induce apoptosis and control cell proliferation (Cook and Haynes 2004; Tinsley, et al. 2009; Zhu, et al. 2005). A recent study suggests that the effects of the PDE5i Zaprinast® may control cell proliferation in human cultured prostatic stromal cells, in a time-of-exposure- and dose-dependent way (Cook and Haynes 2004).
Even though some studies have shown that increased cGMP through PDE5A inhibition controls cell proliferation, other studies have produced contradictory data. PDE5is are also capable of inhibiting PDE6, which may play an important role in cell cycle arrest (Bazhin, et al. 2010; Cote 2004; Ma and Wang 2007; Wang, et al. 2004). Tadalafil®, a drug that is widely used for erectile dysfunction is, in addition to its PDE5i role, the strongest PDE11A inhibitor (Washington and Shindel 2010). PDE11A-inactivating mutations and sequence have been linked to predisposition to PCa, testicular germ cell cancer, and adrenal tumors (Faucz, et al. 2011; Horvath, et al. 2006a; Horvath, et al. 2006b; Horvath, et al. 2009; Libe, et al. 2011).
In this study, we sequenced the PDE exome in PCa samples; our data show higher frequency of PDE sequence variants as compared to the 1000 genomes database. Furthermore, patients with PDE sequence variants had up-regulated cAMP signaling in their PCa tissues.
Materials and Methods
Patients & DNA preparation
We collected frozen tumors tissues and blood from 48 males of white, mixed European descent. The Biobank of A. C. Camargo Cancer Center, Brazil provided the samples (Campos, et al. 2012; Olivieri, et al. 2014). Patients’ stage of disease, age, PSA and Gleason scores can be found on the Supplementary Table 1. Written informed consent was obtained from all participants, and the institutional review boards of the participating centers approved the study. Biopsy samples were obtained at the moment of PCa diagnosis. Peripheral DNA was extracted from blood samples; tumor tissue DNA was extracted from the frozen biopsy samples from all patients following standard procedures. DNA from 16 (out of the 48) biopsy tissues was subjected to targeted sequencing using Agilent's SureSelect SOLiD4 plataform (Life Technologies). Germline DNA from peripheral blood was used to confirm the origin of the novel mutations. Selective variants identified in the group of 16 were further studied in the remaining 32 PCa patients by Sanger sequencing.
Library capture and sequencing
For targeted sequencing, we selected 203 genes that included all the known phosphodiesterases, and molecules related directly and through networks to cAMP and cGMP signaling. Genomic DNA (3μg) from each of the 16 affected tissues was used for sequencing library preparation. Target enrichment was performed using the Agilent's SureSelect solution-based capture assay (Gnirke, et al. 2009); libraries were prepared following the manufacturer's protocols (SureSelect Target Enrichment System for the Applied Biosystems SOLiD System). Paired-end sequencing (50nt read length) was performed on the SOLiD4 platform (Life Technologies), according to the manufacturer protocol.
Data analysis
Briefly, high quality trimmed reads were mapped and aligned to the human reference genome (hg18) using Bioscope version 1.3. The variants were called using GATK, and annotated through SeattleSeq v5.07. All variants considered for further study were visually examined using the Integrative Genomics Viewer (IGV) (Robinson, et al. 2011).
Sanger sequencing
To validate novel variants, we performed Sanger sequencing using a 3130xl Genetic Analyzer (Applied Biosystems). To verify the origin of the novel (not previously reported) variations, Sanger sequencing was performed on both somatic and peripheral DNA. Furthermore, PDE5A variants were studied in all 48 PCa patients. Primer sequences and PCR conditions are available upon request.
Statistics
The p-value and odds ratio was used to determine significant differences between the frequencies of single-nucleotide variants (SNVs) among the patients and 379 Europeans (EUR) and 1092 individuals of all ethnicities (ALL). Both European and general population data were collected from the 1000 Genome Project (Genomes Project, et al. 2010), which was extracted through the Ensembl Genome Browser (http://www.ensembl.org/index.html). To determine significant differences among groups, a two-way analysis was performed, with 95% confidence and calculated through Fisher's exact test and Bonferroni correction using the IBM SPSS software 20. SNVs with p <0.05 when analyzed individually and lost its statistical power after the Bonferroni correction were kept separately in a new table since our sample size is small and their statistical significance might change with a larger sample size. Immunohistochemical statistical analysis was performed through T-test with similar parameters used for Fisher. Furthermore, we performed the Mann–Whitney–Wilcoxon test, which is a nonparametric test of the null hypothesis that two populations are the same against an alternative hypothesis that patients tend to have a greater amount of variations than controls, with significance value of p < 0.1. Both T-test and Mann–Whitney–Wilcoxon were performed through RStudio (v.0.98) and R (v.3.1.1).
Immunohistochemical (IHC) Analysis
IHC was performed on 3 patients with high number of PDE variants, 2 patients with low number of PDE variants and on 3 age-matched control tissues obtained from other surgeries and/or necropsies. For this experiment, three areas of each slide were used for comparison of each control and each patient's with high number of PDE variants's tissue; expression was graded for each reading (9 for the patients with high number of PDE variants and 9 for the controls) without knowledge of the origin of the tissue. Tissues from only 2 patients with low number of PDE variants were available; therefore they were not included for the statistical analysis.
We studied the expression of cAMP-responsive element binding protein (CREB) and its phosphorylated status (pCREB). Slides from PCas were deparaffinized and rehydrated through a series of alcohol solutions (100%, 95%, 70%, 50% and 1XPBS). Followed by heat-induced antigen retrieval, whereby slides were boiled in Antigen Unmasking Solution (Citric Acid Based, pH 6.0; Vector Laboratories), for 30 min in a standard rice cooker. All slides were blocked with 10% normal goat serum (NGS, JacksonImmunoResearch) made in 1XPBS for one hour at room temperature and then sections were incubated with the following primary antibodies: rabbit anti-CREB and mouse anti-pCREB (both from Cell Signaling, Danvers, MA; dilution 1:100) overnight at 4°C. The next day, slides were washed in 1XPBS, 3x15 mins at room temperature. All slides were then incubated with Alexa Fluor Goat anti-rabbit 488 and Alexa Fluo Goat anti-mouse 555 (Life Technologies, CA) for 1-2 hours. Again, slides were washed in 1XPBS, 3x15 mins at room temperature. Cover slips were affixed to each slide with ProLong® Gold Antifade Reagent with DAPI (Life Technologies, CA). For negative control, a section was incubated under identical conditions with no primary antibody. Fluorescence was analyzed with a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) and images acquired using the Olympus DMP7 camera using the CellSens image acquisition software. For each channel, separate images were acquired and all images were collected at the same exposure. Following image acquisition, fluorescent staining for pCREB and CREB was quantified blindly, using ImageJ imaging software (Schneider, et al. 2012). By delimitating the area, the mean fluorescent amplitude was determined from the integrated pixel intensity for the whole area on both the 488nm and 555nm emission channels separately.
To determine the level of CREB phosphorylation, a ratio of the mean fluorescent amplitude measured by the excitation of Alexa Fluor 555 bound to pCREB was divided by the mean fluorescent amplitude measured by the excitation of Alexa Fluor 488 bound to CREB for all slides. Relative pCREB/CREB ratios were reported for each area and for each group as mean±S.E.M. Differences were tested for statistical significance by using the T-test and the level of significance was set at p< 0.05.
Results
DNA Sequencing
Analysis of the sequencing datasets revealed an average of 184kbs covered region per sample. We identified 106 known SNVs and 4 novel variants within the PDE gene family among the 16 randomly selected biopsies DNA. Distribution of the variants was assessed between our group and the general population, taking into account the ethnicity (Supplementary Table 2). Because our patients were of mixed European-Caucasian descent, only those control individuals (EUR) were considered for comparison.
From these 106 SNVs, 3 were significantly more frequent among PCa patients as compared to the controls (p <0.05/106, after correction for multiple trials) (Table 1), and the remaining 19 lost its statistical power when analyzed within the group (Table 2). Most of these known SNVs are absent or extremely rare (≤1%) in the EUR, with the exception of rs3733526, which has minor allele frequency (MAF<20.1%). Among these SNVs, 6 were significantly overrepresented in PCa patients compared to both ALL and EUR populations.
Table 1.
3 significant associated SNVs with prostate cancer.
| Gene | SNV | Location / Distance to Splice | SIFT / PPH2 | Population | Alelic p-value / OR (CI) |
|---|---|---|---|---|---|
| PDE2A | rs577536 | c.2112+3 A>G | * | EUR | p= 6.1*10^(-5) OR - 1.10 (95% CI. 0.98-1.23) |
| 3 bps | * | ALL | p=0.028 OR - 0.19 (95% CI. 0.05-0.67) | ||
| PDE11A | rs75461311 | Intron | * | EUR | p=1.4*10^(-4) OR - 26.9 (95% CI. 6.40-113.0) |
| > 10 bps | * | ALL | p=0.029 OR - 3.80 (95% CI. 1.30-11.1) | ||
| PDE11A | rs79903863 |
p.Glu396Lys
p.Glu840Lys |
Damaging | EUR | p=6.1*10^(-5) OR - 1.10 (95% CI. 0.98-1.23) |
| > 10 bps | Benign | ALL | p=1.1*10^(-5) OR - 0.00 (95% CI. 0.00-0.04) |
Unmeasurable
**: Absent data; EUR: European population; ALL: All 1092 individuals from 1000 Genome Project; OR: odds ratio; CI: confidence interval.
Table 2.
19 SNVs that lost statistical power after the correction.
| Gene | SNV | Location / Distance to Splice | SIFT / PPH2 | Population | Alelic p-value / OR (CI) |
|---|---|---|---|---|---|
| PDE1C | rs115957738 | p.Asp122 = > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.184 OR - 0.18 (95% CI. 0.02-1.46) | |||
| PDE1C | rs116449276 | Intron > 10 bps | * | EUR | p=0.001 OR - 1.06 (95% CI. 0.97-1.16) |
| * | ALL | p=0.008 OR - 0.05 (95% CI. 0.01-0.27) | |||
| PDE1C | rs76290787 | p.Ser175 = > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.264 OR - 0.28 (95% CI. 0.03-2.20) | |||
| PDE1C | rs78021925 | Intron > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.452 OR - 1.72 (95% CI. 0.23-12.9) | |||
| PDE2A | rs392818 | utr-3 > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.483 OR - 0.63 (95% CI. 0.08-4.77) | |||
| PDE2A | rs11824084 | utr-3 > 10 bps | * | EUR | p=0.001 OR - 1.06 (95% CI. 0.97-1.16) |
| * | ALL | p=0.098 OR - 0.24 (95% CI. 0.05-1.06) | |||
| PDE2A | rs4944557 | p.Asp572 = 9 bps | * | EUR | p=0.001 OR - 1.06 (95% CI. 0.97-1.16) |
| * | ALL | p=0.381 OR - 0.63 (95% CI. 0.14-2.70) | |||
| PDE4B | rs34492439 | p.Val388 = > 10 bps | * | EUR | p=0.003 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p= ** | |||
| PDE4B | rs114408173 | Intron > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.172 OR - 0.17 (95% CI. 0.02-1.35) | |||
| PDE4B | rs7534999 | c.604-3 C>T 3 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.587 OR - 0.86 (95% CI. 0.11-6.41) | |||
| PDE5A | rs3733526 | p.Ala93Val > 10 bps | Tolerated | EUR | p=0.052 OR - 0.51 (95% CI. 0.26-0.99) |
| Benign | ALL | p=0.080 OR - 0.55 (95% CI. 0.29-1.04) | |||
| PDE5A | rs114886951 | p.Ile778Thr 2 bps | Tolerated | EUR | p=0.034 OR - 16.1 (95% CI. 1.44-179.0) |
| Damaging | ALL | p=0.005 OR - 46.4 (95% CI. 4.17-516.0) | |||
| PDE6A | rs113309832 | Intron > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.295 OR - 0.32 (95% CI. 0.04-2.52) | |||
| PDE6B | rs61734864 | p.Val835 = 2 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.231 OR - 4.11 (95% CI. 0.53-31.8) | |||
| PDE6B | rs73058408 | c.1722+5 G>A 5 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.014 OR - 1.03 (95% CI. 0.96-1.09) | |||
| PDE6C | rs193276411 | Intron > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.148 OR - 0.14 (95% CI. 0.01-1.14) | |||
| PDE6C | rs76999928 | p.Asp157Glu 10 bps | Tolerated | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| Damaging | ALL | p=0.295 OR - 3.03 (95% CI. 0.39-23.1) | |||
| PDE8A | rs148254104 | p.Leu350 = > 10 bps | * | EUR | p=0.011 OR - 0.00 (95% CI. 0.00-0.08) |
| * | ALL | p= ** | |||
| PDE8B | rs182775622 | p.Ala375 = > 10 bps | * | EUR | p=0.040 OR - 1.03 (95% CI. 0.96-1.09) |
| * | ALL | p=0.028 OR - 0.01 (95% CI. 0.00-0.23) |
Unmeasurable
Absent data; EUR: European population; ALL: All 1092 individuals from 1000 Genome Project; OR: odds ratio; CI: confidence interval.
To determine the somatic or germline nature of the novel PDE variants, we performed Sanger sequencing in both tumor and blood-derived DNA. This analysis identified two variants that were present in the tumor tissue only. One was a novel non-synonymous (ns) substitution in the PDE4B (Table 3). The second was a novel PDE10A intronic variant located near the splice site of the exon 5 (c.347-9 A>C) identified in 3 out of 16 samples (Table 3). In silico analysis of potential splice alterations resulting from this variant was performed using five different web-based tools: SplicePort (Dogan, et al. 2007), ASSP (Wang and Marin 2006), Sroogle (Schwartz, et al. 2008), NetGene2 (Hebsgaard, et al. 1996) and BDGP (Reese, et al. 1997). They all predicted a significant change in the splice acceptor. Furthermore, bioinformatics analysis of this variant using the MutationTaster software consider this variation as disease causing (prob: 0.999999995965687) due increased chances of splice site shifting and evolutionary conservation (Schwarz, et al. 2014). The remaining two novel variants were a nsSNV in PDE7B (p. Met239Ile) and an intronic variant in PDE6C (Table 3). Even though the PDE7B variation was near the splice site, no significant change in the splicing regions was predicted through in silico modeling. All the previously reported variants were originally described in studies of germline DNA; therefore, we did not anticipate seeing them in the somatic setting, as we did in our study.
Table 3.
Four novel mutations in prostate cancer
| Gene | Chr | Position (hg18) | Genotype | Frequency | Distance to Splice | Location | SIFT | PPH2 | MutationTaster | Sample origin |
|---|---|---|---|---|---|---|---|---|---|---|
| PDE10A | chr6 | 165782464 | 0/1 | 3 Patients | 9 | c.347-10 A>C | * | * | Disease Causing | Somatic |
| PDE4B | chr1 | 66231291 | 0/1 | 1 Patient | 123 | p.Asn38Lys | Tolerated | Benign | Polymorphism | Somatic |
| PDE6C | chr10 | 95408566 | 0/1 | 1 Patient | 80 | Intronic | * | * | Polymorphism | Germline |
| PDE7B | chr6 | 136536633 | 0/1 | 1 Patient | 6 | p.Met239Ile | Tolerated | Benign | Disease Causing | Germline |
Unmeasurable; Chr: chromosome.
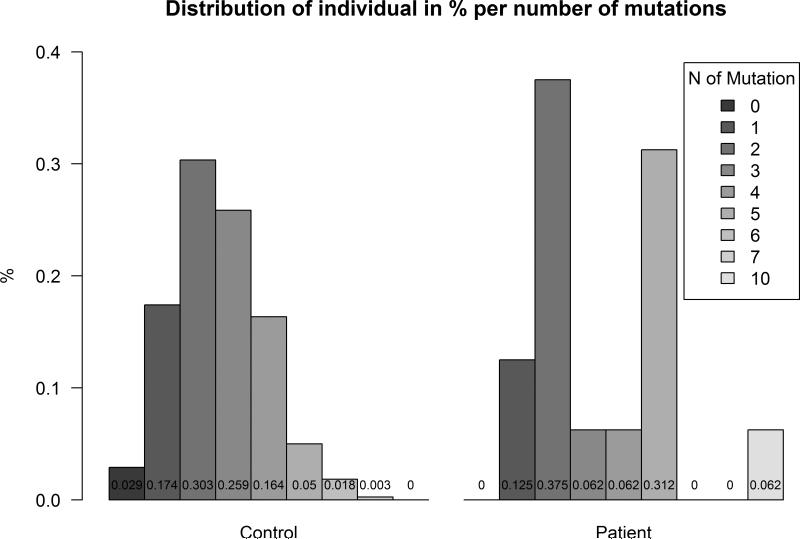
Among the 106 known SNVs, 19 were nsSNVs. Each nsSNV (n=21; including the two novel ones) was counted in each individual from both control (n=379, x̄=2.59, SE=0.07) and patient (n=16, x̄=3.5, SE=0.58) groups. The average number of PDE nsSNV in patients was 35% higher than in the general EUR population with a significant p-value of 0.07996 (<0.1; Mann–Whitney–Wilcoxon test) (Figure 1 and Table 4).
Figure 1.
PDE11A sequence variants and patients: side-by-side barplot expressing the percent of individuals distributed by the number of nonsynonymous variants encountered in each sample.
Table 4.
Presence of mutations, nsSNV or significant associated in each of the 16 tumor samples run through SOLiD
| Novel | PDE1C | PDE2A | PDE4A | PDE4B | PDE4C | PDE5A | PDE6A | PDE6B | PDE6C | PDE7A | PDE8A | PDE8B | PDE11A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DT01 | @rs115957738 |
@rs577536 @rs11824084 |
#rs114886951 | *rs115775983 | *rs701865 |
#rs79903863 @rs75461311 |
||||||||
| DT02 |
@rs4944557 @rs392818 |
*rs2229228 |
@rs193276411 *rs701865 |
@rs75461311 | ||||||||||
| DT03 | PDE6C |
@rs76290787 @rs78021925 |
@rs11824084 | @rs114408173 | *rs2229228 | *rs701865 | @rs75461311 | |||||||
| DT04 | *rs1051738 | |||||||||||||
| DT07 | PDE10A PDE7B |
@rs4944557 | *rs2229228 | *rs701865 | *rs115599001 | #rs79903863 | ||||||||
| DT08 | *rs2229228 | *rs701865 | *rs115599001 |
#rs79903863 *rs75672043 |
||||||||||
| DT10 | *rs2229228 | *rs701865 | *rs11557049 | *rs115599001 |
@rs75461311 *rs77597060 |
|||||||||
| DT12 | PDE10A | *rs75398902 | *rs2229228 | *rs17400325 | ||||||||||
| DT18 | #rs3733526 | *rs701865 | ||||||||||||
| DT19 | PDE4B | #rs76999928 | ||||||||||||
| DT21 | *rs61736729 | *rs2229228 |
#rs3733526 *rs17051276 |
*rs701865 | *rs75127279 | |||||||||
| DT22 | *rs1051738 | #rs3733526 | *rs701865 | *rs11557049 | ||||||||||
| DT32 | *rs2229228 | *rs701865 | ||||||||||||
| DT35 | @rs577536 | *rs701865 | @rs148254104 | |||||||||||
| DT38 | @rs116449276 |
@rs34492439 @rs7534999 |
*rs2229228 | @rs113309832 |
@rs73058408 @rs61734864 |
*rs11557049 | ||||||||
| DT45 | PDE10A |
@rs116449276 *rs61745829 |
*rs1051738 | *rs2229228 |
#rs3733526 *rs17051276 |
*rs115775983 |
*rs45522236 *rs701865 |
@rs182775622 |
*rs75672043 *rs17400325 |
Nonsynonymous SNV
Significant and nonsynonymous SNV
Significant synonymous or intronic SNV.
IHC Analysis
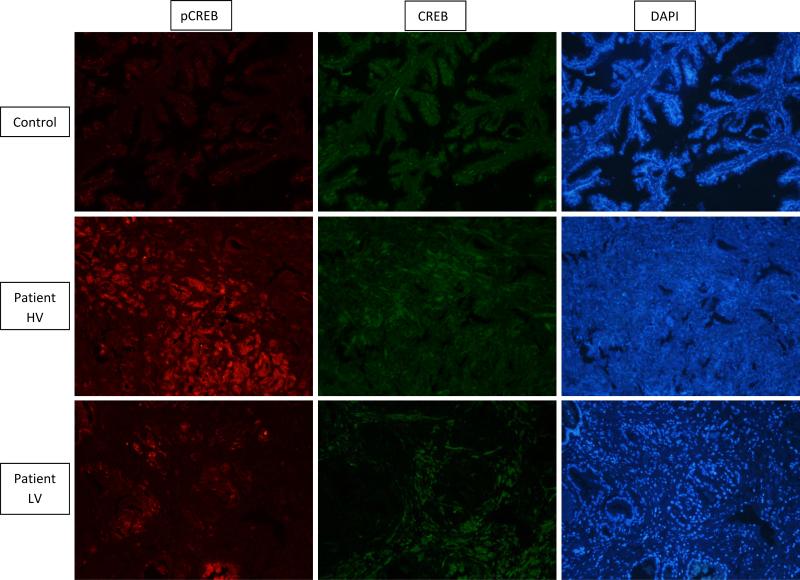
Patients carrying high number of PDE variants had a highly significant increase of pCREB (p-value<0.001), whereas, CREB levels seemed to be approximately the same between controls and patients (a representative sample from 1 patient with high number of PDE variants, 1 patient with low number of PDE variants and 1 control is shown in Figure 2). Thus, the pCREB to CREB ratio (pCREB/CREB) was significantly higher in patients (0.97± 0.03) vs. controls (0.52± 0.03; p-value<0.001) (Table 5).
Figure 2.
Immunohistochemistry (IHC) for the CREB (green) and pCREB (red) proteins in prostate tissue (10x magnification). Primary antibodies of rabbit anti-CREB and mouse anti-pCREB (dilution 1:100) and secondary antibodies goat anti-rabbit 488 and goat anti-mouse 555 where used to achieve this image. Higher levels of pCREB (red) are observed in PCa tissue when compared to a control; there does not appear to be a difference in unphoshorylated CREB (green). Patient (HV) – High number of PDE variants; Patient (LV) – Low number of PDE variants.
Table 5.
IHC statistic result for pCREB/CREB ratio
| p-value | CI - 95% | Mean Patient |
SE Patients | Mean Control |
SE Control | |
|---|---|---|---|---|---|---|
| Ratio | 2.97E-08 | −0.54 to −0.35 | 0.97 | 0.03 | 0.52 | 0.03 |
| PCREB | 1.88E-05 | −23.51 to −11.51 | 31.45 | 1.62 | 13.95 | 2.29 |
| CREB | 0.4418 | −17.37 to 8.28 | 32.44 | 1.17 | 27.89 | 5.52 |
SE: standard error; CI: confidence interval.
Discussion
In this work, we describe the presence of four novel variants in the PDE genes that might be associated with PCa development and/or progression. We also report 3 known SNVs to be significantly more frequent in PCa patients; an additional total of 19 (p-value<0.05 when analyzed individually) lost statistical power after the correction. In addition, we report for the first time 35% higher occurrence of nsSNVs (p-value=0.07996) in patients as compared to controls. As an index of function of these variants, the pCREB/CREB ratio was significantly higher in patients with high number of ns PDE variants (p < 0.001).
Our group has previously reported on the possible involvement of PDE11A in PCa pathogenesis (Faucz et al. 2011). Recent findings from other groups support these observations. To date, three PDEs, PDE4 (Henderson, et al. 2014; Kashiwagi, et al. 2012; Sarwar, et al. 2014), PDE5, and PDE11 (Chavez, et al. 2013; Hamilton, et al. 2013) have been associated with PCa.
According to Sarwar et al. (2014), PDE4 inhibitors (PDE4i) play important roles in the AR/PSA expression pathway (Sarwar et al. 2014). It was discovered that the use of Rolipram®, a selective PDE4 inhibitor, enhances the production of cAMP/PKA and increases AR and PSA expression significantly (Sarwar et al. 2014). Prostatic tissue is indeed highly sensitive to cAMP (Faucz et al. 2011), and both PKA activity and cAMP levels have been associated with PCa (Chung, et al. 2009; Desiniotis, et al. 2010; Dimitriadis, et al. 2008; Sarwar et al. 2014). Our finding in this study that a novel, somatic nonsynonymous PDE4B variant (p.Asn38Lys) is present in PCa patients, may explain the down-regulation of PDE4B in PCa that has been described by others (Kashiwagi et al. 2012).
PDE5A does not seem to be influenced by the AR (Yang, et al. 2009). A handful of recent studies have provided new insight on the possible therapeutic effects of PDE5i in PCA. Chavez et al (2013) showed that men with erectile dysfunction treated with PDE5i tended to have reduced chance (p<0.0001) of developing PCa when compared to those who did not receive treatment. Two additional groups have reported that the compound found in green tea (Epigallocatechin Gallate – EGCG) together with PDE5i, might stimulate apoptosis in PCa cells (Kumazoe, et al. 2013; Yang and Wang 2013). Even though the rs114886951 lost its statistical power after Bonferroni correction, in silico analysis of this SNV seems to show a promising therapeutic target. This variation is located in the 100% splice site domain, in silico analysis shows complete loss of splice. Furthermore, this amino acid is highly conserved among different species (Mutationtaster) (Schwarz et al. 2014). In our work this variant was present in two individuals within the patient group (4.2%), whereas it was found in only one individual within the 1,092 used in the project genome (0.0009%, p = 0.034; OR = 16.1, 95%; CI = 1.44–179). The higher reported frequency of this SNV was by the CSAgilent project (0.015%) whereas the ExAC project showed a similar frequency to the 1000 genome (0.001228%). It is noteworthy that this nsSNV (p.Ile778Thr) is located at the end of the catalytic domain of PDE5, within an alpha-helix secondary structure of the protein core and it may destabilize the protein structure, in agreement to the in silico prediction for a damaging effect by SIFT (Ryan, et al. 2009).
Even though PDE11A is capable of binding both cGMP and cAMP, it is important to highlight that PDE11A and PDE5A structures are quite similar (Fawcett, et al. 2000). Both PDEs are responsible for the inhibition of 86% of the free cGMP in PCa cell lines (Hamilton et al. 2013), and together they are responsible for reducing the intracellular levels of PKG, a protein kinase involved in cell proliferation, the MAP kinase pathway, migration and neo-vessel growth in some types of endothelial cells (Hood and Granger 1998; Pyriochou, et al. 2007). Tadalafil®, a PDE5i, binds relatively strongly to PDE11 and reduces its activity (Pomara and Morelli 2005). While Chavez et al (2013) showed that PDE5i inhibitors may have a protective effect from PCa, studies of PDE11A inactivation suggest opposite effects in various tumor settings (Boikos, et al. 2008; Faucz et al. 2011; Horvath et al. 2006a; Horvath et al. 2006b; Horvath et al. 2009; Libe, et al. 2008; Libe et al. 2011; Vezzosi, et al. 2012). According to Faucz et al (2011), patients with PCa tended to have a higher frequency of PDE11A-inactivating variants when compared to controls (p-value < 0.001; OR = 3.81, 95%; CI = 1.86–7.81). The present work confirms these findings and also adds two significantly associated variants that are overrepresented in the PCa patients: rs79903863 (p-value ≈ 6.1*10^(-5); OR = 1.10; 95% CI = 0.98-1.23) and rs75461311 (p-value ≈ 1.4*10^(-4); OR = 26.9; 95% CI = 6.40-113).
Together with PDE11, PDE2 enzymes are both capable of degrading cAMP and cGMP at the same rate. Although there is no bibliographical references linking these PDEs to PCa, both are expressed in the prostate and have already been studied for their possible association with cancer (Abusnina, et al. 2011; Durand, et al. 2011; Lukk, et al. 2010; Morita, et al. 2013; Traka, et al. 2008; Wheeler, et al. 2005; Zhang, et al. 2012). With similar results to our findings in PDE11A, our statistical analysis found one significant SNV in PDE2A rs577536 (p-value ≈ 6.1*10^(-5); OR = 1.10; 95% CI = 0.98-1.23).
PDE10A was the second gene in which we found an interesting, frequent (≈19%) and novel somatic mutation. Like most of PDEs outlined above, there is no report of PDE10A genetic variants in any type of cancer. A recent review described the use of PDE10 inhibitors for schizophrenia and neuropsychiatric disorders (Azevedo et al. 2014). Although PDE10A is mainly expressed in the brain, it is also present in the testis and prostate (Fujishige, et al. 1999; Wheeler et al. 2005). Furthermore, in silico analysis of this novel variant shows a possible alteration in the splice site of the fifth exon. This exon is responsible for encoding the first 17% of the PDE10 first GAF domain. GAF's function in PDE10 remains controversial. It has been suggested that these domains binds to, and are activated by cAMP, which paradoxically results in potent competitive inhibition of cGMP turnover by cAMP (Jager, et al. 2012). Even though this splice site variation is not located between the two 100% conserved cleavage site, others variations with a similar distance has already been reported to cause mRNA changes and complete exon skipping (Fernandez Alanis, et al. 2012). Furthermore, this site could be explain as a possible adenine branch site (Pagani and Baralle 2004), however, further studies should be conducted to verify this hypothesis.
PDE7B has been explored for treatment of neurological diseases. Unlike most PDEs, PDE7B expression is minimal or absent in the prostate (Gardner, et al. 2000; Hetman, et al. 2000); however, it is activated through the cAMP/PKA/CREB pathway. The novel variant p.Met239Ile, is located 6 bp from the most nearby exon boundary, and its effects in prostate cells are worthy of in vitro studies.
Interestingly, we found in our patients three previously described variants that shown association with testicular or adrenal tissues. The PDE11A variants rs17400325 and rs75127279 were found in three patients with bilateral testicular germ cell tumors (Horvath et al. 2006b). While the PDE8B variant rs115599001 was found in one patient with adrenal cancer (Rothenbuhler, et al. 2012) and in three of our patients, with a tendency of significance when compared with the ALL population (p=0.021 OR – 0.09 (95% CI. 0.02-0.44).
Finally, analysis of the pCREB/CREB ratio has been shown to be a good indicator of the amount of cAMP present in any tissue. Kaname et al. showed that an increase in PDE4A and PDE4B levels might cause low cAMP accumulation, which resulted in a decrease in the pCREB/CREB ratio (Kaname, et al. 2014). A different study showed that PDE4i is capable of reversing this effect through up-regulation of cAMP and increasing pCREB/CREB ratio (Guo, et al. 2014). Our IHC experiment showed an increase of the ratio (1.86x higher when compared to control) similar to the results obtained by the group that used PDE4i. Kumar et al. (2007) had previously reported similar results in the prostate and suggested its potential role as a prognostic marker.
How the inhibitors selectively bind the conserved PDE catalytic domains is unknown for most PDEs (Huai, et al. 2003). A single report suggest that the Rolipram® inhibitor selectivity on PDE4 is determined by the chemical nature of amino acids and also describes a relationship between the insensitivity of this drug with PDE7A due variants (Huai et al. 2003). Maybe this same principle can be expanded to other inhibitor drugs, bringing different outcomes depending in each tissue and disease. In addition, higher intracellular levels of cGMP have been reported to “crossover” and activate this kinase (Cornwell, et al. 1994; Hood and Granger 1998). Over-expression of PKA has already been associated with PCa (Desiniotis et al. 2010; Merkle and Hoffmann 2011), and other types of cancers (Almeida, et al. 2012; Horvath, et al. 2010; Horvath, et al. 2008; Horvath, et al. 2006a; Libe et al. 2011; Stratakis, et al. 2010).
We conclude that various PDE genetic variants may be associated with predisposition and or progression to PCa. Understanding the molecular signaling basis of this association could be key to the development of novel therapeutic approaches for this common form of cancer.
Supplementary Material
Figure 3.
Ratio difference between control and patient groups: A boxplot of the results for the analysis of fluorescence with a Leica TCS SP5 laser scanning confocal microscope. The results showed increased pCREB and pCREB/CREB ratio. Patients had a mean ratio of 0.97 with a standard error of 0.03, whereas controls had a mean of 0.52 with a standard error of 0.03. The boxplot also shows the presence of two outliers in the control group and one in the patients group.
Acknowledgements
We want to knowledge AC Camargo Biobank, Brazil, for providing the samples that were used in this study.
Funding
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development intramural National Institutes of Health Project Z01-HD-000642-04 (to C.A.S.); in part, by a grant from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Process: 311166/2011-3 - PQ-2 (to F.R.F.); and in part, by a grant from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Process: 5406/10-2 – 2011 (to R.B.A.).
Footnotes
Declaration of interest
The authors have no competing interests to disclose.
References
- Abusnina A, Keravis T, Yougbare I, Bronner C, Lugnier C. Anti-proliferative effect of curcumin on melanoma cells is mediated by PDE1A inhibition that regulates the epigenetic integrator UHRF1. Mol Nutr Food Res. 2011;55:1677–1689. doi: 10.1002/mnfr.201100307. [DOI] [PubMed] [Google Scholar]
- Almeida MQ, Azevedo MF, Xekouki P, Bimpaki EI, Horvath A, Collins MT, Karaviti LP, Jeha GS, Bhattacharyya N, Cheadle C, et al. Activation of cyclic AMP signaling leads to different pathway alterations in lesions of the adrenal cortex caused by germline PRKAR1A defects versus those due to somatic GNAS mutations. J Clin Endocrinol Metab. 2012;97:E687–693. doi: 10.1210/jc.2011-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo MF, Faucz FR, Bimpaki E, Horvath A, Levy I, de Alexandre RB, Ahmad F, Manganiello V, Stratakis CA. Clinical and molecular genetics of the phosphodiesterases (PDEs). Endocr Rev. 2014;35:195–233. doi: 10.1210/er.2013-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, MacKenzie SJ, McPhee I, Houslay MD. Sub-family selective actions in the ability of Erk2 MAP kinase to phosphorylate and regulate the activity of PDE4 cyclic AMP-specific phosphodiesterases. Br J Pharmacol. 2000;131:811–819. doi: 10.1038/sj.bjp.0703636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazhin AV, Tambor V, Dikov B, Philippov PP, Schadendorf D, Eichmuller SB. cGMP-phosphodiesterase 6, transducin and Wnt5a/Frizzled-2-signaling control cGMP and Ca(2+) homeostasis in melanoma cells. Cell Mol Life Sci. 2010;67:817–828. doi: 10.1007/s00018-009-0214-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Boikos SA, Horvath A, Heyerdahl S, Stein E, Robinson-White A, Bossis I, Bertherat J, Carney JA, Stratakis CA. Phosphodiesterase 11A expression in the adrenal cortex, primary pigmented nodular adrenocortical disease, and other corticotropin-independent lesions. Horm Metab Res. 2008;40:347–353. doi: 10.1055/s-2008-1076694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AH, Silva AA, Mota LD, Olivieri ER, Prescinoti VC, Patrao D, Camargo LP, Brentani H, Carraro DM, Brentani RR, et al. The value of a tumor bank in the development of cancer research in Brazil: 13 years of experience at the a C camargo hospital. Biopreserv Biobank. 2012;10:168–173. doi: 10.1089/bio.2011.0032. [DOI] [PubMed] [Google Scholar]
- Chavez AH, Scott Coffield K, Hasan Rajab M, Jo C. Incidence rate of prostate cancer in men treated for erectile dysfunction with phosphodiesterase type 5 inhibitors: retrospective analysis. Asian J Androl. 2013;15:246–248. doi: 10.1038/aja.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Furihata M, Tamura K, Uemura M, Daigo Y, Nasu Y, Miki T, Shuin T, Fujioka T, Nakamura Y, et al. Overexpressing PKIB in prostate cancer promotes its aggressiveness by linking between PKA and Akt pathways. Oncogene. 2009;28:2849–2859. doi: 10.1038/onc.2009.144. [DOI] [PubMed] [Google Scholar]
- Cook AL, Haynes JM. Protein kinase G II-mediated proliferative effects in human cultured prostatic stromal cells. Cell Signal. 2004;16:253–261. doi: 10.1016/s0898-6568(03)00134-7. [DOI] [PubMed] [Google Scholar]
- Cornwell TL, Arnold E, Boerth NJ, Lincoln TM. Inhibition of smooth muscle cell growth by nitric oxide and activation of cAMP-dependent protein kinase by cGMP. Am J Physiol. 1994;267:C1405–1413. doi: 10.1152/ajpcell.1994.267.5.C1405. [DOI] [PubMed] [Google Scholar]
- Cote RH. Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int J Impot Res. 2004;16(Suppl 1):S28–33. doi: 10.1038/sj.ijir.3901212. [DOI] [PubMed] [Google Scholar]
- Desiniotis A, Schafer G, Klocker H, Eder IE. Enhanced antiproliferative and proapoptotic effects on prostate cancer cells by simultaneously inhibiting androgen receptor and cAMP-dependent protein kinase A. Int J Cancer. 2010;126:775–789. doi: 10.1002/ijc.24806. [DOI] [PubMed] [Google Scholar]
- Dimitriadis F, Giannakis D, Pardalidis N, Zikopoulos K, Paraskevaidis E, Giotitsas N, Kalaboki V, Tsounapi P, Baltogiannis D, Georgiou I, et al. Effects of phosphodiesterase-5 inhibitors on sperm parameters and fertilizing capacity. Asian J Androl. 2008;10:115–133. doi: 10.1111/j.1745-7262.2008.00373.x. [DOI] [PubMed] [Google Scholar]
- Dogan RI, Getoor L, Wilbur WJ, Mount SM. SplicePort--an interactive splice-site analysis tool. Nucleic Acids Res. 2007;35:W285–291. doi: 10.1093/nar/gkm407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand J, Lampron A, Mazzuco TL, Chapman A, Bourdeau I. Characterization of differential gene expression in adrenocortical tumors harboring beta-catenin (CTNNB1) mutations. J Clin Endocrinol Metab. 2011;96:E1206–1211. doi: 10.1210/jc.2010-2143. [DOI] [PubMed] [Google Scholar]
- Faucz FR, Horvath A, Rothenbuhler A, Almeida MQ, Libe R, Raffin-Sanson ML, Bertherat J, Carraro DM, Soares FA, Molina Gde C, et al. Phosphodiesterase 11A (PDE11A) genetic variants may increase susceptibility to prostatic cancer. J Clin Endocrinol Metab. 2011;96:E135–140. doi: 10.1210/jc.2010-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, Hetman J, Beavo JA, Phillips SC. Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc Natl Acad Sci U S A. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Alanis E, Pinotti M, Dal Mas A, Balestra D, Cavallari N, Rogalska ME, Bernardi F, Pagani F. An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Hum Mol Genet. 2012;21:2389–2398. doi: 10.1093/hmg/dds045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishige K, Kotera J, Michibata H, Yuasa K, Takebayashi S, Okumura K, Omori K. Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A). J Biol Chem. 1999;274:18438–18445. doi: 10.1074/jbc.274.26.18438. [DOI] [PubMed] [Google Scholar]
- Gardner C, Robas N, Cawkill D, Fidock M. Cloning and characterization of the human and mouse PDE7B, a novel cAMP-specific cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 2000;272:186–192. doi: 10.1006/bbrc.2000.2743. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Lin P, Zhao X, Zhang J, Wei X, Wang Q, Wang C. Etazolate abrogates the lipopolysaccharide (LPS)-induced downregulation of the cAMP/pCREB/BDNF signaling, neuroinflammatory response and depressive-like behavior in mice. Neuroscience. 2014;263:1–14. doi: 10.1016/j.neuroscience.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Halpin DM. ABCD of the phosphodiesterase family: interaction and differential activity in COPD. Int J Chron Obstruct Pulmon Dis. 2008;3:543–561. doi: 10.2147/copd.s1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TK, Hu N, Kolomitro K, Bell EN, Maurice DH, Graham CH, Siemens DR. Potential therapeutic applications of phosphodiesterase inhibition in prostate cancer. World J Urol. 2013;31:325–330. doi: 10.1007/s00345-012-0848-7. [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DJ, Byrne A, Dulla K, Jenster G, Hoffmann R, Baillie GS, Houslay MD. The cAMP phosphodiesterase-4D7 (PDE4D7) is downregulated in androgen-independent prostate cancer cells and mediates proliferation by compartmentalising cAMP at the plasma membrane of VCaP prostate cancer cells. Br J Cancer. 2014;110:1278–1287. doi: 10.1038/bjc.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman JM, Soderling SH, Glavas NA, Beavo JA. Cloning and characterization of PDE7B, a cAMP-specific phosphodiesterase. Proc Natl Acad Sci U S A. 2000;97:472–476. doi: 10.1073/pnas.97.1.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Wilkinson IR, McCallum JF, Engels P, Houslay MD. cAMP-specific phosphodiesterase HSPDE4D3 mutants which mimic activation and changes in rolipram inhibition triggered by protein kinase A phosphorylation of Ser-54: generation of a molecular model. Biochem J. 1998;333(Pt 1):139–149. doi: 10.1042/bj3330139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J, Granger HJ. Protein kinase G mediates vascular endothelial growth factor-induced Raf-1 activation and proliferation in human endothelial cells. J Biol Chem. 1998;273:23504–23508. doi: 10.1074/jbc.273.36.23504. [DOI] [PubMed] [Google Scholar]
- Horvath A, Bertherat J, Groussin L, Guillaud-Bataille M, Tsang K, Cazabat L, Libe R, Remmers E, Rene-Corail F, Faucz FR, et al. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase A (PRKAR1A): an update. Hum Mutat. 2010;31:369–379. doi: 10.1002/humu.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Boikos S, Giatzakis C, Robinson-White A, Groussin L, Griffin KJ, Stein E, Levine E, Delimpasi G, Hsiao HP, et al. A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat Genet. 2006a;38:794–800. doi: 10.1038/ng1809. [DOI] [PubMed] [Google Scholar]
- Horvath A, Bossis I, Giatzakis C, Levine E, Weinberg F, Meoli E, Robinson-White A, Siegel J, Soni P, Groussin L, et al. Large deletions of the PRKAR1A gene in Carney complex. Clin Cancer Res. 2008;14:388–395. doi: 10.1158/1078-0432.CCR-07-1155. [DOI] [PubMed] [Google Scholar]
- Horvath A, Giatzakis C, Robinson-White A, Boikos S, Levine E, Griffin K, Stein E, Kamvissi V, Soni P, Bossis I, et al. Adrenal hyperplasia and adenomas are associated with inhibition of phosphodiesterase 11A in carriers of PDE11A sequence variants that are frequent in the population. Cancer Res. 2006b;66:11571–11575. doi: 10.1158/0008-5472.CAN-06-2914. [DOI] [PubMed] [Google Scholar]
- Horvath A, Korde L, Greene MH, Libe R, Osorio P, Faucz FR, Raffin-Sanson ML, Tsang KM, Drori-Herishanu L, Patronas Y, et al. Functional phosphodiesterase 11A mutations may modify the risk of familial and bilateral testicular germ cell tumors. Cancer Res. 2009;69:5301–5306. doi: 10.1158/0008-5472.CAN-09-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Mathyakina L, Vong Q, Baxendale V, Pang AL, Chan WY, Stratakis CA. Serial analysis of gene expression in adrenocortical hyperplasia caused by a germline PRKAR1A mutation. J Clin Endocrinol Metab. 2006a;91:584–596. doi: 10.1210/jc.2005-1301. [DOI] [PubMed] [Google Scholar]
- Hotston M, Shukla N, Bloor J, Persad R, Jeremy JY. Pre-clinical evidence for the use of phosphodiesterase-5 inhibitors for treating benign prostatic hyperplasia and lower urinary tract symptoms. BJU Int. 2006;98:1331–1332. doi: 10.1111/j.1464-410X.2006.06628_5.x. [DOI] [PubMed] [Google Scholar]
- Huai Q, Wang H, Sun Y, Kim HY, Liu Y, Ke H. Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Jager R, Russwurm C, Schwede F, Genieser HG, Koesling D, Russwurm M. Activation of PDE10 and PDE11 phosphodiesterases. J Biol Chem. 2012;287:1210–1219. doi: 10.1074/jbc.M111.263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaname T, Ki CS, Niikawa N, Baillie GS, Day JP, Yamamura KI, Ohta T, Nishimura G, Mastuura N, Kim OH, et al. Heterozygous mutations in cyclic AMP phosphodiesterase-4D (PDE4D) and protein kinase A (PKA) provide new insights into the molecular pathology of acrodysostosis. Cell Signal. 2014;26:2446–2459. doi: 10.1016/j.cellsig.2014.07.025. [DOI] [PubMed] [Google Scholar]
- Kashiwagi E, Shiota M, Yokomizo A, Itsumi M, Inokuchi J, Uchiumi T, Naito S. Downregulation of phosphodiesterase 4B (PDE4B) activates protein kinase A and contributes to the progression of prostate cancer. Prostate. 2012;72:741–751. doi: 10.1002/pros.21478. [DOI] [PubMed] [Google Scholar]
- Kodimuthali A, Jabaris SS, Pal M. Recent advances on phosphodiesterase 4 inhibitors for the treatment of asthma and chronic obstructive pulmonary disease. J Med Chem. 2008;51:5471–5489. doi: 10.1021/jm800582j. [DOI] [PubMed] [Google Scholar]
- Kumazoe M, Sugihara K, Tsukamoto S, Huang Y, Tsurudome Y, Suzuki T, Suemasu Y, Ueda N, Yamashita S, Kim Y, et al. 67-kDa laminin receptor increases cGMP to induce cancer-selective apoptosis. J Clin Invest. 2013;123:787–799. doi: 10.1172/JCI64768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libe R, Fratticci A, Coste J, Tissier F, Horvath A, Ragazzon B, Rene-Corail F, Groussin L, Bertagna X, Raffin-Sanson ML, et al. Phosphodiesterase 11A (PDE11A) and genetic predisposition to adrenocortical tumors. Clin Cancer Res. 2008;14:4016–4024. doi: 10.1158/1078-0432.CCR-08-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libe R, Horvath A, Vezzosi D, Fratticci A, Coste J, Perlemoine K, Ragazzon B, Guillaud-Bataille M, Groussin L, Clauser E, et al. Frequent phosphodiesterase 11A gene (PDE11A) defects in patients with Carney complex (CNC) caused by PRKAR1A mutations: PDE11A may contribute to adrenal and testicular tumors in CNC as a modifier of the phenotype. J Clin Endocrinol Metab. 2011;96:E208–214. doi: 10.1210/jc.2010-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wang HY. Mitogen-activated protein kinase p38 regulates the Wnt/cyclic GMP/Ca2+ non-canonical pathway. J Biol Chem. 2007;282:28980–28990. doi: 10.1074/jbc.M702840200. [DOI] [PubMed] [Google Scholar]
- MacKenzie SJ, Baillie GS, McPhee I, Bolger GB, Houslay MD. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D cAMP-specific phosphodiesterases. The involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J Biol Chem. 2000;275:16609–16617. doi: 10.1074/jbc.275.22.16609. [DOI] [PubMed] [Google Scholar]
- Merkle D, Hoffmann R. Roles of cAMP and cAMP-dependent protein kinase in the progression of prostate cancer: cross-talk with the androgen receptor. Cell Signal. 2011;23:507–515. doi: 10.1016/j.cellsig.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Morita H, Murata T, Shimizu K, Okumura K, Inui M, Tagawa T. Characterization of phosphodiesterase 2A in human malignant melanoma PMP cells. Oncol Rep. 2013;29:1275–1284. doi: 10.3892/or.2013.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri EH, Franco Lde A, Pereira RG, Mota LD, Campos AH, Carraro DM. Biobanking practice: RNA storage at low concentration affects integrity. Biopreserv Biobank. 2014;12:46–52. doi: 10.1089/bio.2013.0056. [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Pan CQ, Sudol M, Sheetz M, Low BC. Modularity and functional plasticity of scaffold proteins as p(l)acemakers in cell signaling. Cell Signal. 2012;24:2143–2165. doi: 10.1016/j.cellsig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Pomara G, Morelli G. Inhibition of phosphodiesterase 11 (PDE11) impacts on sperm quality. Int J Impot Res. 2005;17:385–386. doi: 10.1038/sj.ijir.3901304. author reply 387. [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Campbell DG, Morrice NA, Mackintosh C. Phosphodiesterase 3A binds to 14-3-3 proteins in response to PMA-induced phosphorylation of Ser428. Biochem J. 2005;392:163–172. doi: 10.1042/BJ20051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyriochou A, Zhou Z, Koika V, Petrou C, Cordopatis P, Sessa WC, Papapetropoulos A. The phosphodiesterase 5 inhibitor sildenafil stimulates angiogenesis through a protein kinase G/MAPK pathway. J Cell Physiol. 2007;211:197–204. doi: 10.1002/jcp.20929. [DOI] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenbuhler A, Horvath A, Libe R, Faucz FR, Fratticci A, Raffin Sanson ML, Vezzosi D, Azevedo M, Levy I, Almeida MQ, et al. Identification of novel genetic variants in phosphodiesterase 8B (PDE8B), a cAMP-specific phosphodiesterase highly expressed in the adrenal cortex, in a cohort of patients with adrenal tumours. Clin Endocrinol (Oxf) 2012;77:195–199. doi: 10.1111/j.1365-2265.2012.04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwar M, Sandberg S, Abrahamsson PA, Persson JL. Protein kinase A (PKA) pathway is functionally linked to androgen receptor (AR) in the progression of prostate cancer. Urol Oncol. 2014;32:25 e21–12. doi: 10.1016/j.urolonc.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Shakur Y, Holst LS, Landstrom TR, Movsesian M, Degerman E, Manganiello V. Regulation and function of the cyclic nucleotide phosphodiesterase (PDE3) gene family. Prog Nucleic Acid Res Mol Biol. 2001;66:241–277. doi: 10.1016/s0079-6603(00)66031-2. [DOI] [PubMed] [Google Scholar]
- Stratakis CA, Tichomirowa MA, Boikos S, Azevedo MF, Lodish M, Martari M, Verma S, Daly AF, Raygada M, Keil MF, et al. The role of germline AIP, MEN1, PRKAR1A, CDKN1B and CDKN2C mutations in causing pituitary adenomas in a large cohort of children, adolescents, and patients with genetic syndromes. Clin Genet. 2010;78:457–463. doi: 10.1111/j.1399-0004.2010.01406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley HN, Gary BD, Keeton AB, Zhang W, Abadi AH, Reynolds RC, Piazza GA. Sulindac sulfide selectively inhibits growth and induces apoptosis of human breast tumor cells by phosphodiesterase 5 inhibition, elevation of cyclic GMP, and activation of protein kinase G. Mol Cancer Ther. 2009;8:3331–3340. doi: 10.1158/1535-7163.MCT-09-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Gasper AV, Melchini A, Bacon JR, Needs PW, Frost V, Chantry A, Jones AM, Ortori CA, Barrett DA, et al. Broccoli consumption interacts with GSTM1 to perturb oncogenic signalling pathways in the prostate. PLoS One. 2008;3:e2568. doi: 10.1371/journal.pone.0002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsertsvadze A, Fink HA, Yazdi F, MacDonald R, Bella AJ, Ansari MT, Garritty C, Soares-Weiser K, Daniel R, Sampson M, et al. Oral phosphodiesterase-5 inhibitors and hormonal treatments for erectile dysfunction: a systematic review and meta-analysis. Ann Intern Med. 2009;151:650–661. doi: 10.7326/0003-4819-151-9-200911030-00150. [DOI] [PubMed] [Google Scholar]
- Vezzosi D, Libe R, Baudry C, Rizk-Rabin M, Horvath A, Levy I, Rene-Corail F, Ragazzon B, Stratakis CA, Vandecasteele G, et al. Phosphodiesterase 11A (PDE11A) gene defects in patients with acth-independent macronodular adrenal hyperplasia (AIMAH): functional variants may contribute to genetic susceptibility of bilateral adrenal tumors. J Clin Endocrinol Metab. 2012;97:E2063–2069. doi: 10.1210/jc.2012-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Phosphodiesterase-5 inhibitors and benign prostatic hyperplasia. Curr Opin Urol. 2010;20:49–54. doi: 10.1097/MOU.0b013e328333ac68. [DOI] [PubMed] [Google Scholar]
- Wang H, Lee Y, Malbon CC. PDE6 is an effector for the Wnt/Ca2+/cGMP-signalling pathway in development. Biochem Soc Trans. 2004;32:792–796. doi: 10.1042/BST0320792. [DOI] [PubMed] [Google Scholar]
- Wang M, Marin A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Washington SL, 3rd, Shindel AW. A once-daily dose of tadalafil for erectile dysfunction: compliance and efficacy. Drug Des Devel Ther. 2010;4:159–171. doi: 10.2147/dddt.s9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, Ayyagari RR, Wheeler GL, Weiss RM. Regulation of cyclic nucleotides in the urinary tract. J Smooth Muscle Res. 2005;41:1–21. doi: 10.1540/jsmr.41.1. [DOI] [PubMed] [Google Scholar]
- Wong P, Lawrentschuk N, Bolton DM. Phosphodiesterase 5 inhibitors in the management of benign prostatic hyperplasia and erectile dysfunction: the best of both worlds. Curr Opin Urol. 2009;19:7–12. doi: 10.1097/MOU.0b013e328316c357. [DOI] [PubMed] [Google Scholar]
- Yang CS, Wang H. Cancer therapy combination: green tea and a phosphodiesterase 5 inhibitor? J Clin Invest. 2013;123:556–558. doi: 10.1172/JCI67589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Huang YC, Lin G, Wang G, Hung S, Dai YT, Sun ZY, Lue TF, Lin CS. Lack of direct androgen regulation of PDE5 expression. Biochem Biophys Res Commun. 2009;380:758–762. doi: 10.1016/j.bbrc.2009.01.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Du J, Liu L, Chen X, Yang F, Jin Q. Inhibitory effects and underlying mechanism of 7-hydroxyflavone phosphate ester in HeLa cells. PLoS One. 2012;7:e36652. doi: 10.1371/journal.pone.0036652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao AZ, Huan JN, Gupta S, Pal R, Sahu A. A phosphatidylinositol 3-kinase phosphodiesterase 3B-cyclic AMP pathway in hypothalamic action of leptin on feeding. Nat Neurosci. 2002;5:727–728. doi: 10.1038/nn885. [DOI] [PubMed] [Google Scholar]
- Zhu B, Vemavarapu L, Thompson WJ, Strada SJ. Suppression of cyclic GMP-specific phosphodiesterase 5 promotes apoptosis and inhibits growth in HT29 cells. J Cell Biochem. 2005;94:336–350. doi: 10.1002/jcb.20286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.