Abstract
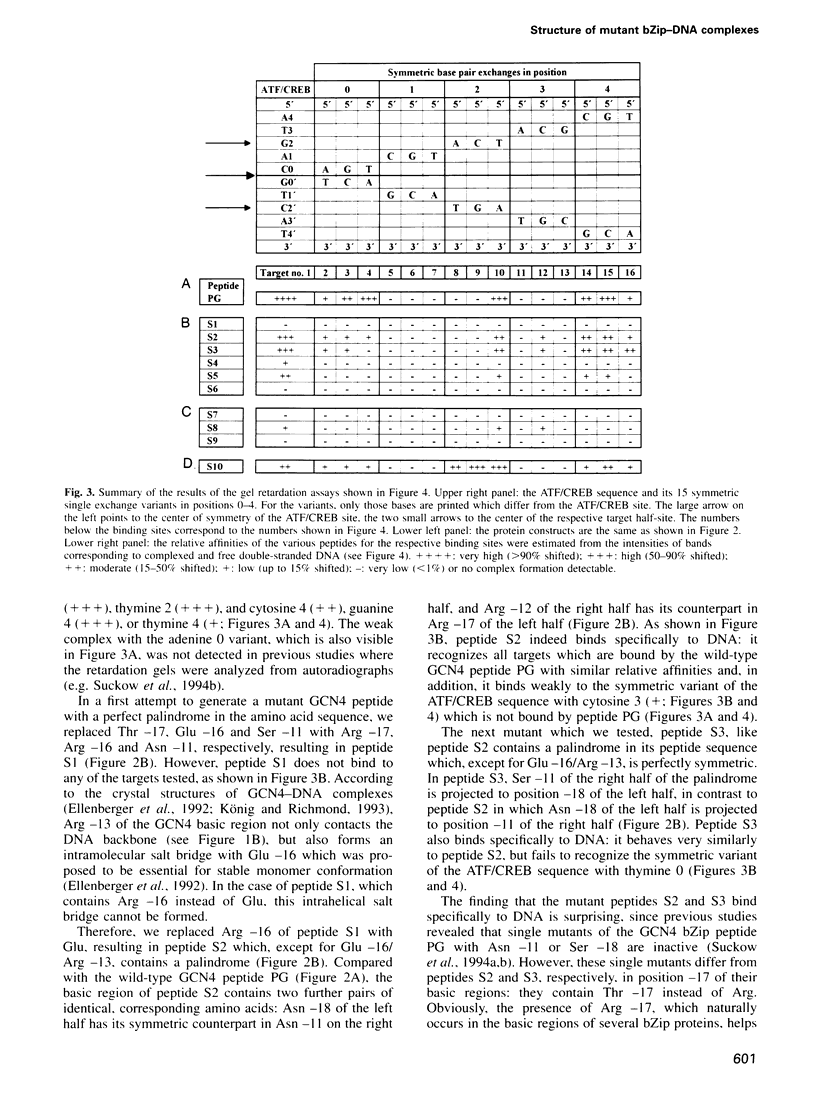
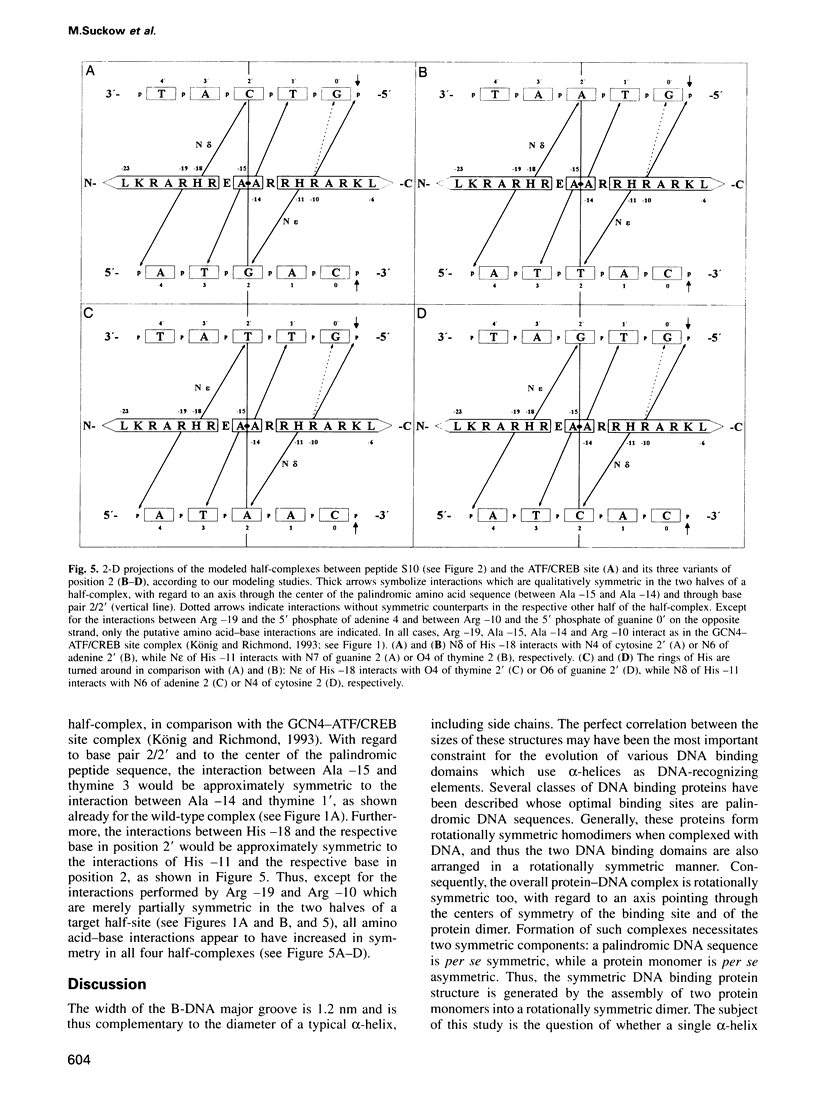
The complex between the yeast transcriptional activator GCN4 and the palindromic ATF/CREB site 5'- A4T3G2A1C0*G0'T1'C2'A3'T4'-3' shows dyad symmetry. The basic region of GCN4 contains a segment of 18 amino acids with a partially palindromic sequence: N-LKRARNTEA*ARRSRARKL-C. Symmetric residues are underlined. Apart from the ATF/CREB site, GCN4 also binds well to the symmetric variants with guanine in position 4 (5'-G4T3G2A1C0*G0'T1'C2'A3'C4'-3') or thymine in position 0 (5'-A4T3G2A1T0*A0'T1'C2'A3'T4'-3'). The half-sites of these sequences can be regarded as short pseudo-palindromes with central guanine 2/cytosine 2' base pairs. We investigated whether the geometry of the peptide of the basic region of GCN4 could be functionally related to the pseudo-palindromic character of some target half-sites. Since inspection of the X-ray structures of GCN4-DNA complexes reveals that several amino acid-DNA interactions are symmetric within the wild-type half-complexes, we introduced mutations into a GCN4 bZip peptide that improve the symmetry of the peptide. We found that most of the constructs retain specific DNA recognition. For one mutant, we conclude that it is not only capable of forming DNA complexes showing the well-known overall dyad symmetry, but that the protein-DNA interface of each half-complex can be divided further into two quasi-identical, quasi-symmetric substructures.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Agre P., Johnson P. F., McKnight S. L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989 Nov 17;246(4932):922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Glover J. N., Harrison S. C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995 Jan 19;373(6511):257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez G. A., Yamamoto K. K., Fischer W. H., Karr D., Menzel P., Biggs W., 3rd, Vale W. W., Montminy M. R. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989 Feb 23;337(6209):749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Liu F., Coukos W. J., Green M. R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989 Dec;3(12B):2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hurst H. C. Transcription factors. 1: bZIP proteins. Protein Profile. 1994;1(2):123–168. [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature. 1989 Aug 17;340(6234):568–571. doi: 10.1038/340568a0. [DOI] [PubMed] [Google Scholar]
- König P., Richmond T. J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol. 1993 Sep 5;233(1):139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Sakura H., Kanei-Ishii C., Sudo T., Yoshimura T., Fujisawa J., Yoshida M., Ishii S. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J. 1989 Jul;8(7):2023–2028. doi: 10.1002/j.1460-2075.1989.tb03610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón A., Wolberger C., Harrison S. C. Structure of phage 434 Cro protein at 2.35 A resolution. J Mol Biol. 1989 Jan 5;205(1):179–188. doi: 10.1016/0022-2836(89)90374-4. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Hoess R. H., DeGrado W. F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990 Aug 17;249(4970):774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Oas T. G., McIntosh L. P., O'Shea E. K., Dahlquist F. W., Kim P. S. Secondary structure of a leucine zipper determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1990 Mar 27;29(12):2891–2894. doi: 10.1021/bi00464a001. [DOI] [PubMed] [Google Scholar]
- PAULING L., COREY R. B., BRANSON H. R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci U S A. 1951 Apr;37(4):205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. GAL4 transcription factor is not a "zinc finger" but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M. A., Choi K. Y., Zalkin H., Brennan R. G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science. 1994 Nov 4;266(5186):763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sellers J. W., Vincent A. C., Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990 Oct;10(10):5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow M., Madan A., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. Creating new DNA binding specificities in the yeast transcriptional activator GCN4 by combining selected amino acid substitutions. Nucleic Acids Res. 1994 Jun 25;22(12):2198–2208. doi: 10.1093/nar/22.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow M., Schwamborn K., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. Replacement of invariant bZip residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 1994 Oct 25;22(21):4395–4404. doi: 10.1093/nar/22.21.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow M., von Wilcken-Bergmann B., Müller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993 Mar;12(3):1193–1200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow M., von Wilcken-Bergmann B., Müller-Hill B. The DNA binding specificity of the basic region of the yeast transcriptional activator GCN4 can be changed by substitution of a single amino acid. Nucleic Acids Res. 1993 May 11;21(9):2081–2086. doi: 10.1093/nar/21.9.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]