Summary
Little information is available about the natural cycle of foot-and-mouth disease (FMD) in the absence of control measures such as vaccination. Cameroon presents a unique opportunity for epidemiological studies because FMD vaccination is not practiced. We carried out a prospective study including serological, antigenic and genetic aspects of FMD virus (FMDV) infections among different livestock production systems in the Far North of Cameroon to gain insight into the natural ecology of the virus. We found serological evidence of FMDV infection in over 75% of the animals sampled with no significant differences of prevalence observed among the sampled groups (i.e. market, sedentary, transboundary trade and mobile). We also found antibodies reactive to five of the seven FMDV serotypes (A, O, SAT1, SAT2 and SAT3) among the animals sampled. Finally, we were able to genetically characterize viruses obtained from clinical and subclinical FMD infections in Cameroon. Serotype O viruses grouped into two topotypes (West and East Africa). SAT2 viruses grouped with viruses from Central and Northern Africa, notably within the sublineage causing the large epidemic in Northern Africa in 2012, suggesting a common origin for these viruses. This research will guide future interventions for the control of FMD such as improved diagnostics, guidance for vaccine formulation and epidemiological understanding in support of the progressive control of FMD in Cameroon.
Keywords: foot-and-mouth disease virus, Cameroon, Africa, phylogeny, serotyping, SAT2
Introduction
Foot-and-mouth disease (FMD) is an economically devastating disease of livestock affecting cloven-hoofed domestic and wild animals worldwide. Foot-and-mouth disease is one of the main diseases limiting animal production and trade, contributing towards food insecurity in regions with a high demand for animal protein for an increasing human population (Knight-Jones and Rushton, 2013). The lack of infrastructure, economic resources and vaccines tailored to their condition renders many developing countries particularly vulnerable to the spread and devastating effects of the disease. In Southern Africa, more than 75% of livestock are raised under communal smallholder systems and these sustain the livelihoods of vulnerable groups such as women and children (Animal Production, 2012). These groups are the most negatively impacted by animal diseases, in particular transboundary animal diseases (TADs) such as FMD which not only limits livestock productivity and well-being, but also the opportunities for regional trade of livestock and livestock products (Hammond, 2011).
The causal agent of FMD is a highly variable small RNA virus and is a member of the family Picornaviridae. As with other picornaviruses, FMDV is non-enveloped and contains a positive sense RNA genome within an icosahedral capsid composed of four structural viral proteins (SP) named VP1, VP2, VP3 and VP4. In addition to the structural proteins, during viral infection, a number of non-structural viral proteins (NSP) are also made in the infected cells.
Based on the serological response to the capsid proteins, FMDV has been classified into seven serotypes denoted as O, A, C, SAT1, SAT2, SAT3 and Asia1. Within Africa, serotypes O, A, SAT1, SAT2 and SAT3 are present, with only sporadic reports of SAT (South African Territory) viruses in the northern part of Africa in 2000, 2003 and 2012 (Ahmed et al., 2012; Lockhart et al., 2012). These serotypes have further been classified into topotypes, which are based on geographic region as well as VP1 sequencing. There is no lasting cross-protection between serotypes and animals having an immune response against one serotype (either by vaccination or infection) will not be protected against the other serotypes (Grubman and Baxt, 2004). Furthermore, there are multiple strains within some serotypes that fail to cross-protect and vaccination programs require vaccines that match the circulating viral strains (Hammond, 2011).
Most animals survive infection by FMDV, although it can be lethal in young animals due to myocarditis (Grubman and Baxt, 2004). In ruminants (e.g. cattle, sheep, goats), clinical signs include excess salivation and nasal discharge due to the virus infecting epithelial cells in the mouth and nose and causing painful vesicular lesions (Holliman, 2005). Vesicles also appear on the epithelia of coronary bands, inter-digital spaces and teats in lactating animals. After rupturing, vesicles form into painful ulcerous erosions. The clinical disease can cause weight loss and drop in milk production and lameness. After resolution of clinical signs, ruminants can become persistent carriers, which is defined as having virus present (usually detected in their oesophageal-pharyngeal fluid) beyond 28 days post-infection (Zhang and Alexandersen, 2004). Virus infection results in a strong serological response against all viral proteins both structural (SP) and non-structural protein (NSP). Vaccines composed of chemically inactivated virus have been available for over 70 years and good-quality vaccines are largely devoid of NSP and induce antibodies mainly to SP (Doel, 2003). Therefore, antibodies to NSP are usually associated with viral infection, whereas the presence of antibodies only to SP is usually associated with vaccination (provided the use of good-quality vaccines) (Doel, 2003).
Despite years of FMD research, many questions remain in terms of the virus ecology and maintenance during inter-epidemics. Studies to understand viral ecology in natural conditions are difficult to carry out especially in countries with vaccination programs; therefore, few of these studies exist. Cameroon presents a unique study opportunity because there has never been a formal FMD control program and therefore vaccination against FMD is not practiced (Bronsvoort et al., 2004a,b). Previous work on FMD in Cameroon has focused on the Adamawa region during a twelve-month inter-epidemic period between 2000 and 2001, which is estimated to have two to three million head of cattle (Bronsvoort et al., 2004a). Using a cross-sectional sampling design of FMD incidence, Bronsvoort et al. (2004a, 2004b) developed a questionnaire for herdsmen in the Adamawa region. The herdsmen reported a 57.9% (50.4–65.4%) incidence of FMD in cattle during the previous 12 months. The molecular epidemiology of the disease was also investigated with both serotype SAT2 and A being present (Bronsvoort et al., 2004b). The SAT2 isolates appeared to be genetically similar with a phylogenetic distance of <6% (based on 357 base pairs in VP1 coding region) and were closely related to isolates from Eritrea and Saudi Arabia. Serotype A isolates were more diverse with a phylogenetic distance of up to 11% and forming its own monophyletic group (based on 537 base pairs in VP1 coding region). These serotype A viruses were most closely related to historical samples from Cameroon. In this same paper, the authors state that the lack of serotype O was surprising although it was found in swine being brought to slaughter. These serotype O viruses could not be clustered with any recent outbreaks.
The current report is part of a prospective study of animal diseases in sedentary and mobile herds in the Far North region of Cameroon (Healy Profitos et al., 2013; Moritz et al., 2013a). This area is a narrow region of Cameroon bordered by Chad to the east and Nigeria to the west and is considered to be one of the more neglected portions of the country in terms of basic veterinary services (R. Garabed, personal observation). The local livestock population is a mix of sedentary (village) and mobile (seasonally transhumant) animals. In general, the sedentary herds are smaller but more numerous and may contain sheep, goats, cattle and pigs, while the mobile herds are larger but fewer in number and contain cattle, sheep and goats. These two production systems share pasture and water resources as well as trade networks (markets) and veterinary services. The veterinary services in the region are primarily concerned with annual vaccination campaigns against contagious bovine pleuropneumonia, hemorrhagic septicaemia, black leg, and anthrax and regulation of the livestock markets. There are no large confined feeding operations for ungulates in the study region and herders do not appear to raise their animals with a goal of export. Most animals are consumed within the community or kept as a durable capital asset reserve to be sold at markets as needed to buy necessities. However, there are several transboundary cattle trade routes that cross the region. For the most part, cattle originate in the east (Chad and Sudan have been reported as origins for cattle found along these routes) and are moved on foot across the Far North of Cameroon on their way to being sold in Nigeria. During the ongoing study, we have not observed any west to east transboundary transport of cattle in the region. Although the trade routes are well defined and traders are not supposed to buy and sell cattle along their path, there appears to be some co-mingling between these cattle and the local cattle population.
The ongoing study has not documented the use of FMD vaccinations in animals of this region. We found that vaccines for ungulates in the region are distributed by the veterinary services (Ministry of Agriculture, Fisheries and Animal Industries, MINEPIA) and the veterinary services have not had a supply of FMD vaccine of any type in memory. This finding is supported in surveys of herders and drug sellers, who are not aware of the existence of an FMD vaccine and report never having used or sold one.
The objective of this study is to determine the serotype (s) of FMDV naturally circulating in the Far North region of Cameroon using serological evidence of infections in cattle and viruses obtained from clinical or subclinical FMD infections in Cameroon. This information will then be used to understand the antigenic and genetic relationships of FMDV strains circulating among the different livestock production systems in the Far North of Cameroon in the absence of FMD vaccination.
Materials and Methods
Sample collection
For the purposes of this study, a herd was considered to be the smallest homogeneously mixing unit of animals, which in practice meant that portions of sedentary herds that were mobile for the dry season were not counted as part of the herd. The sampling scheme for the study is summarized in Table 1. Sampling was carried out differently for each of four herd types (sedentary, mobile, market and transboundary), which are defined here: sedentary herds were located in one village and grazed around that village throughout the year with herders living in permanent structures; mobile herds had no fixed location and were seasonally transhumant with herders living in temporary shelters; market herds were artificial groupings of animals present for sale at local cattle markets on a sampling day; transboundary trade cattle were sampled at borders and local veterinary control stations where they were stopped in transit between the eastern and western borders of Cameroon.
Table 1.
Schematic of sampling
| Routine sampling |
Additional sampling | |||
|---|---|---|---|---|
| Group | Mobile herds | Sedentary herds | Transboundary trade routes/ market |
Reports of clinical disease |
| Number of herds | 15 | 15 | Variable | Variable |
| Animals/herd | 5/herda,b | 5/herda,b | 10-30/sampling day | Up to 10 infected individuals |
| Sample frequency | 2x year (rainy season/ dry season) |
2 x year (rainy season/ dry season) |
4x year(2x rainy season/2x dry season) |
Sampling at report |
| Samples/activity | Serum, probang, survey | Serum, probang, survey | Serum, probang, survey | Serum, probang, abbreviated survey, lesion swab/tissue sample/vesicular fluid |
The same five cattle were followed at each visit. Cattle lost to follow-up for any reason were replaced with their oldest descendant still present in the herd or another close relative if no direct descendants were remaining in the herd. Additionally, newly purchased cattle were tested and followed from the first visit after entry into the herd until they left the herd.
As part of a larger project, all animals (including small ruminants and all cattle over 1 year of age) for each herd were sampled once between 2011 and 2014. This paper includes samples from the first three herds.
The fifteen sedentary herds were chosen in pairs (and one triple) to represent different ecological and cultural areas in the Far North. The villages within pairs were one village with substantial contact with mobile herds and one with less contact with mobile herds. Contact with mobile herds was based on the assessment of a local expert in herder mobility. Fifteen mobile herds were selected randomly from a list of mobile herds visiting the region between 2008 and 2009 (Moritz et al., 2013b). These mobile and sedentary sentinel herds were sampled for serology and probang (oropharyngeal fluid) regularly once during the dry season (January–May) and once during the wet season (June–August) each year and the same five cattle were sampled each time. In a few instances, an animal that was sampled departed from the herd (died, sold, stolen or lent to another herd). These animals were replaced in the regular sampling by their oldest offspring still present in the herd or another close relative if no offspring were present.
In addition, these thirty sentinel herders were given cellular telephones and asked to report all outbreaks of clinical FMD. Upon a report of FMD, additional samples were taken from up to ten clinical or recovering animals (all reports were from cattle). Most outbreak samples originated in the Far North region; however, some viral samples were collected from outbreaks in the adjacent North region. This complex sampling method resulted in three types of data for mobile and sedentary herds – routine biannual samples repeated on the same animals; samples from reports of clinical FMD; and one-time whole herd samples.
The four largest cattle markets (Maroua, Bogo, Mour-voudaye and Mazara) in the study area were sampled twice during each season and 30 non-clinical cattle were sampled at each visit taking care that different areas of the market were sampled, because cattle for sale in the markets were arranged based on locations of origin. Similarly, four locations along transboundary trade routes (Yagoua, Guirvidig, Kousseri and Petté) were visited twice during each season and 10 non-clinical cattle passing that day were sampled. We had intended to sample any market or transboundary trade cattle showing signs of FMD in addition to these regular samples, but we did not observe any clinical animals on the days we visited.
After collection in the field by local para-veterinarians employed by the project and following a protocol approved by the Ohio State University Institutional Animal Care and Use Committee (protocol #2010A0018), the samples were transported to the Laboritoire National Veterinaire (LANAVET), in Garoua, and stored at −70°C until shipment to Plum Island Animal Disease Center (PIADC-ARS) in New York, USA, in two batches. The first batch of samples received at PIADC (739 probang, 15 swabs, 1 tissue and 1 footwash) was collected in transport media and shipped in dry liquid nitrogen transport tanks. These samples were collected in 2010 and 2011. The second batch of samples included probangs (n = 391) and tissue samples (n = 29), which were collected in 2011–2012. Instead of transport media, RNA-Later®, (Sigma-Aldrich, St. Louis, MO, USA) was used for sample preservation in this second batch and shipped under refrigeration rather than liquid nitrogen. Blood samples (n = 844) were also collected from cattle during the study. The blood was centrifuged and the serum was collected within 4 h of the sampling. The serum samples were then refrigerated until they were sent to LANAVET and stored at −70°C. Upon receipt at PIADC, all samples were stored at −70°C until processed.
Different subsets of the total serum samples received were used for various types of analyses. The first subset (called ‘first shipment’) included samples received in the first shipment (n = 434) and was used to assess type of exposure (i.e. vaccination versus natural infection) by determining structural protein (SP) by serum neutralization test (SNT) and/or non-structural protein (NSP) reactivity by 3-ABC ELISA. This data set included only the first sample taken from each animal. The second subset (called ‘local cattle’) was used to test correlation between the age and number of serotypes the cattle were serologically reactive against. This data subset included mobile and sedentary herds only (cattle known to be local to the region) and included only the first sample from each animal (n = 293). Market and transboundary trade cattle were excluded because the ages obtained for these cattle were not reliable. The third subset (called ‘all cattle’) was used for the cross-serotype reactivity analysis. This data subset included all herd types and multiple samples from the same cattle (n = 844, with 100 animals having duplicate samples and 25 animals having triplicate samples). This third whole data set included animals sampled at the following temporal intervals: 5 February, 2010 to 3 April, 2010 (dry season, n = 252), 23 June, 2010 to 23 August, 2010 (wet season, n = 283), 18 October, 2010 to 29 December, 2010 (outbreak sampling, n = 34), 21 January, 2011 to 4 February, 2011 (dry season, n = 151) and 1 January, 2012 to 8 February, 2012 (dry season, n = 124).
Non structural protein ELISA
A subset of 434 serum samples (first shipment) were inactivated at 56°C for 45 mins. An initial 1 : 5 dilution was made of each serum sample before testing by ELISA. To determine the presence of antibodies against the non-structural proteins (NSP) of FMDV, the serum samples were tested using a 3ABC ELISA kit (Prio Check®; Prionics, Lelystadt, The Netherlands: Product No: 7610450) following the manufacturer’s instructions.
Antibodies against structural protein measured by serum neutralization test (SNT)
Field serum samples received during 2010–2012 (n = 844) were tested for FMDV antibodies against serotypes SAT1, SAT2, SAT3, A and O using the serum neutralization test described in the OIE manual (Vallat and Edwards, 2012). The viruses used to neutralize the sera were FMDV SAT1/SW/3/49 (of South West Africa), SAT2/ZIM/5/81, SAT3/ZIM/2/83 and A22/Iraq/24/64. The choice of viruses used for neutralizations was based on the genetically closest virus available in our virus repository. These were obtained as p1 or p2 in BHK-21 cells from the pathogen repository at PIADC, USA, except for SAT1/SW/3/49, which was first propagated in lamb kidney cells and then propagated on BHK-21 cells. O/CAR/59/2010, a virus isolated during this study, was used as the Serotype O neutralizing virus. All viruses were propagated once on LFBK cells and titrated to determine the tissue culture infective dose at 50% (TCID50) before SNT was carried out.
For each test, as per OIE recommendations for SNT, a back titration was conducted to insure a challenge virus titre within an accepted range of log 1.5–2.5 and TCID50 was obtained (Vallat and Edwards, 2012). Positive control sera and cell controls were run on each plate. To the neutralization plate, each of the heat-inactivated test sera was serially diluted twofold in a 96-well plate in duplicate. Then previously titrated virus (102 TCID50) was added to each well and the plates were incubated for 1 h at 37°C. After incubation, 4.5 × 105 LFBK cells suspended in MEM media (Gibco Cat#2013-03: containing 10% bovine serum and 1% antibiotics and antimycotics) were added to each well. Plates were incubated at 37°C for 48–72 h in an atmosphere of 5% carbon dioxide. The plates were fixed by adding histochoic (Amresco Cat#H108-4G) containing crystal violet and ethanol.
The test was accepted when the back titration was within the acceptable range and the positive standard serum was within twofold of its expected titre. A serum neutralization titre of higher than log10 1.6 was considered positive and was determined by using the Kaerber method (Kaerber, 1931).
Molecular characterization of foot and mouth disease virus
FMDV RNA detection
Samples received in transport media and those received in RNA later were screened for FMDV RNA by rRT-PCR as previously described (Arzt et al., 2010; Pacheco et al., 2010). Briefly, RNA was extracted using Ambion’s Mag-Max-96 viral RNA isolation kit (Ambion, Cat #1836, Austin, TX, USA) on a King Fisher-96 Magnetic Particle Processor (Thermo-Fisher, Hudson, NH, USA). RNA was analysed by rRT-PCR on the ABI 7000 (Applied Biosystems, Life Technologies Inc., Carlsbad, CA, USA) following the previously described protocol (Callahan et al., 2002). Samples were considered positive when Ct values were <40.
Virus isolation
Samples received in transport medium that tested positive by rRT-PCR were treated with Trichlorotrifluoroethane (TTE) dissociate virus–antibody complexes and increase the chance of isolating infectious virus in tissue culture (Pacheco et al., 2010). Tissue samples were macerated with stainless steel beads (Qiagen, Cat#69989) and MEM-25 mm Hepes. Samples were then shaken in a TissueLyser bead beater (Qiagen, Venlo, Limburg, the Netherlands). The supernatant was processed through a Spin-X tube to filter the sample (Spin-X, Costa, Sigma-Aldrich). The filtrate (50 µl) was used for RNA extraction and screened by rRT-PCR as described previously. The remaining sample was stored at −70°C until further use.
Virus isolation was carried out using LFBK αVβ6, a bovine kidney cell line highly sensitive to all FMDV serotypes (Swaney, 1988; LaRocco et al., 2013). In brief, 80% confluent LFBK αVβ6 cells in flasks (T-25) were rinsed with serum free media and inoculated with 150 µl of the processed tissue or probang sample. One flask was set up for each sample. After 1 h of adsorption at 37°C on a rocker plate, 5 ml of cell maintenance media with 1% serum was added to each flask. Inoculated flasks were incubated at 37°C for 48 h. Supernatants from samples showing cytopathic effect (CPE) were collected and stored at −70°C for further testing. Samples not showing CPE were subjected to three blind passages in LFBK αVβ6 cells before they were deemed negative.
Phylogenetic analyses
For the tissue and probang samples that were positive by rRT-PCR, the viral RNA was used for sequencing (Pacheco et al., 2010). The RT-PCR products were generated using SuperScript®III One-Step RT-PCR System with Platinum® Taq High Fidelity (Invitrogen, Carlsbad, CA, USA). The amplification primers were designed to universally amplify the entire P1 region of FMDV (Xu et al., 2006). The RT-PCR products were sequenced using the di-deoxy termination method (Big dye terminator; Life Technologies) and analysed as previously described (Pauszek et al., 2011). Additional internal sequencing primers specific to the Cameroon isolates were used to obtain the complete VP1 sequence. Chromatograms were viewed using Sequencher® v4.8 (GeneCodes, Ann Arbor, MI, USA) and a consensus sequence was assembled for each virus VP1 region. The sequences were aligned using ClustalW algorithm (Larkin et al., 2007) implemented in Mega5© (Tamura et al., 2011).
For both the serotype SAT2 and serotype O, phylogenetic trees, the evolutionary parameters were estimated using ModelTest v3.7 and the hierarchical likelihood ratio test (hLRT) (Posada and Crandall, 1998). The most likely model was found to be TRN+I+G for serotype O and HKY+I+G for serotype SAT2. PAUP* 4.0b10 was used to ascertain a likelihood tree that was mid-point rooted for serotype O and rooted by UGA/MBF4/2002 for serotype SAT2 (Swofford, 2002). Topotypes were determined by grouping with previously identified prototype strains (Knowles and Samuel, 2003).
Statistical analysis
To examine relationships between (i) age and seropositivity, and (ii) management type and seropositivity, SNT results (positive versus not positive and number of serotypes) from the local cattle subset (n = 293) were used to calculate descriptive statistics and contingency tables in SPSS (IBM SPSS Statistics version 21, IBM, 2012, Armonk, NY, USA). Serotype-specific results from the SNTs of 844 cattle samples (all cattle) were tested for pairwise independence using Pearson’s chi-square test; temporal subsets were tested for pairwise independence using Fisher’s exact test, with consistency of results compared across all five subsets. Tests for independence were conducted first by using the raw log values for SNT titre (see supporting information) and second by including uncertainty introduced by the inherent variability in SNT results. To consider the uncertainty, titre values were re-sampled 1 000 000 times with replacement from a uniform distribution with a mean of the raw titre values and a variance of log10 0.3. The same chi-squared and Fisher’s exact statistics were calculated for each set of re-sampled values and the number of times the null hypothesis of independence was not rejected was recorded. If re-sampled tests resulted in a P-value greater than the significance value of 0.05 more than half of the time, that is, in at least 500 000 of the iterations, we considered the results of the test to be consistent within the known variability of the SNT test. The analysis was conducted using R version 2.15 (R Team, 2012).
Results
Serological response to non-structural and structural viral proteins
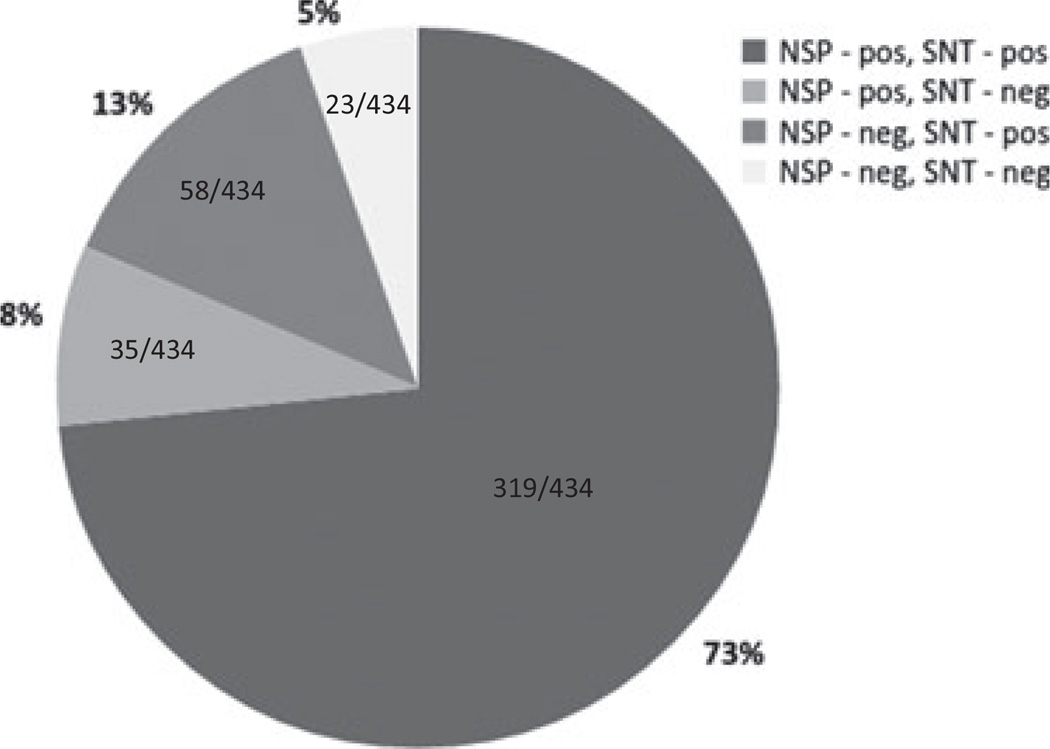
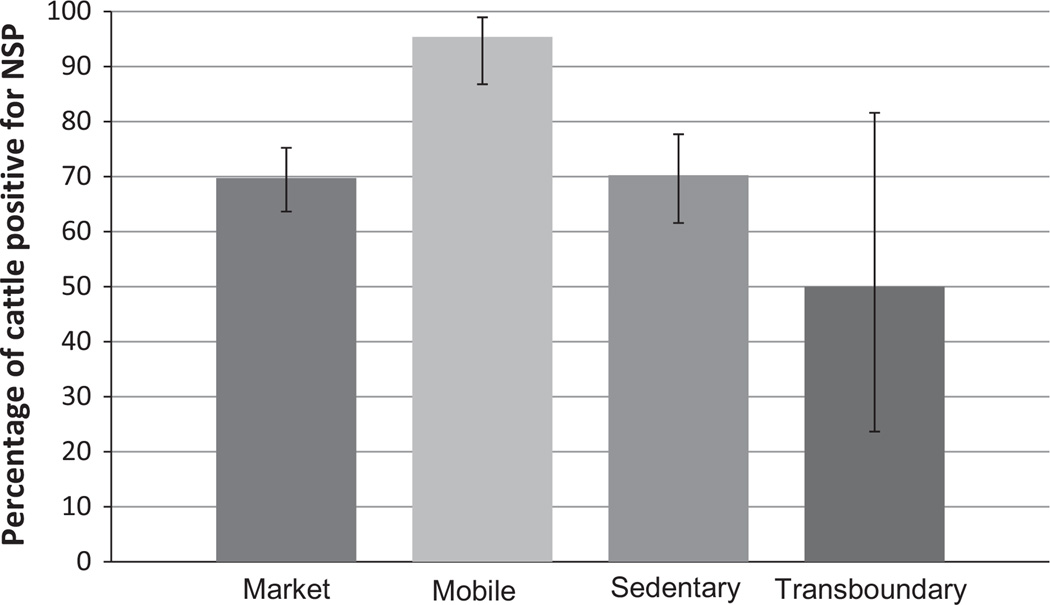
Among the first shipment animals (434 tested by SNT and NSP ELISA), 73% (318/434) were positive for both neutralizing (structural protein) and non-structural protein (3ABC) antibodies, which is consistent with natural infection by FMDV (Fig. 1). A proportion [13% (58/434)] of animals had neutralizing antibodies and non-detectable NSP antibodies, a serological status usually associated with vaccination (Brocchi et al., 2006). The distribution of these serological reactions was even among groups except for the mobile group with a smaller proportion of animals having neutralizing antibodies and non-detectable NSP antibodies. NSP-seropositivity (indicative of previous FMDV infection) was highest among the mobile group (Fig. 2). There was also a small percentage [8% (35/434)] that had NSP antibodies in the absence of neutralizing activity (Fig. 1). Five percent (23/434) of the animals did not have antibodies against either non-structural or structural proteins.
Fig. 1.
Percent positivity to NSP-EUSA and SNT in sera from cattle (n = 434) sampled between 2010 through 2011 in Cameroon. A serum titre above log 1.6 TCID50 is considered positive on SNT. For the NSP ELISA samples showing percent inhibition (PI) above or equal to 50% are considered positive.
Fig. 2.
Distribution of animals by management type with serological non-structural viral proteins (NSP) positive results, which could be consistent with active or past infection of FMDV. The total number of animals per management type was the following: 238 for market, 65 for mobile, 121 for sedentary and 10 for transboundary. The error bars are the 95% confidence interval with no correction for clustering within herds or on sampling days.
Serotype specific serology
Serum neutralization tests against five FMDV serotypes (A, O, SAT1, SAT2 and SAT3) were carried out on each serum. We tested the cross-reactivity among the control sera and viruses being used. No cross-reactivity was seen among serotypes with all neutralization values being <log 1.3 (the lowest serum dilution analysed).
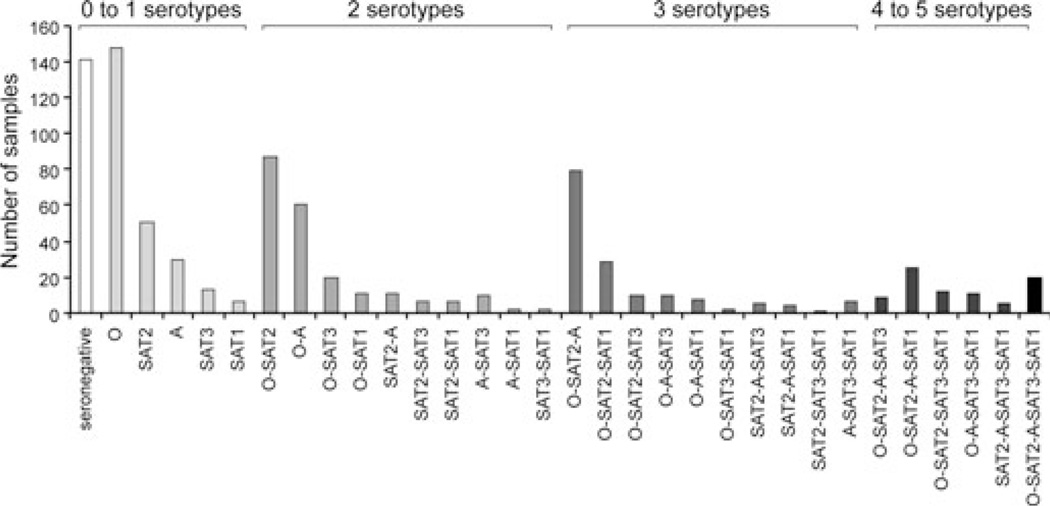
The distribution of animals with serological reactions against multiple serotypes is shown in Fig. 3. Most cattle sera were reactive to either one or two serotypes and this result did not vary with management type (market, mobile or sedentary group). Some animals had antibodies against all five serotypes tested (Fig. 3). The highest serological prevalence was for serotype O, followed by serotypes SAT2 and A (Fig. 3). This result must be taken with care as serotype O was the only one tested with a local circulating viral strain. However, there was a clear dominance of serotypes O, SAT2 and A either as single or multiple serological reactions (Fig. 3). There was a small number of animals with single reactivity to serotypes SAT1 and SAT3, suggesting the presence of these serotypes in Cameroon. We tested if the number of positive serotype reactivity increased with age. A weak but significant positive correlation was found between the number of serotypes and age (Pearson’s correlation = 0.387; P-value < 0.000) (local cattle, n = 293). A small margin of the local cattle (15%; n = 44) was negative for neutralizing antibodies against all serotypes on their first test. These neutralization seronegative cattle appeared to be slightly younger than sero-positive animals (mean age 2.20 years lower for seronegatives; 95% CI 1.18–3.22) and were more likely to be in sedentary herds (relative risk for seropositivity mobile versus sedentary 1.12; 95% CI 1.02–1.24).
Fig. 3.
Distribution of cattle with serological response (neutralizing activity) to single or multiple FMDV serotypes. The data were collected from 2010 to 2012, represent 844 samples from 694 unique cattle and include both subclinical and clinical animals.
To evaluate potential associations between seropositivity to individual serotypes, we carried out chi-square tests for independence including uncertainty in the test results, on five subsets of data representing different collection dates (all cattle, n = 844). When correlation was indicated by significant P-values in more than half of the iterations, the subset was considered correlated. The procedure was repeated and the cumulative number of subsets showing correlation between serotypes was recorded in Table 2. Scores of zero indicate independence across all time periods; conversely, scores of five indicate correlation across all time periods. Scores ranging from one to four indicate the number of subsets for which correlation was observed. Trivially, each serotype displayed correlation with itself across all five subsets as is indicated on the diagonal of the results table. Serotypes SAT1 and SAT3 were correlated in four of five subsets. Serotypes SAT3 and A, SAT2 and O, and A and O were correlated in three subsets. Seropositivity to serotypes SAT1 and SAT2, SAT1 and A, and SAT1 and O was correlated in two subsets. SAT2 and A and SAT3 and O were correlated in one subset and SAT 2 and SAT3 were never correlated (Table 2).
Table 2.
Frequency of correlated titre results among five different temporal sampling subsets (dry season 2010, wet season 2010, outbreak sampling 2010, dry season 2011 and dry season 2012). Correlation was determined by iterated Fisher’s exact tests for independence incorporating uncertainty by re-sampling from the known test variability
| SAT1 | SAT2 | SAT3 | O | A | |
|---|---|---|---|---|---|
| SAT1 | 5 | 2 | 4 | 2 | 2 |
| SAT2 | 2 | 5 | 0 | 3 | 1 |
| SAT3 | 4 | 0 | 5 | 1 | 3 |
| O | 2 | 3 | 1 | 5 | 3 |
| A | 2 | 1 | 3 | 3 | 5 |
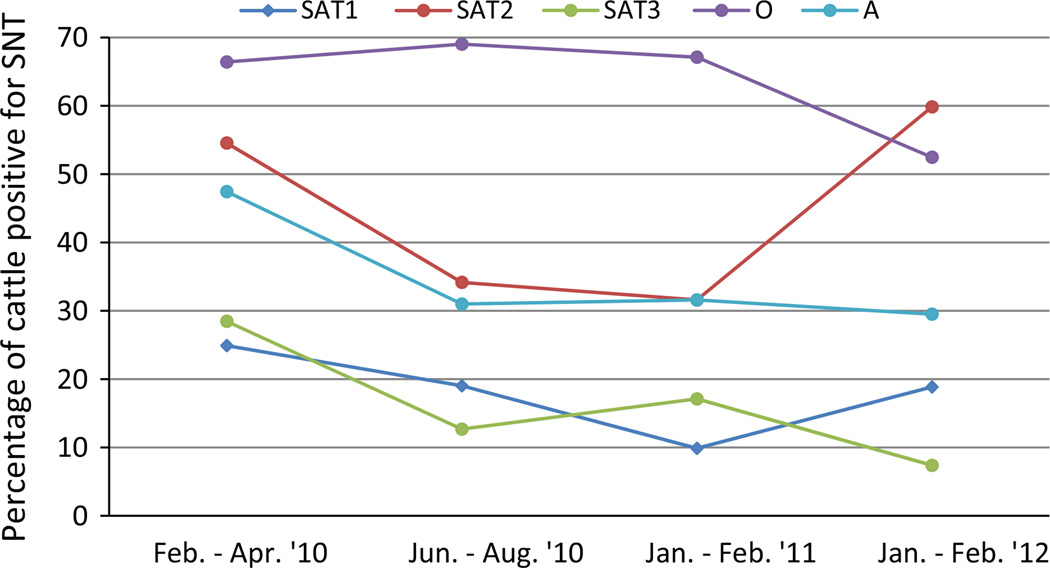
Analysing the serological prevalence by sampling period showed that until 2011, the highest serological prevalence was for serotype O. This pattern changed with SAT2 becoming more prevalent than serotype O by the dry (winter) season of 2011–2012 (Fig. 4).
Fig. 4.
FMDV serotypes prevalent in Cameroon from 2010 to 2012 in cattle. A total of 844 serum neutralization values were analysed. Serum neutralization tests with values above log 1.6 TCID50 were considered to be positive. These samples were taken during routine and outbreak sampling. Standard error was <0.09% in all cases (without controlling for correlation within herds or sampling days).
Viral detection and characterization
Probang and/or clinical samples were received in two separate shipments. The first set were collected and stored in transport media, which allows for virus isolation. From this first set of probang samples, 29 of 739 (3.92%) were positive by rRT-PCR and two viruses were isolated and determined to be of serotype O topotype East African group 3 (Table 3 and Fig. 5a). Three of 15 oral swabs were positive by rRT-PCR and both a tissue and foot-wash samples were negative. The low number of rRT-PCR-positive samples and even fewer samples yielding infectious virus could have been due to virus degradation during transfer from the field. Therefore, the second set of samples (391 probangs) was collected and shipped in RNA-Later®, which prevented virus isolation, but resulted in a greater number of rRT-PCR-positive samples and subsequently viral sequences. There were 39 rRT-PCR-positive probang samples (10%) in this second shipment. Interestingly, only 3% of tongue tissues (4/14) received were positive by rRT-PCR, while 79% of foot lesions (11/14) were positive. There did not seem to be a difference in percent rRT-PCR positive between the different herd types.
Table 3.
VP1 genetic sequences obtained during this investigation
| Genbank ID | Collection date | Herd type | Sample type | Last FMD outbreak | Serotype |
|---|---|---|---|---|---|
| CAR/59/2010 | 6-February-10 | Sedentary | Probang in VTM* | None | O |
| CAR/61/2010 | 6-February-10 | Sedentary | Probang in VTM* | January 2010 | O |
| CAR/766/2012 | 31-January-12 | Sedentary | Probang in RNALater | January 2012 | O |
| CAR/769/2012 | 31-January-12 | Sedentary | Probang in RNALater | January 2012 | O |
| CAR/773/2012 | 31-January-12 | Sedentary | Probang in RNALater | January 2012 | O |
| CAR/791/2012 | 31-January-12 | Sedentary | Tissue in RNA Later | January 2012 | O |
| CAR/777/2012 | 31-January-12 | Sedentary | Probang in RNALater | January 2012 | O |
| CAR/786/2012 | 31-January-12 | Sedentary | Probang in RNALater | January 2012 | O |
| CAR/776/2012 | 31-January-12 | Sedentary | Probang in RNALater | January 2012 | O |
| CAR/820/2012 | 6-February-12 | Mobile | Probang in RNALater | January 2012 and December 2011 | O |
| CAR/909/2012 | 3-February-12 | Response to outbreak | Probang in RNALater | February 2012 | SAT 2 |
| CAR/910/2012 | 3-February-12 | Response to outbreak | Probang in RNALater | February 2012 | SAT 2 |
| CAR/911/2012 | 3-February-12 | Response to outbreak | Probang in RNALater | February 2012 | SAT 2 |
Virus transfer media.
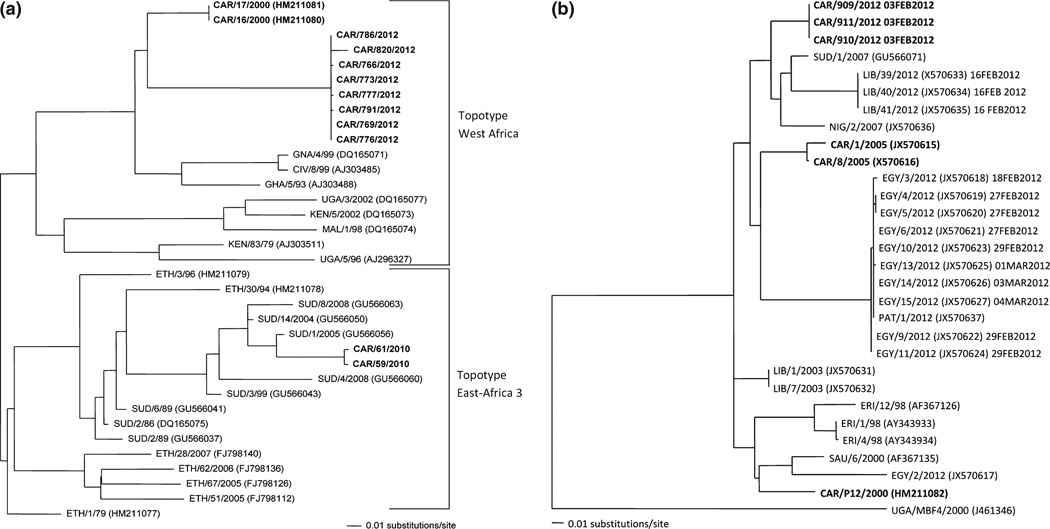
Fig. 5.
Maximum likelihood phylogenetic trees of Cameroon serotype O (a) or SAT2 isolates (b). The phlyogenetic trees are based on 633 nucleotides of VP1. The mid-point tree shown in (a) highlights the serotype O sequences from Cameroon (CAR). West Africa (WA) topotype Cameroon viruses are from 2000 and 2012 and the East Africa (EA) topotype viruses are from 2010. The serotype SAT2 phylogenetic tree (b) above is rooted by UG/V MBF4/2002. Cameroon (CAR) samples are in bold. This phylogenetic tree is of the complete VP1 region. Two recent FMDV outbreaks are of interest: Libya and Egypt shown in blue and green, respectively. The dates of when these samples as well as the SAT2 viruses sequenced for this study have been added. Trees were generated by maximum likelihood algorithm (within PAUP) using the evolutionary model TRN+I+G, no bootstrap values are generated by this method.
Among all the rRT-PCR-positive samples, we were able to obtain sequence data from 13 samples representing serotypes O and SAT2 (Table 3). These and other available sequences in GenBank were used to reconstruct the phylogenies for the two serotypes found. Phylogenetic analysis of serotype O sequences showed that the 2010 virus grouped within EA-3 (East Africa-3), while three 2012 viruses grouped with topotype WA (West Africa) (Fig. 5a).
The phylogeny of serotype SAT2 showed Cameroon viruses from 2005 were distinct from those circulating in 2012. Interestingly, the three Cameroon SAT2 isolates from 2012 grouped within the same lineage causing the SAT2 incursion into Northern African, specifically in Libya (Fig. 5b).
Discussion
To the authors’ knowledge, vaccination is not carried out in the region of Cameroon where this study occurred. To confirm the infection or vaccination status of the animals included in this study, we tested their sera for the presence of antibodies to non-structural proteins (3ABC ELISA), usually indicative of active FMDV infection. Virus neutralizing activity, which reflects presence of antibodies to the viral capsid (structural proteins), can be induced either by virus infection or by vaccination. The presence of neutralizing antibodies and the absence of NSP antibodies could therefore suggest vaccination without infection. The current data indicate that 13% of cattle (58/434) were SNT positive and NSP negative. This might suggest that the animals were exposed to structural proteins and not nonstructural proteins of FMDV (i.e. vaccination). This result was unexpected as all other survey data suggested that no vaccination is practiced in the study region. One possible explanation for this finding is that these animals were, in fact, vaccinated. This may especially be true for animals that were tested at the market for these animals may have come from other countries where vaccination occurs. However, when comparing the different types of herds (market, sedentary, mobile and transboundary), the percentage of animals being SNT positive and NSP negative were similar among all groups with a slightly lower percentage for the mobile group. Another, more likely explanation for this result is that there are differences in the level of detection of the serological tests used: ELISA versus SNT. A study comparing various NSP ELISA tests showed that the ELISA used in our study had a sensitivity of 92.5 (86.3–97.4) and a specificity of 84.0 (77.6–90.1) (Goris et al., 2007). It is likely that the SNT test used here has a higher sensitivity than NSP ELISA. However, conclusions regarding these serological reactions cannot be made with our data and is beyond the scope of this work. No animals <1 year of age were sampled, so it is unlikely that maternal immunity is confounding these results.
We found antibody reactivities against five serotypes of FMDV through serological surveillance. However, only two serotypes (O and SAT2) were found among the viral samples tested in this study and a previous viral isolate of serotype A has been reported in Cameroon (Bronsvoort et al., 2004b). In the absence of viral isolates for SAT1 and SAT3 serotypes in Cameroon, it is difficult to determine the origin of the serological reactivity observed. The presence of animals with single serological reactivity to either SAT1 or SAT3, shown in Fig. 3, supports the presence of these serotypes. To determine the level of cross-reactivity among serotypes, we tested for correlations between serotype-specific seropositivity using chi-square and Fisher’s exact tests in data subsets each representing a different sampling period as shown in Fig. 4. Correlations between SAT1 and SAT3 seropositivity existed in four of five data subsets according to sampling date, indicating a high level of correlation. This could suggest long-term co-circulation or systematic diagnostic cross-reactivity due to heterotypic immune responses or recombination events. On the other hand, we carried out a similar analysis but using only sera with single serotype reactivity and found SAT1 and SAT3 peaks did not correspond with those of the other serotypes, indicating that these reactivities are not the result of cross-serotype reactions between SAT1 or SAT3 and the other serotypes (Figure S1). Although cross-reactivity among monospecific sera raised against each of the five serotypes against each other was not observed in the lab, at the positive cut-off level, we could not rule out heterotypic immune responses or recombination across serotypes. Heterotypic immune responses have previously been observed when infection with one serotype causes increased titre values against a different serotype, and may even result in the highest titre values corresponding to a serotype different than the one causing the active infection (Hedger et al., 1982). An alternate explanation for the observed serological correlation is the potential existence of field viruses containing antigenic epitopes of more than one serotype (e.g. SAT2 and SAT3 or SAT1 and SAT2). Recombination events are known to occur among FMDV serotypes (Jackson et al., 2007). Although not reported to date for SAT serotypes, recent reports document the occurrence of inter-serotypic recombination between serotypes A and Asia 1, resulting in altered antigenic characteristics (Jamal et al., 2011). It is hoped that with added surveillance currently being undertaken in Cameroon, additional strains will be isolated and allow more conclusive evidence as to the presence of the SAT1 and SAT3 serotypes in Cameroon.
Fisher’s exact tests showed correlation between serotypes at specific time intervals, which suggest co-circulating sub- types with patterns that vary through time. For example, correlation indicating possible co-circulation of SAT1 and SAT2 was observed during the first and second time intervals, between February 2010 and August 2010. Similarly, co-circulation of SAT3 and A was implicated by correlation of seropositive results during the second, fourth and fifth time periods, which include data sampled between February 2010 and April 2010, January 2011 to February 2011, and January 2012 to February 2012. In this endemic area, there may be multiple serotypes of FMDV co-circulating.
Serum neutralization tests also showed that some individual animal sera showed measurable neutralizing activity against up to five FMDV serotypes. However, most animals, regardless of their age or their management type (i.e. mobile, market, sedentary, etc.) had SNT titres to one, two or three serotypes. We tested the hypothesis that serological reactivity to multiple serotypes increased with age. There was a weak positive correlation between the age of the animals and the number of serotypes to which they reacted. This weak correlation suggests that other epidemiological factors such as time of infection and longevity of antibody titres or cross-reaction among serotypes might be at play. The detection of seropositivity against multiple serotypes in individual animals raises several intriguing questions pertaining to FMDV pathogenesis, particularly regarding the possibilities of sequential versus concomitant infection with different viruses.
Two different serotype O topotypes, WA and EA-3, were found in Cameroon. The EA-3 topotype had been found in Sudan previously, but finding this topotype outside of Eastern Africa is a reminder that FMDV strains are not geographically restricted and have the potential to spread throughout Africa. The east to west transboundary trade in this region suggests a potential route for transmission. SAT2 was the second most prevalent serotype throughout the study. Furthermore, in the dry season 2012 sampling period, SAT2 surpassed serotype O in prevalence. Interestingly, this serotype made an incursion into Northern Africa causing extensive outbreaks in Libya and Egypt in 2012. Coincidentally, SAT2 viruses isolated during an outbreak in the North Region of Cameroon (Garoua-Boulaye) in 2012 grouped closely with the viruses from northern Africa, suggesting a common origin among these viruses. Our data indicate the widespread distribution of the SAT2 strain, found in Libya and Egypt, in Western Africa.
Further knowledge of FMDV circulating in endemic regions, such as Cameroon, is critical to understanding viral ecology and preventing and controlling FMDV in endemic regions (Ferguson et al., 2013). Furthermore, viral strains circulating in Western Africa could cause outbreaks in disease free countries, greatly affecting their economy or cause severe outbreaks in regions were animals are not protected against specific FMDV strains. This scenario highlights the importance of further surveillance of FMD in Africa and the need to better understand the movement of people, animals and viruses.
In conclusion, the data presented herein indicate that within the Far North region of Cameroon, where no known vaccination occurs, five different serotypes of FMDV are circulating: serotypes O, A, SAT1, SAT2 and SAT3. We confirmed the presence of serotypes O and SAT2 viruses by virus isolation and sequencing and serotype A was previously confirmed. However, further studies are needed to confirm the presence of serotypes SAT1 and SAT3 in Cameroon. The SAT2 virus found was closely related to that seen in Libya, suggesting a common origin for these outbreaks. This study documents a diverse population of FMDV serotypes and strains within Cameroon that could threaten other regions of Africa. This highlights the importance of carrying out surveillance, not only in Cameroon but also other countries where FMD control programs are currently not in place. Studying FMD in a natural setting without vaccination provides the opportunity for an in-depth understanding of the disease ecology, which is critical information for establishing future control programs.
Acknowledgements
The authors would like to thank Mike Larocco for his technical support, Penny Rempe for administrative assistance, Tim Vojt for help with Table 1 and Figure 3 and Karla Moreno Torres for helpful discussions. This project was funded through an inter-agency agreement with the Science and Technology Directorate of the U.S. Department of Homeland Security under Award Number HSHQDC-12-X-00060, award number DEB-1015908 from the National Science Foundation, award number R24-HD058484 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development awarded to the Ohio State University Initiative in Population Research and a pilot grant awarded by the Public Health Preparedness for Infectious Diseases program at the Ohio State University. Zaheer Ahmed and Anna Ludi are the recipients of a Plum Island Animal Disease Center Research Participation Program fellowship, administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). All opinions expressed in this paper are the author’s and do not necessarily reflect the policies and views of the USDA, DOE, or ORAU/ORISE. LANAVET, The Ohio State University and USDA-ARS FADRU are members of the Global Foot-and-Mouth Disease Research Alliance (GFRA).
References
- Ahmed HA, Salem SA, Habashi AR, Arafa AA, Ag-gour MG, Salem GH, Gaber AS, Selem O, Abdelkader SH, Knowles NJ, Madi M, Valdazo-Gonzalez B, Wadsworth J, Hutchings GH, Mioulet V, Hammond JM, King DP. Emergence of foot-and-mouth disease virus SAT 2 in Egypt during 2012. Transbound. Emerg. Dis. 2012;59:476–481. doi: 10.1111/tbed.12015. [DOI] [PubMed] [Google Scholar]
- Animal Production. [accessed December, 2013];Animal Production-Southern African Development Community. 2012 Available at http://www.sadc.int/fanr/livestock/print/animal_production.php.
- Arzt J, Pacheco JM, Rodriguez LL. The early pathogenesis of foot-and-mouth disease in cattle after aerosol inoculation. Identification of the nasopharynx as the primary site of infection. Vet. Pathol. 2010;47:1048–1063. doi: 10.1177/0300985810372509. [DOI] [PubMed] [Google Scholar]
- Brocchi E, Bergmann IE, Dekker A, Paton DJ, Sam-min DJ, Greiner M, Grazioli S, De Simone F, Yadin H, Haas B, Bulut N, Malirat V, Neitzert E, Goris N, Parida S, Soren-sen K, De Clercq K. Comparative evaluation of six ELISAs for the detection of antibodies to the non-structural proteins of foot-and-mouth disease virus. Vaccine. 2006;24:6966–6979. doi: 10.1016/j.vaccine.2006.04.050. [DOI] [PubMed] [Google Scholar]
- Bronsvoort BM, Nfon C, Hamman SM, Tanya VN, Etching RP, Morgan KL. Risk factors for herdsman-reported foot-and-mouth disease in the Adamawa Province of Cameroon. Prev. Vet. Med. 2004a;66:127–139. doi: 10.1016/j.prevetmed.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Bronsvoort BM, Radford AD, Tanya VN, Nfon C, Etching RP, Morgan KL. Molecular epidemiology of foot-and-mouth disease viruses in the Adamawa province of Cameroon. J. Clin. Microbiol. 2004b;42:2186–2196. doi: 10.1128/JCM.42.5.2186-2196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan JD, Brown F, Osorio FA, Sur JH, Kramer E, Long GW, Lubroth J, Ellis SJ, Shoulars KS, Gaffney KL, Rock DL, Nelson WM. Use of a portable realtime reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 2002;220:1636–1642. doi: 10.2460/javma.2002.220.1636. [DOI] [PubMed] [Google Scholar]
- Doel TR. FMD vaccines. Virus Res. 2003;91:81–99. doi: 10.1016/s0168-1702(02)00261-7. [DOI] [PubMed] [Google Scholar]
- Ferguson KJ, Cleaveland S, Haydon DT, Caron A, Kock RA, Lembo T, Hopcraft JG, Chardonnet B, Nyariki T, Keyyu J, Paton DJ, Kivaria FM. Evaluating the potential for the environmentally sustainable control of foot and mouth disease in sub-Saharan Africa. EcoHealth. 2013;10:314–322. doi: 10.1007/s10393-013-0850-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris N, Praet N, Sammin D, Yadin H, Paton D, Brocchi E, Berkvens D, De Clercq K. Foot-and-mouth disease non-structural protein serology in cattle: use of a Bayesian framework to estimate diagnostic sensitivity and specificity of six ELISA tests and true prevalence in the field. Vaccine. 2007;25:7177–7196. doi: 10.1016/j.vaccine.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Grubman MJ, Baxt B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004;17:465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J. [accessed December, 2013];OIE/FAO FMD Reference Laboratory Network Annual Report. 2011 Available at http://www.wrlfmd.org/ref_labs/ref_lab_reports/OIE-FAO%20FMD%20Ref%20Lab%20Network%20Report%20201l.pdf.
- Healy Profitos JM, Moritz M, Garabed RB. What to Do with Chronically Sick Animals? Pastoralists’ management strategies in the far north region of Cameroon. Pastoral-ism Res. Policy and Practice. 2013;3:8. doi: 10.1186/2041-7136-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedger RS, Barnett BA, Gradwell DV, Travassos Dias P. Serological tests for foot-and-mouth disease in bovine serum samples. Problems of interpretation. Rev. Sci. Tech. 1982;1:387–393. doi: 10.20506/rst.1.2.75. [DOI] [PubMed] [Google Scholar]
- Holliman A. Differential diagnosis of disease causign oral lesions in cattle. In Practice. 2005;27:2–13. [Google Scholar]
- Jackson AL, O’Neill H, Maree F, Blignaut B, Carrillo C, Rodriguez L, Haydon DT. Mosaic structure of foot-and-mouth disease virus genomes. J. Gen. Virol. 2007;88:487–492. doi: 10.1099/vir.0.82555-0. [DOI] [PubMed] [Google Scholar]
- Jamal SM, Ferrari G, Ahmed S, Normann P, Bel-sham GJ. Molecular characterization of serotype Asia-1 foot-and-mouth disease viruses in Pakistan and Afghanistan; emergence of a new genetic Group and evidence for a novel recombinant virus. Infect. Genet. Evol. 2011;8:2049–2062. doi: 10.1016/j.meegid.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kaerber G. Beitrag zur Kollektiven Behendlung Phanrak-ologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 1931;162:480–487. [Google Scholar]
- Knight-Jones TJ, Rushton J. The economic impacts of foot and mouth disease - what are they, how big are they and where do they occur? Prev. Vet. Med. 2013;112:161–173. doi: 10.1016/j.prevetmed.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles NJ, Samuel AR. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 2003;91:65–80. doi: 10.1016/s0168-1702(02)00260-5. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Hig-gins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- LaRocco M, Krug PW, Kramer E, Ahmed Z, Pacheco JM, Duque H, Baxt B, Rodriguez LL. A continuous bovine kidney cell line constitutively expressing bovine alp-havbeta6 integrin has increased susceptibility to foot-and-mouth disease virus. J. Clin. Microbiol. 2013;51:1714–1720. doi: 10.1128/JCM.03370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart C, Sumption K, Lubroth J. Foot-and-mouth disease caused by serotype SAT2 in Egypt and Libya: a regional concern for animal health in North Africa and the Middle East. Empres. Watch. 2012;25:1–7. [Google Scholar]
- Moritz M, Ewing D, Garabed RB. On not knowing zoonotic diseases: Pastoralists’ ethnoveterinary knowledge in the far north region of Cameroon. Hum. Organ. 2013a;72:1. doi: 10.17730/humo.72.1.72672642576gw247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Scholte P, Hamilton IM, Kari S. Pastoral management of common-pool resources in the Chad Basin. Hum. Ecol. 2013b;41:351–365. [Google Scholar]
- Pacheco JM, Arzt J, Rodriguez LL. Early events in the pathogenesis of foot-and-mouth disease in cattle after controlled aerosol exposure. Vet. J. 2010;183:46–53. doi: 10.1016/j.tvjl.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Pauszek SJ, Barrera Jdel C, Goldberg T, Allende R, Rodriguez LL. Genetic and antigenic relationships of vesicular stomatitis viruses from South America. Arch. Virol. 2011;156:1961–1968. doi: 10.1007/s00705-011-1081-1. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Swaney LM. A continuous bovine kidney cell line for routine assays of foot-and-mouth disease virus. Vet. Microbiol. 1988;18:1–14. doi: 10.1016/0378-1135(88)90111-3. [DOI] [PubMed] [Google Scholar]
- Swofford D PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, Massachusettes: Sinauer Associates; 2002. Version, 4 edn. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mot Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team DRC. [accessed May 19, 2012];R: a language and environment for statistical computing. 2012 Available at http://www.R-project.org. [Google Scholar]
- Vallat B, Edwards S. [accessed March 14, 2014];OIE terrestrial manual chapter 2.1.5 foot-and-mouth-disease. 2012 Available at http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.01.05_FMD.pdf.
- Xu L, Hurtle W, Rowland JM, Casteran KA, Bucko SM, Grau FR, Valdazo-Gonzalez B, Knowles NJ, King DP, Beckham TR, Mcintosh MT. Development of a universal RT-PCR for amplifying and sequencing the leader and capsid-coding region of foot-and-mouth disease virus. J. Virol Methods. 2006;189:70–76. doi: 10.1016/j.jviromet.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Alexandersen S. Quantitative analysis of foot-and-mouth disease virus RNA loads in bovine tissues: implications for the site of viral persistence. J. Gen. Virol. 2004;85:2567–2575. doi: 10.1099/vir.0.80011-0. [DOI] [PubMed] [Google Scholar]