Abstract
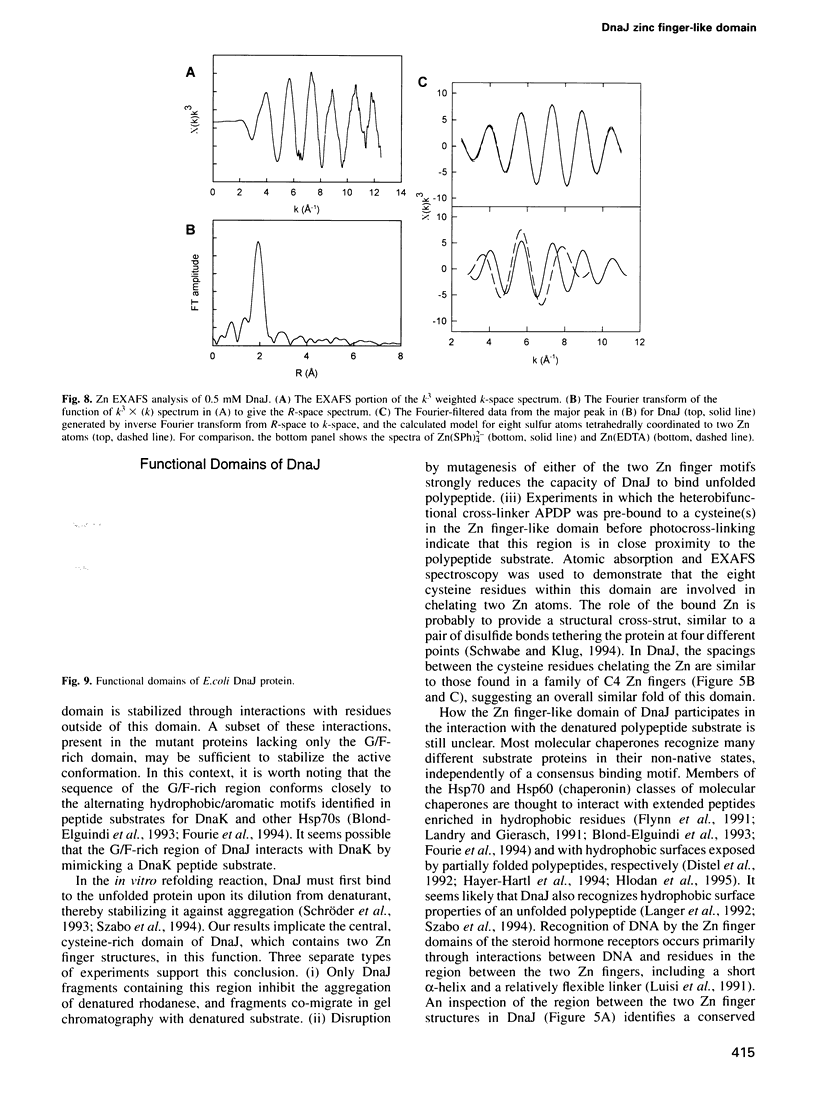
The Escherichia coli heat-shock protein DnaJ cooperates with the Hsp70 homolog DnaK in protein folding in vitro and in vivo. Little is known about the structural features of DnaJ that mediate its interaction with DnaK and unfolded polypeptide. DnaJ contains at least four blocks of sequence representing potential functional domains which have been conserved throughout evolution. In order to understand the role of each of these regions, we have analyzed DnaJ fragments in reactions corresponding to known functions of the intact protein. Both the N-terminal 70 amino acid 'J-domain' and a 35 amino acid glycine-phenylalanine region following it are required for interactions with DnaK. However, only complete DnaJ can cooperate with DnaK and a third protein, GrpE, in refolding denatured firefly luciferase. As demonstrated by atomic absorption and extended X-ray absorption fine structure spectroscopy (EXAFS), the 90 amino acid cysteine-rich region of DnaJ contains two Zn atoms tetrahedrally coordinated to four cysteine residues, resembling their arrangement in the C4 Zn binding domains of certain DNA binding proteins. Interestingly, binding experiments and cross-linking studies indicate that this Zn finger-like domain is required for the DnaJ molecular chaperone to specifically recognize and bind to proteins in their denatured state.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993 Nov 19;75(4):717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Cyr D. M., Douglas M. G. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994 Apr 1;269(13):9798–9804. [PubMed] [Google Scholar]
- Cyr D. M., Langer T., Douglas M. G. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994 Apr;19(4):176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Diakun G. P., Fairall L., Klug A. EXAFS study of the zinc-binding sites in the protein transcription factor IIIA. Nature. 1986 Dec 18;324(6098):698–699. doi: 10.1038/324698a0. [DOI] [PubMed] [Google Scholar]
- Flynn G. C., Pohl J., Flocco M. T., Rothman J. E. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991 Oct 24;353(6346):726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Fourie A. M., Sambrook J. F., Gething M. J. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994 Dec 2;269(48):30470–30478. [PubMed] [Google Scholar]
- Freedman L. P., Luisi B. F., Korszun Z. R., Basavappa R., Sigler P. B., Yamamoto K. R. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988 Aug 11;334(6182):543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- Freeman B. C., Myers M. P., Schumacher R., Morimoto R. I. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995 May 15;14(10):2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J., Nimmesgern E., Ohtsuka K., Hartl F. U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994 Jul 14;370(6485):111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Gragerov A., Nudler E., Komissarova N., Gaitanaris G. A., Gottesman M. E., Nikiforov V. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer-Hartl M. K., Ewbank J. J., Creighton T. E., Hartl F. U. Conformational specificity of the chaperonin GroEL for the compact folding intermediates of alpha-lactalbumin. EMBO J. 1994 Jul 1;13(13):3192–3202. doi: 10.1002/j.1460-2075.1994.tb06618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick J. P., Langer T., Davis T. A., Hartl F. U., Wiedmann M. Control of folding and membrane translocation by binding of the chaperone DnaJ to nascent polypeptides. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10216–10220. doi: 10.1073/pnas.90.21.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R. B., Flanagan J. M., Prestegard J. H. 1H and 15N magnetic resonance assignments, secondary structure, and tertiary fold of Escherichia coli DnaJ(1-78). Biochemistry. 1995 Apr 25;34(16):5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- Hlodan R., Tempst P., Hartl F. U. Binding of defined regions of a polypeptide to GroEL and its implications for chaperonin-mediated protein folding. Nat Struct Biol. 1995 Jul;2(7):587–595. doi: 10.1038/nsb0795-587. [DOI] [PubMed] [Google Scholar]
- Kudlicki W., Odom O. W., Kramer G., Hardesty B. Activation and release of enzymatically inactive, full-length rhodanese that is bound to ribosomes as peptidyl-tRNA. J Biol Chem. 1994 Jun 17;269(24):16549–16553. [PubMed] [Google Scholar]
- Landry S. J., Gierasch L. M. The chaperonin GroEL binds a polypeptide in an alpha-helical conformation. Biochemistry. 1991 Jul 30;30(30):7359–7362. doi: 10.1021/bi00244a001. [DOI] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., Hartl F. U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992 Apr 23;356(6371):683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. R., Graves R. J., de Murcia G., Castaing B., Laval J. Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J Biol Chem. 1993 Apr 25;268(12):9063–9070. [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S. J., Gronenborn A. M. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993 Jul 23;261(5120):438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Palleros D. R., Welch W. J., Fink A. L. Interaction of hsp70 with unfolded proteins: effects of temperature and nucleotides on the kinetics of binding. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5719–5723. doi: 10.1073/pnas.88.13.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C., Bordas J., Foote A. M., Koch M. H., Moody M. F. Zinc-sulfur bonds of aspartate transcarbamylase studied by x-ray absorption spectroscopy. Biochemistry. 1982 Mar 2;21(5):830–834. doi: 10.1021/bi00534a003. [DOI] [PubMed] [Google Scholar]
- Pérez-Alvarado G. C., Miles C., Michelsen J. W., Louis H. A., Winge D. R., Beckerle M. C., Summers M. F. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Biol. 1994 Jun;1(6):388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- Rothblatt J. A., Deshaies R. J., Sanders S. L., Daum G., Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989 Dec;109(6 Pt 1):2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadis S., Hightower L. E. Unfolded proteins stimulate molecular chaperone Hsc70 ATPase by accelerating ADP/ATP exchange. Biochemistry. 1992 Oct 6;31(39):9406–9412. doi: 10.1021/bi00154a012. [DOI] [PubMed] [Google Scholar]
- Sadler I., Chiang A., Kurihara T., Rothblatt J., Way J., Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J Cell Biol. 1989 Dec;109(6 Pt 1):2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H., Langer T., Hartl F. U., Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993 Nov;12(11):4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J. W., Klug A. Zinc mining for protein domains. Nat Struct Biol. 1994 Jun;1(6):345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- Sherman MYu, Goldberg A. L. Involvement of the chaperonin dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 1992 Jan;11(1):71–77. doi: 10.1002/j.1460-2075.1992.tb05029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Szabo A., Langer T., Schröder H., Flanagan J., Bukau B., Hartl F. U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T., Pellecchia M., Wall D., Georgopoulos C., Wüthrich K. NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2-108) containing the highly conserved J domain. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11343–11347. doi: 10.1073/pnas.91.24.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D., Zylicz M., Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994 Feb 18;269(7):5446–5451. [PubMed] [Google Scholar]
- Zylicz M., Ang D., Liberek K., Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989 May;8(5):1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz M., Yamamoto T., McKittrick N., Sell S., Georgopoulos C. Purification and properties of the dnaJ replication protein of Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7591–7598. [PubMed] [Google Scholar]







