Abstract
Breast Cancer (BC) is considered as one of the most important causes of death worldwide. Previous studies showed that apolipoprotein B mRNA- editing catalytic polypeptide-like 3 (APOBEC3) gene deletion significantly increased the risk of BC risk in Chinese and European women. The present study aimed to assess the possible impact of APOBEC3 deletion and the risk of BC in a sample of Iranian population. The APOBEC3 insertion/deletion (I/D) was analyzed in a case- control study including 262 BC patients and 217 healthy women. Polymerase chain reaction (PCR) was used to genotype the variant in APOBEC3 gene. The findings of this study showed that I/D as well as I/D+D/D genotype increased the risk of BC (OR= 1.57, 95% CI= 1.07- 2.31, p= 0.025 and OR= 1.50, 95% CI= 1.03- 2.19, p= 0.037, respectively) in comparison with I/I genotype. In conclusion, our findings suggest that APOBEC3 deletion polymorphism increased the risk of BC in an Iranian population in the southeast of Iran.
Key Words: APOBEC3, breast Cancer, polymorphism, deletion
Every day many women are diagnosed with breast cancer (BC) all over the world. It can be considered the most common cancer amongst females globally and is also the leading cause of cancer-related death in many countries (1). BC is recognized as an important health care problem worldwide affecting approximately 1 million women annually (1-3). BC is also reported as one of the most frequent malignancies among Iranian women, comprising 21.4% of female cancers in this population (4). Interestingly, it was reported that BC affects Iranian women about a decade earlier than Western countries (5) which highlights the importance of research on BC in Iranian population. Several different factors are involved in BC pathogenesis while its exact etiology is complicated and not clearly identified. Our recent publications have provided evidence that genetic factors play important roles in the pathogenesis and progre-ssions of BC in the population of southeast of Iran (6-10).
Somatic mutations in the genome are critical for transformation of normal cells into cancers. The existence of hundreds to thousands of mutations has been shown in most cancers (11-14).
APOBEC3s (A3s) gene cluster is located on chromosome 22q13.1-q13.2 including APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBE-C3E, APOBEC3F, APOBEC3G, and APOBEC3H which are arranged in tandem and play key roles in intracellular defense against viral infection (15, 16). They all have the cytidine deaminase activity and are able to convert cytosine to uracil (15).
Through copy number variation (CNV) genome wide association study (GWAS), Long et al. realized a common (CNV) locus for breast cancer in Chinese women, which was located between the fifth exon of APOBEC3A and the eighth exon of APOBEC3B (17). The finding was also repeated among women of European ancestry (18). It has been shown that a 29.5 Kb deletion occurs between the fifth exon of APOBEC3A and the eighth exon of APOBEC3B, leading to the complete removal of the APOBEC3B coding region. Several studies have found an association between APOBEC3 deletion and risk of various cancers (17-19).
To the best of our knowledge there is no any report regarding the association between APOBEC3 deletion and BC in Iranian women, therefore in the present study we aimed to find out the impact of APOBEC3 deletion on BC risk in a sample of Iranian population.
Material and methods
Patients
This population- based case- control study was accomplished on 262 pathologically confirmed BC patients referred to Ali Ebneh Abitaleb hospital (Iran) from February 2009 until August 21014 and 217 age-matched population-based healthy women with no history of any type of cancer and not related to the patients. Ethical approvals for recruitment were obtained from the local Ethics Committee of Zahedan University of Medical Sciences, and informed consent was obtained from all patients and healthy individuals. Blood samples were collected in EDTA-containing tubes from patients and healthy controls and DNA was extracted using the salting out method. The quality of the isolated DNA was checked by electrophoresis on 1% agarose gel, quantitated spectrophotometrically and stored at -20 °C till further use.
APOBEC3B insertion/deletion genotyping
Genotyping of APOBEC3 insertion/deletion was performed using polymerase chain reaction (PCR) as described by Kidd et al. (20). One pair of deletion and two pairs of insertion (insertions 1 and 2) primers were used to distinguish between the insertion and deletion alleles. The primers sequences used were: Deletion-F: 5`-TAGGTGC-CACCCCGAT-3`, Deletion-R: 5`-TTGAGCAT-AATCTTACTCTTGTAC-3`, Insertion1-F: 5`-TTGGTGCTGCCCCCTC-3`, Insertion1-R: 5`-TAGAGACTGAGGCCCAT-3`, Insertion2-F: 5`-TGTCCCTTTTCAGAGTTTGAGTA-3`, Insertion-2-R: 5`-TGGAGCCAATTAATCACTTCAT-3`. Deletion primers are specific to the deletion sequence configuration and generate a 700 bp PCR product upon amplification. Insertion 1 and Insertion 2 primers amplify only the insertion configuration and produce 490 and 705 bp products, respectively. Insertion and deletion PCR assays were performed separately, the products pooled, and visualized on a standard 2% agarose gel. In addition, each of the samples, which appeared to be homozygous for the deletion, was genotyped using a second set of oligonucleotides for the insertion (Insertion 2).
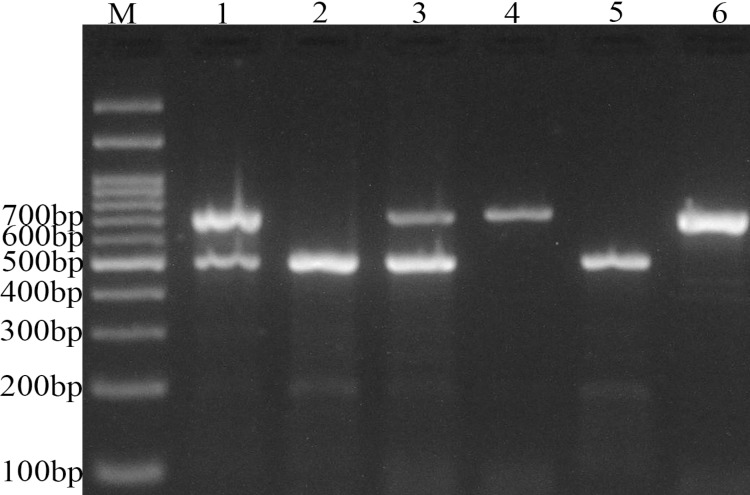
PCR was performed using 2X Prime Taq Premix (Genet Bio, Korea). The amplification procedure consisted of an initial denaturing step for 5 min at 95 °C followed by 34 cycles: for 30 s at 95 °C, 24 s at 61 °C, and 17 s at 72 °C, as well as a final extension step for 10 min at 72 °C. The PCR products were visualized on a 3% agarose gel containing 0.5 µg/ ml of ethidium bromide (Figure 1).
Fig. 1.
Photograph of DNA electophoresis for detection of APOBEC3 I/D variant. M: DNA marker; lanes 1, and 3: I/D; lane 2, and 5: D/D; lanes 4 and 6, I/I genotypes
Statistical analysis
All statistical analyzes were computed using statistical package SPSS 20 software. The differences between the variables were evaluated by chi-square test or independent sample t-test according to the data. The association between genotypes and BC were assessed by computing the odds ratio (OR) and 95% confidence intervals (95% CI) from logistic regression analyses. A p- value < 0.05 was considered as statistically significant. According to our results, sample power was computed for APOBEC3 deletion by comparison of each genotype with the sum of other genotypes by using STATA 10 software (Table 1).
Table 1.
Association between the APOBEC3 gene deletion and breast cancer risk
| APOBEC3 I/D variant |
Cases
n (%) |
Controls
n (%) |
OR (95% CI) | P- value | Study power% |
|---|---|---|---|---|---|
| I/I | 154 (58.8) | 148 (68.2) | 1.00 | - | 53 |
| I/D | 103 (39.3) | 63 (29.0) | 1.57 (1.07-2.31) | 0.025 | 62 |
| D/D | 5 (1.9) | 6 (2.8) | 0.80 (0.24-2.66) | 0.767 | 6 |
| I/D+D/D | 108 (41.2) | 69 (31.8) | 1.50 (1.03-2.19) | 0.037 | 53 |
| Allele | |||||
| I | 411 (78.4) | 359 (82.7) | 1.00 | - | 35 |
| D | 113 (21.6) | 75 (17.3) | 1.32 (0.95-1.82) | 0.102 | 35 |
Results
The study groups included 262 pathologically confirmed BC patients with an average age of 48.9± 11.2 years and 217 healthy women with a mean age of 50.0± 13.0 years. The demographic information of patients group was summarized in Table 1. No significant difference was found between the groups regarding age (p= 0.306). The frequency distribution of the APOBEC3 I/D in BC cases and controls are shown in Table 1. Our results showed that I/D as well as I/D+D/D genotype increased the risk of BC (OR= 1.57, 95% CI= 1.07-2.31, p= 0.025 and OR= 1.50, 95% CI= 1.03-2.19, p= 0.037, respectively) in comparison with I/I genotype.
The D allele frequency in BC and controls were 0.216 and 0.173, respectively. The APOBEC3 D allele was not associated with the risk of BC in comparison with I allele (OR= 1.32, 95% CI= 0.95-1.82, p= 0.102).
The APOBEC3 I/D in controls (χ2= 0.052, p= 0.819) but not in cases (χ2= 6.88, p= 0.008) were in Hardy-Weinberg Equilibrium (HWE).
As shown in table 2, no significant association was observed among APOBEC3 deletion and clinicopathological parameters, including tumor stage, tumor grade, estrogen and progesterone receptors (ER, PgR), tumor size, and human growth factor receptor 2 (HER2).
Table 2.
Association between APOBEC3A I/D polymorphism and clinicopathological characteristics
| Variables | APOBEC3A | P | ||
|---|---|---|---|---|
| I/I | I/D | D/D | ||
| Age (years) | 0.696 | |||
| ≤50 | 84 | 60 | 2 | |
| >50 | 64 | 42 | 3 | |
| Pathological type | 0.138 | |||
| Ductal | 94 | 74 | 5 | |
| Others | 47 | 25 | 0 | |
| Tumor size (cm) | 0.429 | |||
| ≤2 | 35 | 47 | 1 | |
| >2 | 77 | 79 | 7 | |
| TNM Stage | 0.850 | |||
| I | 19 | 22 | 3 | |
| II | 47 | 48 | 5 | |
| III | 32 | 35 | 3 | |
| IV | 13 | 24 | 2 | |
| Grade | 0.437 | |||
| I | 23 | 21 | 2 | |
| II | 64 | 67 | 7 | |
| III | 14 | 30 | 1 | |
| IV | 0 | 0 | 0 | |
| ER status | 0.858 | |||
| Positive | 68 | 78 | 8 | |
| Negative | 37 | 43 | 3 | |
| PgR status | 0.627 | |||
| Positive | 62 | 79 | 6 | |
| Negative | 41 | 42 | 5 | |
| HER2 status | 0.155 | |||
| Positive | 50 | 69 | 9 | |
| Negative | 61 | 59 | 4 | |
Discussion
In the current study we examined the impact of APOBEC3 deletion on BC risk in a sample of Iranian population who were living in southeast of this country. The findings revealed that APOBEC3 deletion significantly increased the risk of breast cancer among a sample of Iranian women. Long et al. performed a genome-wide
association study in Chinese population and found a strong association between common deletion in the APOBEC3 gene and BC (17). Their findings showed the ORs of 1.31 (95% CI= 1.21-1.42) for a one-copy deletion and 1.76 (95% CI= 1.57-1.97) for a two-copy deletion. The result was also replicated among women of European ancestry and the findings showed that the deletion was significantly associated with breast cancer risk, with ORs and 95% CIs of 1.21 (1.02–1.43) for one-copy deletion and 2.29 (1.04–5.06) for two-copy deletion in comparison with no deletion (18).
Qi et al. investigated the possible association between APOBEC3 gene deletion and risk of epithelial ovarian cancer (EOC) in Chinese population (19). They found that one copy deletion as well as two-copy deletion of APOBEC3 significantly increased the risk of EOC in comparison with subjects with no deletions.
Addition evidences suggested that APOBEC3B is involved in mutagenesis of human cancers such as bladder, cervix, lung (adenocarcinoma and squamous cell carcinoma), head and neck as well as BC (21-24). APOBEC3B catalyzes genomic DNA deamination and it is a main mutational source in BC accounting for C-to-T transitions (25). Upregulation of APOBEC3B has been revealed in BC showing that it as an important source of enzymatic mutation and DNA damage that could inactivate the tumor suppressor TP53 gene (25).
We found no significant association between APOBEC3 deletion and clinicopathological parameters of BC patients. To the best of our knowledge, there is no data regarding the impact of APOBEC3 deletion on response to treatment as well as prognosis of BC patients.
In conclusion, our findings indicated that APOBEC3 deletion significantly increased the risk of BC in an Iranian population in southeast of Iran. Large sample sizes in diverse ethnicities are still required to certify our finding.
Acknowledgment
This research was funded as dissertation grant (MSc thesis of MR) from the Deputy for Research, Zahedan University of Medical Sciences.
Conflict of Interests
The authors declared no conflict of interests.
References
- 1.Youlden DR, Cramb SM, Dunn NA, et al. The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 2012;36:237–48. doi: 10.1016/j.canep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Wu JT, Kral JG. The NF-kappaB/IkappaB signaling system: a molecular target in breast cancer therapy. J Surg Res. 2005;123:158–69. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Turashvili G, Bouchal J, Burkadze G, et al. Differentiation of tumours of ductal and lobular origin: II Genomics of invasive ductal and lobular breast carcinomas. . Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2005;149:63–8. doi: 10.5507/bp.2005.006. [DOI] [PubMed] [Google Scholar]
- 4.Babu GR, Samari G, Cohen SP, et al. Breast cancer screening among females in Iran and recommendations for improved practice: a review. Asian Pac J Cancer Prev. 2011;12:1647–55. [PubMed] [Google Scholar]
- 5.Kolahdoozan S, Sadjadi A, Radmard AR, et al. Five common cancers in Iran. Arch Iran Med. 2010;13:143–6. [PubMed] [Google Scholar]
- 6.Omrani M, Hashemi M, Eskandari-Nasab E, et al. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med. 2014;8:259–67. doi: 10.2217/bmm.13.118. [DOI] [PubMed] [Google Scholar]
- 7.Hashemi M, Amininia S, Ebrahimi M, et al. Association between hTERT polymorphisms and the risk of breast cancer in a sample of Southeast Iranian population. BMC Res Notes. 2014;7:895. doi: 10.1186/1756-0500-7-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amininia S, Hashemi M, Ebrahimi M, et al. Association between CCNE1 polymorphisms and the risk of breast cancer in a sample of southeast Iranian population. Med Oncol. 2014;31:189. doi: 10.1007/s12032-014-0189-z. [DOI] [PubMed] [Google Scholar]
- 9.Eskandari-Nasab E, Hashemi M, Ebrahimi M, et al. Evaluation of CCL5 -403 G>A and CCR5 Delta32 gene polymorphisms in patients with breast cancer. Cancer Biomark. 2014;14:343–51. doi: 10.3233/CBM-140411. [DOI] [PubMed] [Google Scholar]
- 10.Eskandari-Nasab E, Hashemi M, Hasani SS, et al. Association between HLA-G 3'UTR 14-bp ins/del polymorphism and susceptibility to breast cancer. Cancer Biomark. 2013;13:253–9. doi: 10.3233/CBM-130364. [DOI] [PubMed] [Google Scholar]
- 11.Stephens P, Edkins S, Davies H, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–2. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 12.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–53. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 16.Wedekind JE, Dance GS, Sowden MP, et al. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–16. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 17.Long J, Delahanty RJ, Li G, et al. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105:573–9. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xuan D, Li G, Cai Q, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013;34:2240–3. doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi G, Xiong H, Zhou C. APOBEC3 deletion polymorphism is associated with epithelial ovarian cancer risk among Chinese women. Tumour Biol. 2014;35:5723–6. doi: 10.1007/s13277-014-1758-7. [DOI] [PubMed] [Google Scholar]
- 20.Kidd JM, Newman TL, Tuzun E, et al. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuong KJ, Loeb LA. APOBEC3B mutagenesis in cancer. Nat Genet. 2013;45:964–5. doi: 10.1038/ng.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–83. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu A, Nagasaki K, Fujimori M, et al. Identification of novel deletion polymorphisms in breast cancer. Int J Oncol. 2008;33:261–70. [PubMed] [Google Scholar]
- 24.Zhang T, Cai J, Chang J, et al. Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet. 2013;22:1262–9. doi: 10.1093/hmg/dds513. [DOI] [PubMed] [Google Scholar]
- 25.Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]