Abstract
Periodontitis (PD) is an inflammatory disease of the periodontal tissues that compromises tooth support and can lead to tooth loss. Although bacterial biofilm is central in disease pathogenesis, host response plays an important role in the progression and severity of PD. Indeed, clinical genetic studies indicate that PD is 50% heritable. In this study, we hypothesized that LPS injections lead to a strain-dependent periodontal bone loss pattern. We utilized five inbred mouse strains that derive the recombinant strains of the hybrid mouse diversity panel (HMDP). Mice received P. gingivalis-LPS injections for six weeks. Micro-CT analysis demonstrated a statistically significant strain-dependent bone loss. The most susceptible strain, C57BL/6J, had a 5-fold higher LPS-induced bone loss compared to the most resistant strain, A/J. More importantly, periodontal bone loss revealed 49% heritability, which closely mimics PD heritability for patients. To further evaluate functional differences that underlie periodontal bone loss, osteoclast numbers of C57BL/6J and A/J mice were measured in vivo and in vitro. In vitro analysis of osteoclastogenic potential showed a higher number of osteoclasts in C57BL/6J compared to A/J mice. In vivo LPS-injections statistically significantly increased osteoclasts numbers in both groups. Importantly, the number of osteoclasts was higher in C57BL/6J vs. A/J mice. These data support a significant role of the genetic framework in LPS-induced periodontal bone loss and the feasibility of utilizing the HMDP to determine the genetic factors that affect periodontal bone loss. Expanding these studies will contribute in predicting patients genetically predisposed to PD and in identifying the biological basis of disease susceptibility.
Keywords: Alveolar bone, animal model, genome, lipopolysaccharide
Introduction
Periodontitis (PD) is “an inflammatory disease of the supporting tissues of the teeth caused by specific microorganisms or groups of specific microorganisms, resulting in progressive destruction of the periodontal ligament and alveolar bone with pocket formation, recession, or both” (1). According to the WHO, PD is a major cause of tooth loss in adults over the age of 40 (2).
Although bacterial biofilm is central in disease pathogenesis, strong evidence supports that the patient’s genetic framework significantly modifies the response of periodontal tissues (3). Polymorphisms in cytokine-, surface receptor-, metabolism-, antigen recognition- and immunity receptor- related genes are associated with PD (3, 4). Moreover, twin studies have provided valuable support for the genetic influence in periodontal disease (5–8), estimating that PD is 50% heritable (6).
The complexity of PD, the heterogeneous genetic composition of patients, and the difficulty to control environmental parameters pose challenges to clinical genetic studies (4, 9), making animal models an attractive complement to human studies. Indeed, mouse studies on experimental periodontitis induced by Porphyromonas gingivalis (P. gingivalis) colonization reveal a strong genetic component in periodontal disease resistance and susceptibility and demonstrate that genetic determinants affect bacterial colonization, as well as periodontal bone levels (10, 11).
These studies provide valuable insight in the heritable aspects of periodontitis as a whole. However, PD is a multifactorial process that involves among others, bacterial colonization, biofilm organization and establishment, inflammatory host response, periodontal bone loss, and decreased tooth support (1). In order to begin dissecting the genetic influence in these pathogenetic disease processes individually, we explored the heritable nature of periodontal bone loss in response to a controlled inflammatory impact, by utilizing the five parental inbred strains of the Hybrid Mouse Diversity Panel (HMDP) (12, 13) and a well-characterized animal model that employs localized LPS delivery to the periodontal tissues (14–17).
Materials and Methods
Mice
Six-week-old male mice (A/J, DBA/2J, C3H/HeJ, BALBc/J, C57BL/6J) were obtained from the Jackson Laboratories (Bar Harbor, ME). In brief, mice were maintained in a temperature and light-controlled environment at UCLA. They were fed a standard chow. All mice were handled according to protocols approved by the Office for Protection of Research Subjects at UCLA and conforms to the ARRIVE guidelines (18).
Inflammatory Bone Loss Model
Mice were anesthetized with 3% isoflurane administered through a nose cone. Under the microscope (Leica Microsystems, Buffalo Grove, IL.), mice received 2 μl (20 μg) of P. gingivalis-LPS (InvivoGen, San Diego, CA) injections in between the 1st and 2nd maxillary molars on both sides of the maxilla, 2 times a week for 6 weeks (Fig. 1A). We utilized a 10 μl Hamilton syringe with a 33 gauge needle (Hamilton Company USA, Reno, NV). Control animals were injected with 2 μl of vehicle (endotoxin-free water) or did not receive injections. This regimen was similar to previously published studies (16). No overt signs of tissue inflammation or soft tissue damage were observed during the course of injections (data not shown). Animals were sacrificed 6 weeks after the first injection. Maxillae were dissected and immersed in 10% buffered formalin for 48 hours.
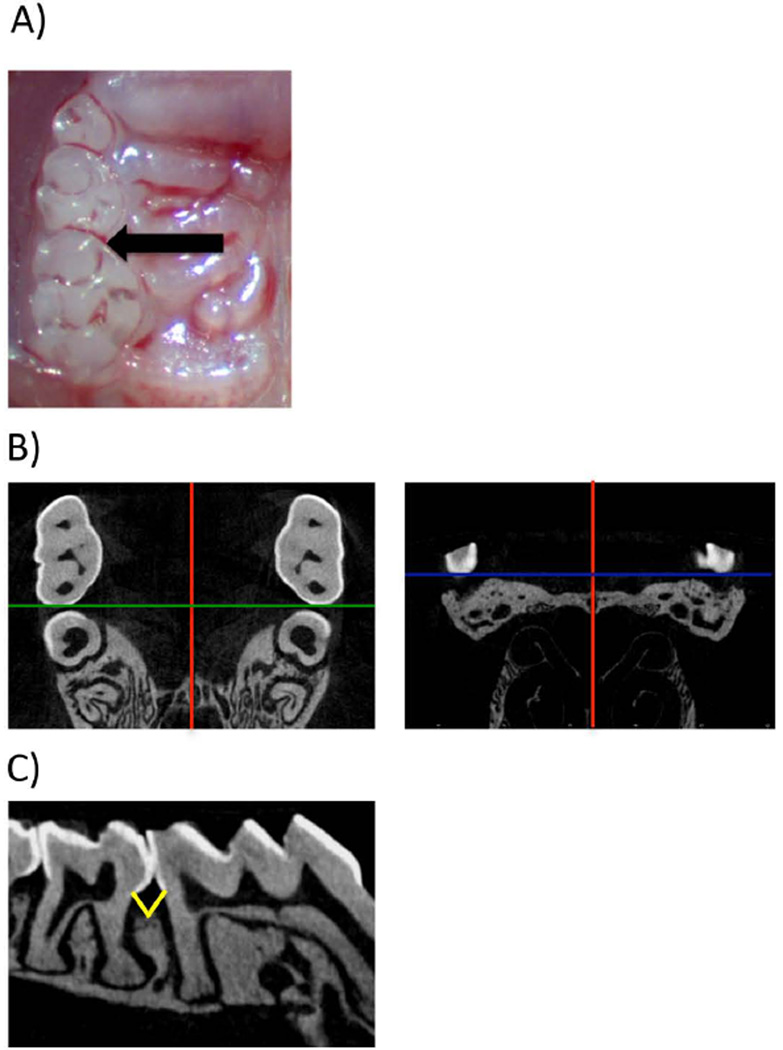
Figure 1.
Injections and Micro-CT image/sample orientation. A) Clinical image with the location of LPS injection. B) Micro-CT data were imported in DICOM format into Dolphin® software and the image volume was oriented in the orthogonal planes such that red line denotes (sagittal plane), green line (coronal), blue line (transverse plane): B1) the axial slices are parallel to the occlusal plane. The intermaxillary suture is parallel to the sagittal plane. C) The distance from the CEJ to the alveolar crest was measured at the sagittal plane intersecting the interproximal molars. Yellow lines depict the measurement that was taken for distal of 1st molar and mesial of 2nd molar.
Micro-CT Analysis
Maxillae were scanned using a μCT scanner (Skyscan 1172, Aartselaar, Belgium) with a voxel size of 10 μm (isotropic voxel) and an x-ray energy of 55 KVp and 181 μA. Each scan was conducted over a period of 21 minutes, with steps of 0.4°. Ten frames were averaged and a 0.5 mm aluminum filter was utilized. Virtual image slices were reconstructed using the cone-beam reconstruction software version 1.5 based on the Feldkamp algorithm.
Volumetric data were converted to DICOM format and were imported into Dolphin® software (Dolphin Imaging, Chatsworth, CA) for further analysis. To quantify the amount of bone loss, the imaged volume was oriented in the coronal (green) and transverse (blue) planes such that the sagittal plane (red) was parallel to the maxillary midline, identified by the intermaxillary suture and the coronal plane intersected the proximal area between the first and second maxillary molars (Fig. 1B). Then, at the sagittal plane crossing the interproximal contact point of the 1st and 2nd molar crowns, the distance between the CEJ and the alveolar crest were measured for the distal surface of the 1st molar and the mesial surface of the 2nd molar just below the contact point and 0.2 mm palatal to the contact point (Fig. 1C).
To quantify the amount of bone loss in the 5 parental strains, the bone level was measured as described above for the right and left sides. Subsequently, the average distance in the control sites was subtracted from the distances on the LPS-injected sites and the remainder represented the net bone loss at the LPS-injected site.
Histology
Maxillae were decalcified in 15% EDTA for 4 weeks. Following decalcification, 5μM-thick sections were cut in the coronal plane using a microtome (McBain Instruments, Chatsworth, CA). Sections were stained with hematoxylin and eosin (H&E) using standard protocols (19). Slices were digitally imaged using Aperio ImageScope model V11.1.2.752 (Vista, CA.)
For osteoclast analysis, cells that presented with ≥2 nuclei, in contact with the bone surface, were classified as osteoclasts (20). Osteoclast numbers were averaged for the right and left side for each mouse. Groups were compared using a Student’s t-test.
Bone Marrow Cell Isolation and in vitro Osteoclast Differentiation
Total bone marrow cells were harvested from femurs and tibias of 4-week-old A/J and C57BL/6J male mice according to Pirih et al (21). In brief, cells were filtered through nylon mesh screens (70 μm BD Falcon, Franklin Lakes, NJ, USA). At day 8, non-adherent cells were enumerated using a hemocytometer with trypan blue, to determine cell viability. Then, non-adherent cells were re-plated at 1.8x105 cells/well in a 24-well plate in osteoclastogenic medium (a-MEM + 10% FBS, 50 ng/mL M-CSF, 80 ng/mL sRANKL), which was replaced at day 3. At day 6, cells were fixed and tartrate resistant acid phosphatase (TRAP) staining was performed using a leukocyte acid phosphatase system (Sigma-Aldrich) according to manufacturers protocol (21). TRAP+ multinucleated cells (osteoclasts) were counted in three different areas of the well, under a light microscope and each well was averaged. Then all 3 wells were averaged. Groups were compared using a Student’s t-test.
Heritability
Heritability of the trait was estimated by fitting the data to the mixed model y=\mu + u + e, where y is a vector of phenotypes, \mu is the mean of the phenotypes, u is a random vector corresponding to the genetic component of the trait and e is a random vector corresponding to the environmental factor. The random vector u is assumed to be normally distributed with mean 0 and covariance matrix \sigma^2_g K where K is a kinship matrix encoding the genetic relationships and the random vector e is assumed to be normally distributed with mean 0 and covariance matrix\sigma^2_e I. If K is the realized relationship matrix (22) then the ratio \sigma^2_g /(\sigma^2_g + \sigma^2_e) is an estimate for the heritability of the trait.
Statistical Analysis
At least 12 animals were utilized per strain (n≥6 animals/group) (n≥24 sites/group). Data among groups were compared by One-Way ANOVA and between groups by Student’s t-test. P values <0.05 were considered significant.
Results
P. gingivalis-LPS Injection Induces Bone Loss in C57BL/6J Mice
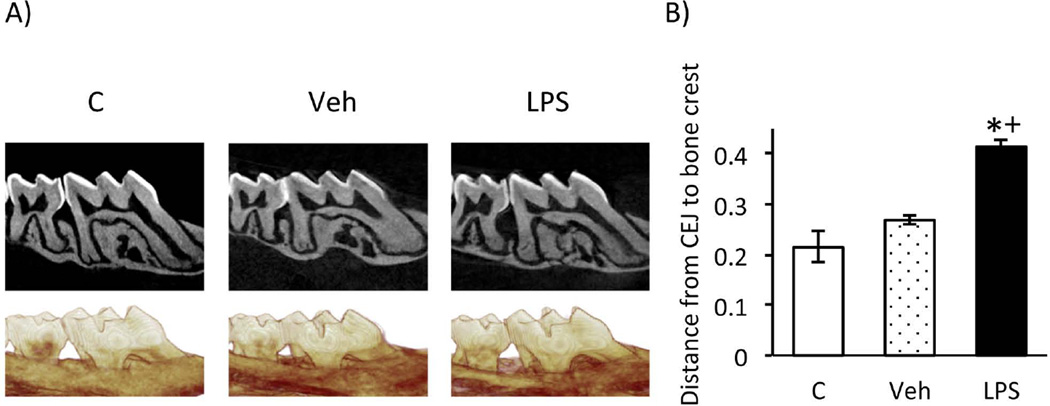
To evaluate PD-bone loss in response to LPS injection, we utilized a well-characterized model of periodontal bone loss through localized LPS delivery to the interdental papillae of maxillary molars in C57BL/6J mice (14–17)(Fig. 1A). Three different treatments were performed a) LPS-injections (between the 1st and 2nd molars on both sides of the maxilla), b) vehicle injections (between the 1st and 2nd molars on both sides of the maxilla), or c) no injections. The micro-CT analysis revealed statistical significant alveolar bone loss at the interproximal space between the 1st-2nd maxillary molars at the LPS-injected sites compared to non-injected or veh-injected sites. No statistical difference was observed between the vehicle injected and non-injected animals (Fig. 2). Since there was no statistical difference in the amount of bone loss comparing the non-injected and the vehicle injected sites (Fig. 2), subsequent experiments were carried out utilizing non-injected sites as controls.
Figure 2.
P.g. LPS induces periodontal bone loss. A) Corrected sagittal and 3D reformatted representative images of non-injected (C), vehicle (veh)- or LPS-injected (LPS) mice. B) Graph of the distance between the CEJ to the alveolar bone level (mm) in non-injected, veh- or LPS-injected sites (average+/− SEM) at the distal of the 1st molar and mesial of the 2nd molar. Statistical analysis was performed by Student’s t-test (n≥24 sites/group). *p≤0.001 compared to control and +p<0.0001 compared to vehicle.
Bone Loss is Strain-Dependent
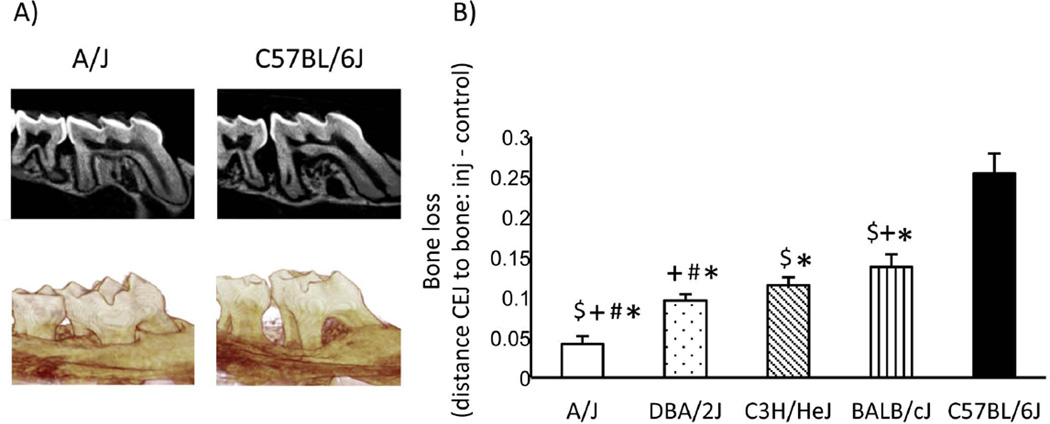
We utilized the P.g. LPS-injection induced inflammatory bone loss model described above to 5 classical inbred strains (BALB/cJ, C3H/HeJ, DBA/2J, A/J, and C57BL/6J), that derive the recombinant inbred strains of the HMDP, to explore genetic contribution of LPS-injection induced bone loss (Fig. 3). Each mouse strain was divided in 2 groups: a) LPS-injected or b) non-injected control. For each strain, bone loss was calculated by subtracting the average CEJ to bone crest distance in the non-injected animals from each LPS-injected site. C57BL/6J was the most susceptible strain to LPS-induced bone loss and presented a 5-fold higher bone loss compared to the most resistant, A/J, strain (Fig. 3B).
Figure 3.
P.g. LPS-induces strain dependent bone loss. A) Corrected sagittal and 3D reformatted representative images of A/J and C57BL/6J LPS-injected mice. B) Graph of periodontal bone loss (mm) of LPS-injected sites subtracted by the respective controls (average+/− SEM) at the distal of the 1st molar and mesial of the 2nd molar. Statistical analysis between groups was performed by Student’s t-test (n≥24 sites/group). p<0.001 * statistically significant compared to C57BL/6J, $ statistically significant compared to DBA/2J, + compared to C3H/HeJ, # compared to BALB/cJ. Significance between BALB/cJ compared to C3H/HeJ is p<0.05.
LPS-Injection-Induced Bone Loss is 49% Heritable
Based on the data presented above, (Fig. 3), heritability was calculated for LPS-induced bone loss in these 5 mouse strains. The heritability estimate for periodontal bone loss in the 5 parental strains of the HMDP was 49%, a value that closely resembles heritability measurements of 50% for PD in patients (6, 23).
C57BL/6J Mice Have Increased Osteoclastogenic Potential Compared to A/J in vitro
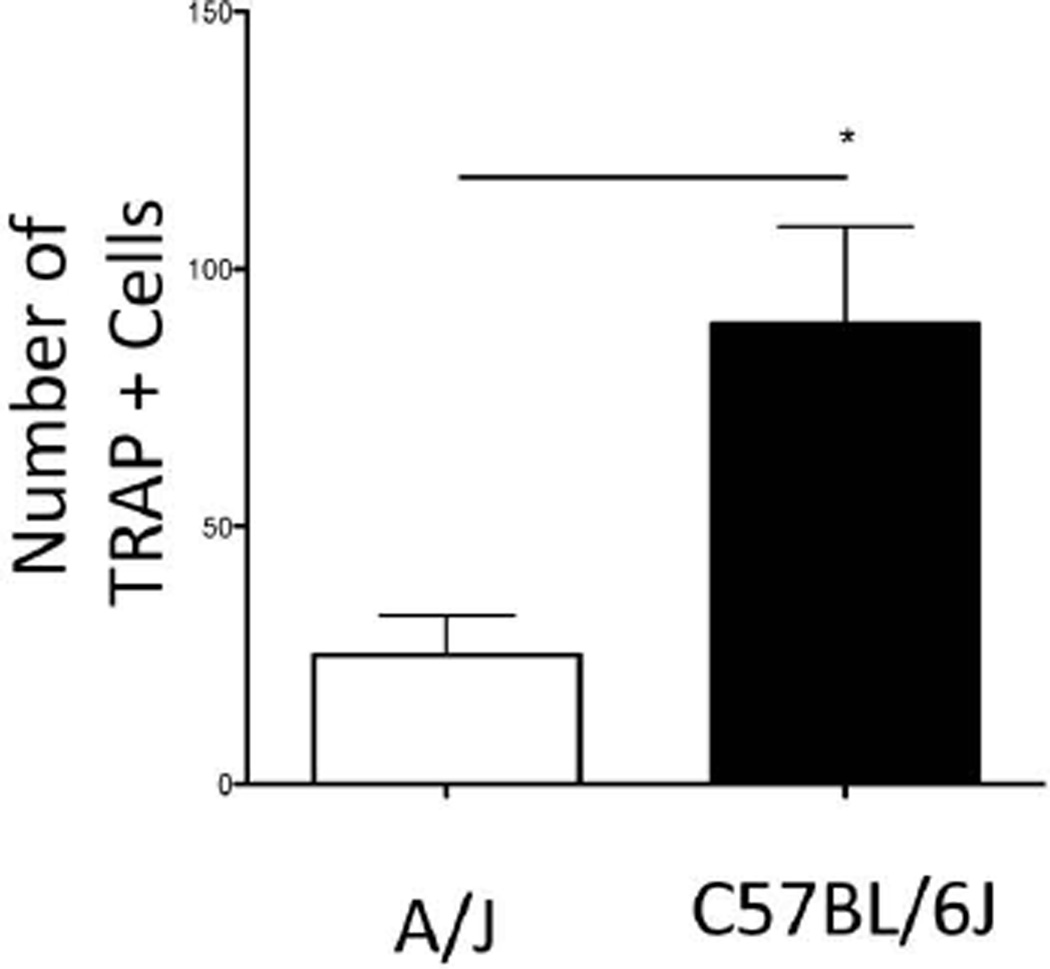
To assess whether the differences in bone loss between the two strains were in part due to inherent differences in osteoclastogenic potential, we evaluated osteoclast differentiation of C57BL/6J and A/J derived bone marrow by performing TRAP staining in vitro. A statistically significant increase in TRAP+ multinucleated cells was observed in the C57BL/6J compared to the A/J cells (Fig. 4).
Figure 4.
C57BL/6J mice have increased osteoclastogenic potential as compared to A/J in vitro. Graph of number of TRAP+ cells. Statistical analysis was performed using a Student’s t-Test. * statistically significant compared to A/J (p<.05).
Osteoclast Numbers Were Higher in C57BL/6J Compared to A/J Mice Following LPS-Injections in Vivo
To identify cellular differences that accompany periodontal bone loss, we evaluated osteoclast numbers of C57BL/6J vs. A/J mice after 5 LPS injections in vivo. LPS injections induced a statistically significant increase in osteoclast numbers in both strains. Importantly, a significantly higher osteoclast number increase was observed in C57BL/6J compared to A/J mice (Fig. 5).
Figure 5.
P.g. LPS-injections increases osteoclasts in C57BL/6J as compared to A/J mice. A) Representative H&E images of A/J control A/J LPS-injections, C57BL/6J control and C57BL/6J 5 LPS-injected. B) Graph of number of osteoclasts in A/J control, A/J LPS-injected, C57BL/6J control, and C57BL/6J LPS-injected (n≥6 mice/group). Statistical analysis between groups was performed using a Student’s t-test, * (p<. 05) *** (P<.001).
Discussion
PD is a polymicrobial infection-driven inflammatory disease that involves complex processes, such as biofilm formation by diverse microbial species, inflammatory response to a multifaceted microbial invasion, and activation of multiple signaling pathways that lead to bone resorption and attachment loss (24). Even though PD is a multifactorial disease, the genetic component is highly significant and estimated to explain 50% of disease burden (8). Moreover, PD heritability involves a large number of genes, each accounting for a small fraction of the disease (25), making GWAS studies an ideal tool to identify genes involved in this trait.
GWAS can be accomplished by human or animal studies, each complementing one another. To date, only a few groups have performed GWAS for PD in humans (26, 27). These studies have identified genes that are likely to be important in periodontitis. However, the main disadvantage of human GWAS is the requirement of large sample size. Therefore, frequently the power is insufficient to detect genes with a small contribution. Mice share structural, functional and genetic traits with humans. Moreover, powerful molecular and genetic tools developed in the past two decades make mice an ideal animal model for the study of complex traits. Mouse GWAS explored diverse conditions such as cardiovascular disease, atherosclerosis, diabetes, inflammatory diseases, hearing, and even behavior (28–33).
Studies performed in inbred mouse strains demonstrated variable bone loss in bacteria-induced periodontitis. In addition, a large variability in bacterial counts recovered among different strains was detected, pointing to a possible role of genetics in bacterial colonization (11). To study the genetic component of periodontal bone response in mice we elected to utilize an inflammatory model, analyzing the host response to a constant bacterial insult, bypassing the genetic influence in microbial colonization. We employed the well-characterized model of periodontal bone loss through the localized LPS delivery in mice to focus on the host response by analyzing bone loss as the outcome measurement (14–17). Moreover, we utilized P. gingivalis-derived LPS for multiple reasons. P. gingivalis, a gram-negative anaerobic rod and member of the “red complex”, is widely recognized as a predominant contributor to chronic PD in humans (24, 34). Additionally, diverse cytokine and chemokine responses of gingival fibroblasts and macrophages have been reported utilizing P. gingivalis vs. E. coli LPS (35). Finally, P. gingivalis infection in mice produces inflammation of the periodontal tissues and associated periodontal bone loss (10, 11).
Herein, utilizing a model of P. gingivalis LPS-induced periodontal bone loss and high-resolution micro-CT, we demonstrated differences in bone loss pattern among 5 classic inbred mouse strains. These differences were expected since the utilization of animal models for evaluating genetic determinants of PD have been proposed (36–38). More recently, oral infection of various inbred mouse strains with human strains of P. gingivalis demonstrates that susceptibility to alveolar bone loss is a genetically modified trait. Some mouse strains were highly susceptible, while others were resistant to alveolar bone loss. Importantly, F1 offsprings of susceptible and resistant strains demonstrated various patterns of heritability, suggesting the existence of recessive and dominant resistance alleles. The importance of exploiting the mouse model to investigate loci associated with susceptibility or resistance to inflammation-induced alveolar bone loss was concluded (10, 11).
More importantly, we detected 49% heritability in bone loss similar to the heritability observed in humans (6). In addition, our data is in agreement with published data in mouse models where alveolar bone loss is a genetically modified trait. (39, 40).
The pathogenesis of periodontitis is complex, involving many different cell types (41–43). The LPS-injection model, as mentioned earlier, bypasses the bacterial colonization process and allows for a more simplified method of studying the inflammatory mediators of this disease. To begin dissecting the mechanisms by which the observed interstrain differences of periodontal bone loss occur, we evaluated the osteoclastogenic potential of A/J and C57BL/6J in vitro. We observed, under supra-physiologic conditions, that C57BL/6J bone marrow cells have a stronger osteoclastogenic potential. To further explore the differences that might mediate periodontal bone loss and how it correlates with our micro-CT findings, we evaluated the number of multinucleated osteoclasts in vivo. Indeed, in vivo, C57BL/6J mice demonstrated a more pronounced inflammatory response with a higher number of osteoclasts after LPS injections when compared to A/J mice. Our results corroborate with studies that demonstrate a hyper-responsiveness to LPS in C57BL/6J mice as compared to A/J mice. The hyper-responsiveness in C57BL/6J mice includes an increase: in vasculitis, in neutrophil numbers, in polymorphonuclear cells and splenocytes followed by LPS treatment (4–6). Moreover, there is an increased production of interleukin-1 by C57BL6/J mice after LPS-injections as compared to A/J mice (4). Additionally, C57BL/6J mice have a lower bone mineral density phenotype compared to A/J (7) further supporting our findings. Clearly the observed differences in osteoclast differentiation and numbers are only part of the pathophysiologic mechanism underlying periodontal bone loss. Immune cell activation, osteoblastic function, cytokine release and extracellular matrix remodeling are all processes that would contribute to the observed interstrain differences. We plan future studies to address variations among the HMDP strains that will shed light to genetic determinants of the periodontal bone loss response.
The HMDP panel consists of 100 commercially available inbred mouse strains selected for systematic genetic analyses of complex traits. These strains were selected with the intent to increase resolution of genetic mapping, offer a renewable resource of inbred mice, and provide for a shared repository for data accumulation that would allow the integration of data across multiple scales including transcriptomic, metabolomic, proteomic, and clinical phenotypes (12). The 100 strains consist of 29 classic inbred strains used for initial association mapping (44, 45) and 71 recombinant inbred (RI) strains (12). The HMDP offers a powerful genetic approach for the study of complex genetic traits. Moreover, the HMDP is currently used to investigate a variety of clinical traits including diet-induced obesity, hearing loss, heart failure, atherosclerosis, bone mineral density and diabetes (12, 46–48). Therefore, determining periodonto-pathogenic LPS-induced bone loss in a mouse model will allow us to expand our studies to perform genome wide association studies (GWAS) utilizing the HMDP. Expanding these studies will contribute in identifying pathways important in disease initiation development; moreover, it will assist in predicting patients that are genetically predisposed to PD and in identifying the biological basis of disease susceptibility. The HMDP offers a powerful genetic approach for the study of complex genetic traits. The HMDP is currently used to investigate a variety of clinical traits including diet-induced obesity, hearing loss, heart failure, atherosclerosis, bone mineral density and diabetes (12, 46–48). We will exploit these powerful mouse genetics approaches to begin unraveling murine genetics affecting periodontal bone loss with an eye towards future translational studies on genetic and environmental regulators of human PD.
Our data supports a significant role of the genetic framework in LPS-induced periodontal bone loss and the feasibility of utilizing the HMDP to explore these genetic factors. Moreover, it corroborates with data in the literature. Expanding these studies will contribute in identifying the biological basis of disease susceptibility. Such understanding would help recognize patients with high-risk or resistance for development of periodontitis and would inform targeted treatment interventions for patients with the disease as we move towards a personalized diagnostic and interventional approach of periodontitis.
Acknowledgements
This work was supported by the NIH/NIDCR DE023901-01, DE 019465- S1 and the UCLA School of Dentistry Seed Grant. SH was supported by the NIH/NIDCR T90 DE022734-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The author(s) declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1.Hinrichs JE, Novak MJ. Classification of Diseases and Conditions Affecting the Periodontium. In: Newman MG, Takei HH, Klokkevold PR, Carranza FA, editors. Carranza's Clinical Periodontology. Elsevier: 2012. pp. 34–64. [Google Scholar]
- 2.Petersen PE. The World Oral Health Report 2003: continuous improvement of oral health in the 21st century--the approach of the WHO Global Oral Health Programme. Community Dent Oral Epidemiol. 2003;31(Suppl 1):3–23. doi: 10.1046/j..2003.com122.x. [DOI] [PubMed] [Google Scholar]
- 3.Kinane DF, Shiba H, Hart TC. The genetic basis of periodontitis. Periodontol 2000. 2005;39:91–117. doi: 10.1111/j.1600-0757.2005.00118.x. [DOI] [PubMed] [Google Scholar]
- 4.Vijayalakshmi R, Geetha A, Ramakrishnan T, Emmadi P. Genetic polymorphisms in periodontal diseases: an overview. Indian journal of dental research : official publication of Indian Society for Dental Research. 2010;21:568–574. doi: 10.4103/0970-9290.74226. [DOI] [PubMed] [Google Scholar]
- 5.Corey LA, Nance WE, Hofstede P, Schenkein HA. Self-reported periodontal disease in a Virginia twin population. J Periodontol. 1993;64:1205–1208. doi: 10.1902/jop.1993.64.12.1205. [DOI] [PubMed] [Google Scholar]
- 6.Michalowicz BS, Diehl SR, Gunsolley JC, et al. Evidence of a substantial genetic basis for risk of adult periodontitis. J Periodontol. 2000;71:1699–1707. doi: 10.1902/jop.2000.71.11.1699. [DOI] [PubMed] [Google Scholar]
- 7.Michalowicz BS. Genetic and heritable risk factors in periodontal disease. J Periodontol. 1994;65:479–488. doi: 10.1902/jop.1994.65.5s.479. [DOI] [PubMed] [Google Scholar]
- 8.Michalowicz BS, Aeppli D, Virag JG, et al. Periodontal findings in adult twins. J Periodontol. 1991;62:293–299. doi: 10.1902/jop.1991.62.5.293. [DOI] [PubMed] [Google Scholar]
- 9.Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists. 2003;14:430–449. doi: 10.1177/154411130301400605. [DOI] [PubMed] [Google Scholar]
- 10.Hart GT, Shaffer DJ, Akilesh S, et al. Quantitative gene expression profiling implicates genes for susceptibility and resistance to alveolar bone loss. Infect Immun. 2004;72:4471–4479. doi: 10.1128/IAI.72.8.4471-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker PJ, Dixon M, Roopenian DC. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect Immun. 2000;68:5864–5868. doi: 10.1128/iai.68.10.5864-5868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett BJ, Farber CR, Orozco L, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome research. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghazalpour A, Rau CD, Farber CR, et al. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23:680–692. doi: 10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Valerio MS, Kirkwood KL. MAPK usage in periodontal disease progression. Journal of signal transduction. 2012;2012:308943. doi: 10.1155/2012/308943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers JE, Li F, Coatney DD, et al. Actinobacillus actinomycetemcomitans lipopolysaccharide-mediated experimental bone loss model for aggressive periodontitis. J Periodontol. 2007;78:550–558. doi: 10.1902/jop.2007.060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartori R, Li F, Kirkwood KL. MAP kinase phosphatase-1 protects against inflammatory bone loss. J Dent Res. 2009;88:1125–1130. doi: 10.1177/0022034509349306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patil C, Rossa C, Jr, Kirkwood KL. Actinobacillus actinomycetemcomitans lipopolysaccharide induces interleukin-6 expression through multiple mitogen-activated protein kinase pathways in periodontal ligament fibroblasts. Oral Microbiol Immunol. 2006;21:392–398. doi: 10.1111/j.1399-302X.2006.00314.x. [DOI] [PubMed] [Google Scholar]
- 18.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowan CM, Shi YY, Aalami OO, et al. Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat Biotechnol. 2004;22:560–567. doi: 10.1038/nbt958. [DOI] [PubMed] [Google Scholar]
- 20.Chaichanasakul T, Kang B, Bezouglaia O, Aghaloo TL, Tetradis S. Diverse Osteoclastogenesis of Bone Marrow From Mandible versus Long Bone. J Periodontol. 2013 doi: 10.1902/jop.2013.130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirih FQ, Michalski MN, Cho SW, et al. Parathyroid hormone mediates hematopoietic cell expansion through interleukin-6. PLoS ONE. 5:e13657. doi: 10.1371/journal.pone.0013657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nature reviews Genetics. 2013;14:139–149. doi: 10.1038/nrg3377. [DOI] [PubMed] [Google Scholar]
- 24.Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes and infection / Institut Pasteur. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshie H, Kobayashi T, Tai H, Galicia JC. The role of genetic polymorphisms in periodontitis. Periodontol 2000. 2007;43:102–132. doi: 10.1111/j.1600-0757.2006.00164.x. [DOI] [PubMed] [Google Scholar]
- 26.Divaris K, Monda KL, North KE, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Human molecular genetics. 2013;22:2312–2324. doi: 10.1093/hmg/ddt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teumer A, Holtfreter B, Volker U, et al. Genome-wide association study of chronic periodontitis in a general German population. J Clin Periodontol. 2013;40:977–985. doi: 10.1111/jcpe.12154. [DOI] [PubMed] [Google Scholar]
- 28.Welch CL. Beyond genome-wide association studies: the usefulness of mouse genetics in understanding the complex etiology of atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32:207–215. doi: 10.1161/ATVBAHA.111.232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mammalian genome : official journal of the International Mammalian Genome Society. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breyer MD, Tchekneva E, Qi Z, Takahashi T, Fogo AB, Harris RC. Examining diabetic nephropathy through the lens of mouse genetics. Current diabetes reports. 2007;7:459–466. doi: 10.1007/s11892-007-0078-3. [DOI] [PubMed] [Google Scholar]
- 31.Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- 32.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White CH, Ohmen JD, Sheth S, et al. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mammalian genome : official journal of the International Mammalian Genome Society. 2009;20:207–213. doi: 10.1007/s00335-009-9178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 35.Jones KJ, Ekhlassi S, Montufar-Solis D, Klein JR, Schaefer JS. Differential cytokine patterns in mouse macrophages and gingival fibroblasts after stimulation with porphyromonas gingivalis or Escherichia coli lipopolysaccharide. J Periodontol. 2010;81:1850–1857. doi: 10.1902/jop.2010.100226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baer PN, Crittenden LB, Jay GE, Jr, Lieberman JE. Studies on periodontal disease in the mouse. II. Genetic and meternal effects. J Dent Res. 1961;40:23–33. doi: 10.1177/00220345610400011901. [DOI] [PubMed] [Google Scholar]
- 37.Baer PN, Lieberman JE. Periodontal disease in six strains of inbred mice. J Dent Res. 1960;39:215–225. doi: 10.1177/00220345600390020301. [DOI] [PubMed] [Google Scholar]
- 38.Baer PN, Lieberman JE. Observation of some genetic characteristics of the periodontium in three strains of inbred mice. Oral Surg Oral Med Oral Pathol. 1959;12:820–829. doi: 10.1016/0030-4220(59)90031-3. [DOI] [PubMed] [Google Scholar]
- 39.Shusterman A, Salyma Y, Nashef A, et al. Genotype is an important determinant factor of host susceptibility to periodontitis in the Collaborative Cross and inbred mouse populations. BMC genetics. 2013;14:68. doi: 10.1186/1471-2156-14-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shusterman A, Durrant C, Mott R, et al. Host susceptibility to periodontitis: mapping murine genomic regions. J Dent Res. 2013;92:438–443. doi: 10.1177/0022034513484039. [DOI] [PubMed] [Google Scholar]
- 41.Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontol 2000. 1997;14:9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 42.Bartold PM, Narayanan AS. Molecular and cell biology of healthy and diseased periodontal tissues. Periodontol 2000. 2006;40:29–49. doi: 10.1111/j.1600-0757.2005.00140.x. [DOI] [PubMed] [Google Scholar]
- 43.Ohlrich EJ, Cullinan MP, Seymour GJ. The immunopathogenesis of periodontal disease. Aust Dent J. 2009;54(Suppl 1):S2–10. doi: 10.1111/j.1834-7819.2009.01139.x. [DOI] [PubMed] [Google Scholar]
- 44.Cervino AC, Darvasi A, Fallahi M, Mader CC, Tsinoremas NF. An integrated in silico gene mapping strategy in inbred mice. Genetics. 2007;175:321–333. doi: 10.1534/genetics.106.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grupe A, Germer S, Usuka J, et al. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- 46.Farber CR, Bennett BJ, Orozco L, et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS genetics. 2011;7:e1002038. doi: 10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghazalpour A, Bennett B, Petyuk VA, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS genetics. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park CC, Gale GD, de Jong S, et al. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC systems biology. 2011;5:43. doi: 10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]