Abstract
Removal of cytosine methylation from the genome is critical for reprogramming and transdifferentiation and plays a central role in our understanding of the fundamental principles of embryo lineage development. One of the major models for studying cytosine demethylation is the mammalian germ line during the primordial germ cell (PGC) stage of embryo development. It is now understood that oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) is required to remove cytosine methylation in a locus-specific manner in PGCs; however, the mechanisms downstream of 5hmC are controversial and hypothesized to involve either active demethylation or replication-coupled loss. In the current study, we used the aorta-gonad-mesonephros (AGM) organ culture model to show that this model recapitulates germ line reprogramming, including 5hmC reorganization and loss of cytosine methylation from Snrpn and H19 imprinting control centers (ICCs). To directly address the hypothesis that cell proliferation is required for cytosine demethylation, we blocked PI3-kinase-dependent PGC proliferation and show that this leads to a G1 and G2/M cell cycle arrest in PGCs, together with retained levels of cytosine methylation at the Snrpn ICC, but not at the H19 ICC. Taken together, the AGM organ culture model is an important tool to evaluate mechanisms of locus-specific demethylation and the role of PI3-kinase-dependent PGC proliferation in the locus-specific removal of cytosine methylation from the genome.
Introduction
The mammalian germline is specified from the proximal epiblast at the time of implantation and is responsible for passing genetic information from parent to child. In recent years, we have come to appreciate the exquisite dynamics of epigenetic remodeling in the germ line not only in the late gestational period and after birth when transposons are repressed in males and females, respectively, but also immediately after specification in the early stages of embryo development. In fact, new lines of research suggest that epigenetic reprogramming, and in particular removal of 5-methylcytosine (5mC) from the newly specified germ line, serves as a protective measure to guard against transmission of epialleles from parent to child [1–4]. This mysterious period of germ line development called the primordial germ cell (PGC) period extends from embryonic (E) day E7.5 to E13.5 of development in the mouse embryo.
Epigenetic reprogramming in PGCs is a two-step process. The first step involves removal of almost all 5mC genome wide, together with depletion of Histone H3 Lysine 9 dimethylation (H3K9me2) and enrichment of H3K27me3 in chromatin [5]. This large-scale epigenetic remodeling occurs within 24–48 h of specification and is called phase I PGC reprogramming. By analyzing PGCs at the conclusion of phase I reprogramming (E9.5–E10.5), it is clear that a small number of interesting loci escape demethylation. These include gonadal-stage germ line genes, such as mouse vasa homologue (mvh), also called dead box polypeptide 4 (ddx4), deleted in Azoospermia-like (Dazl), synaptonemal complex protein 3 (Scp3), various imprinting control centers (ICCs), and sequences of the intracisternal A particle (IAP) repetitive element [6–8]. As PGCs settle in the gonad from E10.5 to E13.5, many of the loci that escaped cytosine demethylation in phase I PGC reprogramming will complete cytosine demethylation in a locus and time-specific manner [9,10]. The second period of PGC reprogramming ending at E13.5 is called phase II PGC reprogramming. Not all epigenetic marks are removed during phase II PGC reprogramming, for example, bivalent domains at developmental regulatory genes and H3K27me3 at retrotransposons persist until E13.5 and cytosine methylation is retained at some IAP sequences [9–13].
The mechanisms that regulate phase II PGC reprogramming of 5mC are under intense investigation. Recent research has implicated Tet methylcytosine dioxygenase 1 (Tet1) and Tet2 in converting 5mC to 5-hydroxymethylcytosine (5hmC) in the PGC genome between E10.5 and E13.5 [14–17]. The Tet enzymes, and in particular Tet1, are proposed to function by targeting removal of cytosine methylation from gonadal-stage meiotic genes of female PGCs as well as imprinted genes and ICCs [18]. Other investigators have proposed that loss of cytosine methylation is due to rapid proliferation coupled with repression of the maintenance methyltransferase 1 (Dnmt1) cofactor, Uhrf1 [19]. These models are not mutually exclusive as oxidation of 5mC to 5hmC could conceivably facilitate replication-coupled demethylation given that Dnmt1 has low affinity for 5hmC [20]. Others have suggested that base excision repair mediated by thymine DNA glycosylase (TDG) is required for demethylation of the insulin growth factor 2 (Igf2) ICC [21]. Still, other groups have reported that poly(ADP-ribose) polymerase (PARP) functions in phase II reprogramming to demethylate the ICC of H19/Igf2, Peg3, and the CpG islands of Scp3 and mvh [22,23]. The function of PARP is thought to involve replication-independent active DNA demethylation involving activation of the base excision repair pathway as well as transcriptional regulation of Tet1 [22,23].
Taken together, understanding the precise mechanisms that regulate DNA demethylation during phase II PGC reprogramming has critical implications in understanding how disease epialleles may arise during PGC reprogramming. Progress in this field has accelerated in the last 2 years; however, the field will benefit from well-characterized in vitro models that accurately recapitulate phase II reprogramming as it occurs in the embryo. Differentiation of PGCs in vitro from ESCs is an important model for phase I PGC reprogramming [24–26]. However, PGCs in this model are arrested in G2 of the cell cycle before initiating phase II reprogramming [24,25]. An alternate possibility to study phase II PGC reprogramming involves the aorta-gonad-mesonephros (AGM) model, which was recently used to examine the role of PARP and base excision repair in phase II PGC reprogramming [23]. However, critical details on the dynamics of 5hmC organization and proliferation-coupled loss of methylation at ICCs in the AGM model remain to be elucidated and must be clarified before widespread use of this model is adopted to study phase II PGC reprogramming.
Materials and Methods
AGM culture
AGMs were isolated from E10.5 embryos. All animal experiments were approved by The UCLA Institutional Animal Care and Use Committee, also known as the Chancellor's Animal Research Committee (ARC). Embryos were obtained from crosses between Oct4-IRES-Gfp (OG) transgenic males and females [27]. CD-1 females were also used where indicated. Embryos were staged by the detection of a vaginal plug (0.5 dpc) and upon dissection of morphological features, such as paddle-shaped forelimb buds, protruding hindlimb buds, as well as 35–40 pairs of somites. AGMs were transferred to a polyethylene tissue culture membrane (BD 353095) in a 24-well plate and grown for 3 days. The media used for the AGM culture contained 20% FBS (Lot: AWG18462; Hyclone), GMEM (Gibco), Penn/Strep, and 1×glutamine (Life Technologies). Media were changed every day by adding 500 μL to the bottom of the tissue culture insert and 20 μL to the top. The PI3-kinase inhibitor, LY294002, (Sigma-Aldrich) was added to the organ culture media at a concentration of 10 μM, and media containing the inhibitor were changed every day using the same volumes as indicated above. LY294002 for this experiment was resuspended using DMSO upon receipt and stored at −20 C. A fresh aliquot was used every time the media were made due to the inhibitor's sensitivity to freeze–thaw cycles. DMSO controls were used to control for the effect of the diluent.
Immunofluorescence
Paraffin-embedded sections were obtained after fixing the AGMs in 4% PFA overnight. Staining was performed using conventional methods with some exceptions in the case of 5hmC [26]. Permeabilization was performed using 0.5% TX-100 in PBS, and washes were performed using 0.2% Tween in PBS. The following antibodies were used: rabbit anti-human Mvh 1:100 (Abcam), mouse anti-human Amh IgG 1:100 (Abd Serotec), mouse anti-mouse 5mC 1:100 (Aviva Biosciences), rabbit anti-mouse 5hmC 1:100 (Active Motif), goat anti-mouse Oct4 1:100, mouse anti-mouse stage-specific embryonic antigen-1 (SSEA-1) 1:100, and goat anti-human Mvh 1:100. Visualization was performed by using isotype-specific secondary antibodies conjugated to fluorophores. Images were captured using a Zeiss LSM 780 confocal microscope. All immunofluorescence and histology experiments were performed on at least n=3 independent biological replicates of gonads or AGMs.
Sorting
AGMs were harvested at day 3 of culture. Media were aspirated from the bottom of the well using a vacuum, and media on top of the membrane were slowly removed through the use of a pipette. The AGM was washed once in calcium and magnesium-free DPBS (Gibco) before addition of 0.05% trypsin (Gibco). Once trypsin was added, the AGM was separated from the membrane by carefully pipetting several times. The AGM as well as the trypsin was then transferred to a 15-mL conical tube containing 3 mL of 0.05% trypsin. This was placed at 37°C for 5 min, followed by gentle flicking and placing it back at 37°C for 5 more min. Trypsin was neutralized with 3 mL of mouse embryonic fibroblast media, and the AGM was further dissociated by pipetting several times. This solution was then centrifuged for 5 min at 1.2 K rpm and resuspended in 1% BSA before sorting. 7AAD (BD) was added at 1:50, and only 7AAD-negative and GFP-positive cells were used for further analysis.
Flow analysis for cell cycle
AGMs were cultured in 10 μM EdU (Life Technologies) for 2 h. The AGMs were then harvested according to the sorting protocol above. Staining of the harvested AGMs was performed according to previous published studies with intracellular staining for Mvh detected with Alexa Flour 488 used to discriminate germ line cells from nongerm cells [28]. Samples were analyzed using the LSRII (BD) flow cytometry machine. To identify the germ cell population, a scatter plot for Alexa Flour 488 nm (log scale) versus side scatter (log scale) was set up. A gate was established around the 488 nm-positive cells (germ cells) as well as the 488 nm-negative cells (somatic cells). Cellular proliferation was then assessed by using these gates and establishing a scatter plot for APC 647 nm (EdU) (log scale) versus PI (Linear scale). A gate was set up around the 647 nm-positive cells, which were the cells passing through the S phase during the period the AGM cultures were exposed to EdU. Gates were then set up around the 647 nm-negative cells at 50 K on the PI linear scale and 100 K to identify cells in G1 and G2, respectively. Further analysis and quantification were performed using FlowJo (version 9.3.13). For detailed information of how to set up the gates for cell cycle analysis, refer to Wakeling et al. [28].
Sexing
To sex the AGMs, the head of the embryo was removed and placed in a separate PCR tube. Twenty microliters of DEPC water was added. This was followed by the mechanical breakdown of the head by pipetting several times. The PCR tubes containing the embryo heads were placed in a PCR machine and incubated for 5 min at 95°C. After heat incubation, the debris as well as the supernatant was transferred to an empty 1.5-mL microcentrifuge tube. This was followed by centrifugation at 13.2 K rpm for 5 min. The supernatant containing the DNA was transferred to a clean 1.5-mL microcentrifuge tube, and 1 μL was used for the PCR reaction. PCR products were run on a 2% gel at 90 V for 1 h. The result is two bands around 300 bp in males, whereas females exhibit only one band equivalent in size to the smaller fragment in males. SMCX-1: CCGCTGCCAAATTCTTTGG. SMCY-1: TGAAGCTTTTGGCTTTGAG.
Bisulfite PCR
DNA was extracted from GFP-positive PGCs at day 3 of culture using the Zymo Quick-gDNA miniprep kit (Zymo). The DNA was subsequently subjected to bisulfite treatment using the EZ DNA methylation kit (Zymo). Gene-specific PCR amplifications were performed by using primers against the Snrpn DMR1 as described in El-Maarri et al. [29], the H19 DMR as described in Kagiwada et al. [19], and IAP as described in Hajkova et al. [6]. PCR products were run on a 1.2% gel and purified using the QIAquick gel extraction kit (Qiagen) and ligated to a pCR-Topo2.1 cloning vector (TOPO cloning kit; Invitrogen). At least 10 clones were picked for analysis and submitted for sequencing.
Snrpn primer sequences:
Snrpn F1: AAA TTT GTG TGA TGT TTG TAA TTA TTT GGG
Snrpn R1: AAA ATC CAC AAA CCC AAC TAA CCT TCC
Snrpn F2: AAT TAT ATT TAT TAT TTT AGA TTG ATA GTG AT
Snrpn R2: TTT ACA AAT CAC TCC TCA AAA CCA A
H19 primer sequences:
H19 F1: AAT GGT TGA ATT TTA GTT TTT GTT TTT ATG GTT
H19 R1: ACC AAT ACA ATC CCA CAT ACT TTA TCA TAA AA
IAP primer sequences:
IAP F1: TTG TGT TTT AAG TGG TAA ATA AAT AAT TTG
IAP R1: CAA AAA AAA CAC ACA AAC CAA AAT
Statistics
Statistical analysis between two groups was performed using a nonparametric t-test. Statistical analysis involving more than two groups was performed using a one-way analysis of variance, followed by Tukey's multiple comparison test with 95% confidence intervals. Correlation of the PGC number with loss of methylation from Snrpn and H19 was performed using linear regression analysis with 95% confidence intervals. In all cases, P<0.05 was considered significant. Data were obtained from at least n=3 independent biological replicates of gonads or AGMs.
Results
PGCs in the AGM organ culture maintain global hypomethylation of 5mC
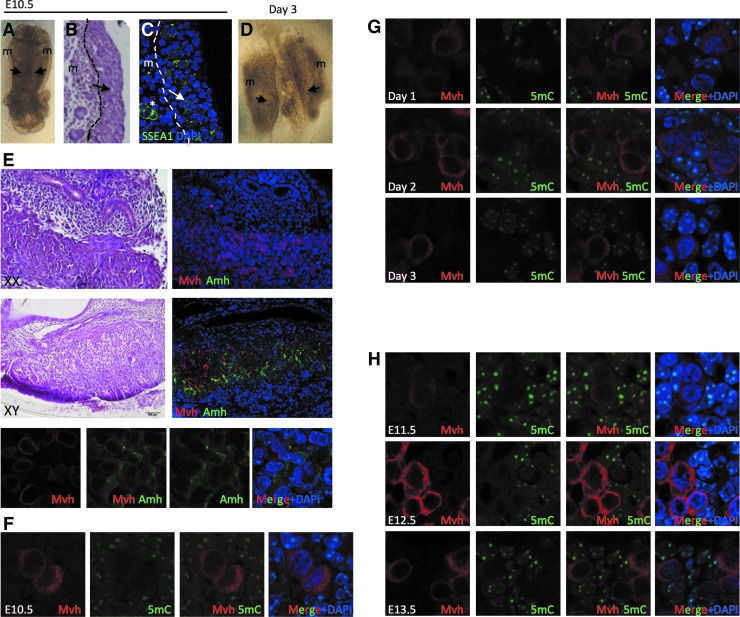
To determine whether the AGM organ culture model recapitulates phase II cytosine demethylation, we first isolated the AGM from CD1 or CD1 Oct4-IRES-Gfp (OG) mice and cultured each AGM individually on transwell inserts for 3–5 days (Fig. 1A). At the time of isolation from the embryo (E10.5), the genital ridge appears as a small medial thickening adjacent to the mesonephros by phase-contrast microscopy (Fig. 1A) and hematoxylin and eosin staining (Fig. 1B). PGCs were identified in the genital ridge and mesonephros (m) by immunofluorescence with SSEA-1 (Fig. 1C, white arrow). By the third day of culture, the genital ridge becomes large and well defined (Fig. 1D). Unlike in vivo gonad development where embryonic testes and ovaries can be distinguished morphologically at E13.5 [30,31], male and female gonads in the organ culture model are indistinguishable from each other, consistent with testicular cords not developing in males when the AGM culture is established at E10.5 before sex determination [32].
FIG. 1.
Primordial germ cells (PGCs) enter the gonad hypomethylated and remain hypomethylated over the next 3 days in vivo and ex vivo. (A) Phase-contrast image of an E10.5 aorta-gonad-mesonephros (AGM). Black arrows point to the genital ridge, (m) refers to mesonephros. (B) Histology of an E10.5 AGM. Dotted line separates the genital ridge (black arrow) from the mesonephros (m). (C) Immunofluorescence of E10.5 AGM for stage-specific embryonic antigen-1 (SSEA-1)-positive PGCs (green), nuclei are detected using DAPI (blue), white star refers to PGCs outside of the genital ridge. Dotted line separates the genital ridge from the mesonephros. (D) Phase-contrast image of an AGM cultured for 3 days. Black arrows point to the genital ridge, m refers to the mesonephros. (E) Histology (left) and immunofluorescence (right) of a female (XX) AGM cultured for 5 days (top) and male (XY) genital ridge (bottom). Bottom panel includes high-power images (63×) of the respective male section. Mvh-positive PGCs (red), Amh-positive sertoli cells (green), and DAPI nuclei (blue). (F) Immunofluorescence of E10.5 AGMs for Mvh-positive PGCs (red) and 5-methyl cytosine (5mC) (green). (G) Days 1–3 AGM organ culture for Mvh-positive PGCs (red) and 5mC (green). (H) E11.5–E13.5 genital ridges. All images were obtained using at least n=3 independent biological replicates of gonads or AGMs.
To determine whether male and female gonads can be distinguished molecularly by the presence or absence of the antimullerian hormone (Amh), which is expressed exclusively in sertoli cells of the male gonad (not Mvh-positive germ cells), we evaluated AGMs at days 1, 2, 3, and 5 of organ culture (shown is day 5, Fig. 1E). We found that AGMs derived from XX embryos never expressed Amh at any time point (Fig. 1E). In contrast, AGMs derived from XY embryos acquire Amh-positive cells between days 1 and 2 of organ culture exclusively in the developing genital ridge, consistent with previous reports in the embryo, demonstrating that AMH first becomes expressed at E12.5 [33]. We also discovered that the majority of PGCs that were identified using Mvh staining in XY gonads were closely associated with the Amh-positive sertoli cells.
Next, we sought to determine whether Mvh-positive PGCs in the genital ridge remain globally hypomethylated within the ex vivo organ culture. We first used immunofluorescence to stain genital ridges at E10.5 and, as expected, the Mvh-positive PGCs have low global levels of 5mC in the nucleus (Fig. 1F). This is in contrast to the surrounding somatic cells of the genital ridge, which exhibit discrete foci of 5mC that correlate with DAPI-positive foci (Fig. 1F). Next, we stained E10.5 cultured AGMs on days 1–3 of organ culture (Fig. 1G). Our results show that 5mC remains globally depleted in the Mvh-positive PGCs over the 3 days of culture (Fig. 1G). Furthermore, analysis of Mvh and 5mC by immunofluorescence in gonads from the embryo at E11.5–E13.5 reveals staining patterns that are indistinguishable from the results obtained in the organ culture (Fig. 1H). Taken together, the E10.5 AGM organ culture model over the first 3 days of culture exhibits the same hypomethylated characteristics of Mvh-positive PGCs found in the embryo from E11.5 to E13.5, despite the absence of cord formation in males.
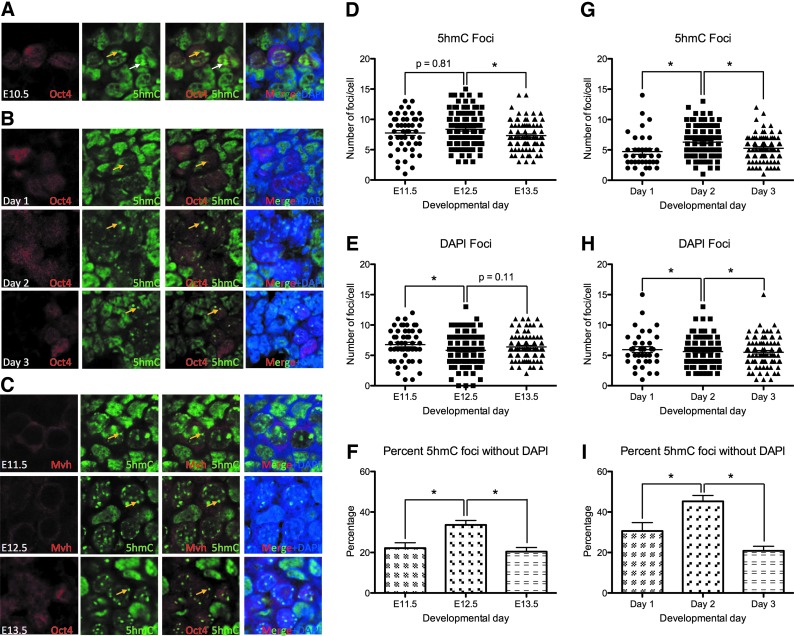
PGCs in the AGM organ culture develop 5hmC foci within 24 h of culture
Although the PGC genome exhibits low levels of 5mC from E10.5 to E13.5, previous studies have revealed that 5hmC becomes enriched in genomic territories at E10.5, resolving to discrete 5hmC foci in the majority of PGCs by E11.5 [17]. To test this in the organ culture model, we first evaluated 5hmC at E10.5 and show that SSEA-1-positive PGCs exhibit both territories (white arrow) and foci (yellow arrow) of 5hmC (Fig. 2A). However, after just 1 day of AGM organ culture, we find that almost all Oct4-positive PGCs exhibit discrete foci of 5hmC and these foci are maintained at days 2 and 3 of organ culture (yellow arrows, Fig. 2B). Analysis of the surrounding somatic cells shows that 5hmC is uniformly enriched in the somatic cell nuclei from days 1–3 of organ culture without obvious enrichment at DAPI foci (Fig. 2B).
FIG. 2.
PGCs during the 3 days of in vitro AGM organ culture recapitulate the nuclear rearrangement of 5-hydroxymethylcytosine (5hmC) identified in PGCs in the embryo. (A) Immunofluorescence of E10.5 AGMs for Oct4-positive PGCs (red), 5hmC (green), and DAPI (blue). (B) AGMs cultured for 3 days to identify Oct4-positive PGCs (red), 5hmC (green), and DAPI (blue). (C) E11.5–E13.5 genital ridges stained for Mvh or Oct4-positive PGCs (red), as indicated, and 5hmC (green) and DAPI (blue). All images were obtained by using at least n=3 independent biological replicates of gonads or AGMs. (D) Quantification of 5hmC foci in individual PGC nuclei between E11.5 and E13.5. (E) Quantification of DAPI foci in individual PGC nuclei between E11.5 and E13.5. (F) Quantification of 5hmC foci that are negative for DAPI in individual PGC nuclei between E11.5 and E13.5. (G) Quantification of 5hmC foci in individual PGC nuclei during AGM organ culture on days 1–3. (H) Quantification of DAPI foci in organ culture PGCs between days 1–3. (I) Quantification of 5hmC foci that are negative for DAPI in individual PGC nuclei during organ culture on days 1–3. Statistical data were obtained by using at least n=3 independent biological replicates of gonads or AGMs. *Refers to statistical significance P<0.05. (A–C) Yellow arrow indicates 5hmC foci, white arrows indicate 5hmC territories.
To prove that the localization of 5hmC in the organ culture model recapitulates results in the embryo, we stained embryonic gonads at E11.5, E12.5, and E13.5 for 5hmC, together with the PGC markers, Oct4 and Mvh, and show that similar to the AGM model, most PGCs contain 5hmC foci starting at E11.5 and these foci are identified in all PGC nuclei through to E13.5 (yellow arrows, Fig. 2C). Similarly, the somatic cells of the gonad in vivo from E11.5 to E13.5 have 5hmC present throughout the nucleus in uniform levels, without specific enrichment in DAPI-positive foci. Next, we quantified the numbers of 5hmC foci (Fig. 2D) and DAPI foci (Fig. 2E) per PGC nucleus from E11.5 to E13.5 and show that PGCs in the embryo have an average 8 5hmC foci and 6–7 DAPI foci per nucleus. In the organ culture model, the average number of 5hmC foci per PGC nucleus was slightly lower at 5–6 (Fig. 2G). However, the average number of DAPI foci was similar to PGCs in the embryo at 6–7 foci per nucleus (Fig. 2H).
Given that the numbers of 5hmC and DAPI foci are relatively stable from E11.5 to E13.5, we quantified the percentage of 5hmC-positive foci that do not correspond to DAPI foci and show that 21% of 5hmC foci at E11.5 do not correspond to DAPI foci, whereas, at E12.5, this increases to around 35%. By E13.5, the percentage of 5hmC foci that are DAPI negative is restored to the levels seen at E11.5 (Fig. 2F). Similarly. in the organ culture model, this trend is also observed (Fig. 2I). Taken together, our data indicate that the organization of 5hmC in PGC nuclei in the organ culture model recapitulates the nuclear rearrangement that occurs in vivo from E11.5 to E13.5 with 5hmC foci uncoupling from DAPI-dense chromocenters in PGCs.
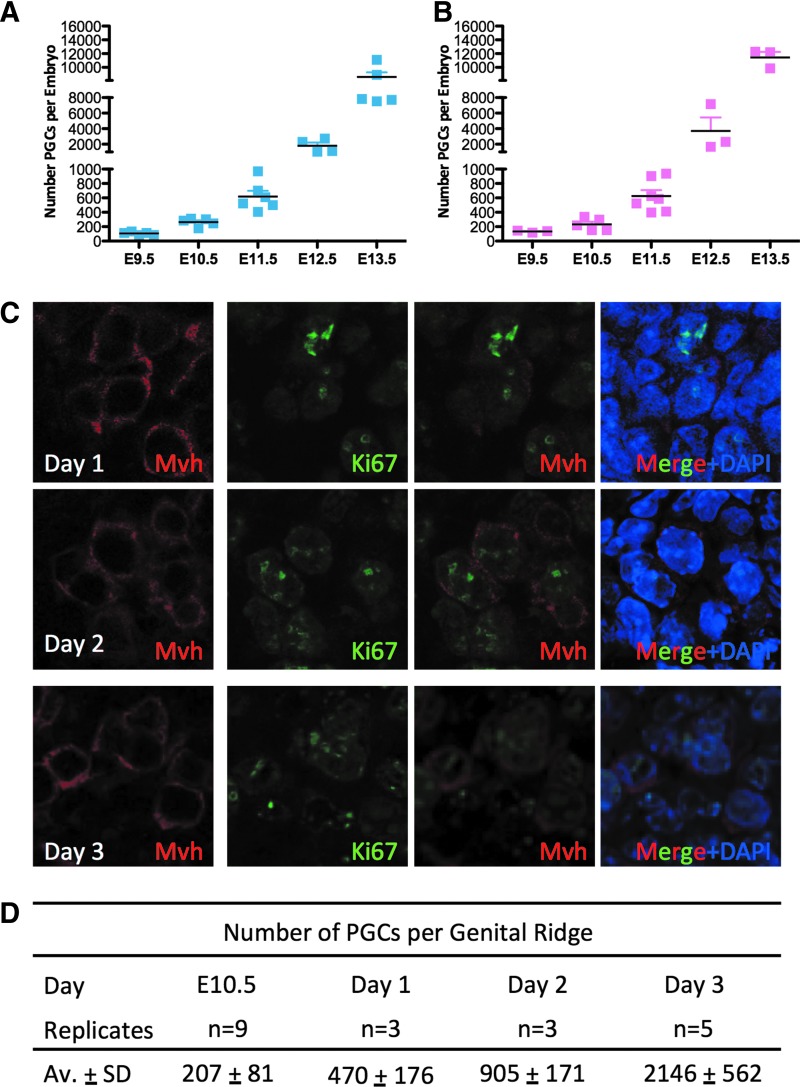
PGC number in the AGM organ culture increases by 10-fold over the first 3 days of culture
Given that phase II reprogramming is associated with proliferation [19], we next evaluated the PGC number in vivo and in the AGM organ culture model. First, we evaluated the total number of GFP-positive PGCs sorted from male and female OG embryos starting at E9.5 through to E13.5 (Fig. 3A, B). Our data show that the PGC number increases with each embryonic day of development in both males (Fig. 3A) and females (Fig. 3B). However, our data show that the largest increase in PGC number occurs between E11.5 and E13.5 in both sexes. Assuming a constant rate of apoptosis over the 3-day window, we estimate that the average doubling time of PGCs from E11.5 to E13.5 in the OG mouse strain is 11.8 h.
FIG. 3.
PGCs in the in vitro cultured AGMs are Ki67 positive and increase in number during 3 days of culture. (A) Number of OG-positive PGCs in male and (B) female embryos at the indicated time points. (C) Immunofluorescence of in vitro cultured AGM from days 1–3 for Mvh-positive PGCs (red), Ki67 (green), and DAPI (blue). (D) Average number of OG-positive PGCs in cultured AGMs at days 1–3 of culture. All images were obtained by using at least n=3 independent biological replicates of gonads or AGMs.
To determine whether Mvh-positive PGCs in the organ culture are proliferative, we first stained for Ki67, a marker of cells that are in cycle (Fig. 3C). We show that all Mvh-positive PGCs are in cycle and Ki67 positive from days 1–3. To count the total number of PGCs, we used the OG mouse from Fig. 3A and B and counted the total number of OCT4+ PGCs in the genital ridges at days 1–3 of organ culture. Given that we did not find a statistically significant difference in the number of male and female PGCs in vivo, we pooled cultured AGMs at each time point and performed FACS to count the PGC number (Fig. 3D). Consistent with the Ki67 staining, indicating that PGCs are in cycle, our data reveal that after 1 day of organ culture, the average number of PGCs more than doubled from 207 to 470 PGCs per AGM. Then, from days 1 to 2, the PGC number doubles again to an average of 905 PGCs per AGM. Finally, at day 3, we recovered an average of 2,146 PGCs per AGM. Taken together, using the E10.5 AGM organ culture model, we discovered that the total number of PGCs increases substantially in the first 3 days of culture, but not to the extent of PGCs in the embryo during the same time period. This suggests that proliferation occurs in PGCs in the E10.5 AGM organ culture, but expansion of the germ line ex vivo in the AGM model is not as robust. Given that we did not detect an increase in 7AAD-positive (dead) PGCs in the AGM culture at any time point, we hypothesize that the reduced number of PGCs found in the organ culture at day 3 relative to the E13.5 embryo is not due to an increase in apoptosis, but rather due to fewer cell divisions taking place over the span of 3 days.
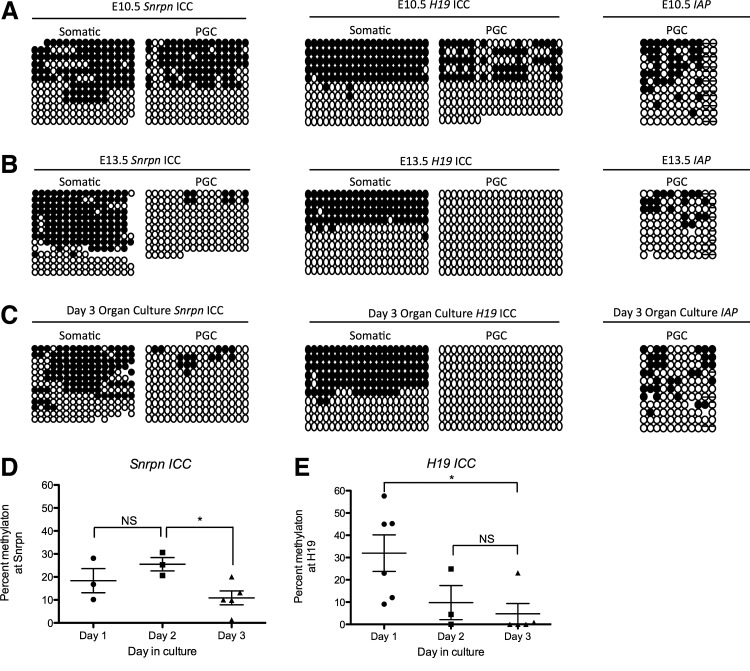
Loss of methylation from the Snrpn ICC and H19 ICC occurs within the first 3 days of organ culture
Given that the number of PGCs at day 3 in the organ culture is lower than what is expected when comparing the number of PGCs after 3 days of development in the embryo, we next sought to evaluate the demethylation dynamics of the Snrpn ICC and H19 ICC in the AGM organ culture given the hypothesis that demethylation may be linked to cell division. Using the OG mouse, we first sorted GFP-positive PGCs at E10.5 and confirmed previous reports [6] that cytosines in a CpG sequence context at the Snrpn and H19 ICCs are methylated at E10.5 similar to GFP-negative somatic cells (Fig. 4A). In contrast, at E13.5 cytosine, methylation at the Snrpn and H19 ICCs in the GFP-positive PGCs is erased (Fig. 4B), whereas the GFP-negative somatic cells still exhibit methylated and unmethylated alleles at these ICCs (Fig. 4B). As a positive control, we show that IAP is methylated in GFP-positive PGCs at E10.5 (Fig. 4A) and does not fully demethylate in PGCs at E13.5, consistent with previous reports [7].
FIG. 4.
Imprint erasure at the Snrpn and H19 imprinting control centers (ICCs) happens within 3 days in the organ culture model. (A) Bisulfite PCR of the Snrpn and H19 ICCs, as well as IAP, comparing somatic cells (left) and OG-sorted PGCs (right) from the embryo at E10.5, (B) E13.5, (C), and after 3 days of AGM organ culture. Black circles, methylated cytosines; white circles, unmethylated cytosines. (D) Quantification of average cytosine methylation in OG-sorted PGCs at the Snrpn ICC from the in vitro cultured AGM at days 1–3. (E) Quantification of average cytosine methylation in OG-sorted PGCs at the H19 ICC from the in vitro cultured AGM at days 1–3. * Refers to statistical significance P<0.05.
Next, we turned to the E10.5 AGM organ culture model and monitored the Snrpn ICC, H19 ICC, and IAP demethylation in GFP-positive-sorted PGCs at day 3 of organ culture (Fig. 4C). Our data reveal that by day 3, methylation at the Snrpn ICC and H19 ICC is erased in the GFP-positive PGCs, whereas GFP-negative somatic cells still exhibit methylated and unmethylated alleles (Fig. 4C). In contrast, similar to PGCs in the embryo, IAP does not completely demethylate in the germ line (Fig. 4C).
To evaluate the timing of CpG erasure in the organ culture model, we evaluated methylation at the Snrpn ICC and H19 ICC at days 1–3 of culture (Fig. 4D, E, respectively). We discovered that the average percent CpG methylation at the Snrpn ICC is around ∼20% at days 1 and 2 of AGM culture. However, by day 3, the percentage of methylated cytosines in the CpG sequence context of the Snrpn ICC is significantly reduced to around 10% (Fig. 4D). Similarly, the paternally methylated H19 ICC also undergoes a significant decrease in cytosine methylation within the first 3 days of organ culture (Fig. 4E). However, compared with the Snrpn ICC, where a significant decrease in DNA methylation was observed between days 2 and 3 of AGM culture, a significant decrease in cytosine methylation at the H19 ICC was observed between days 1 and 3.
Loss of cytosine methylation from the Snrpn ICC in PGCs in the AGM culture strongly correlates with increasing cell number
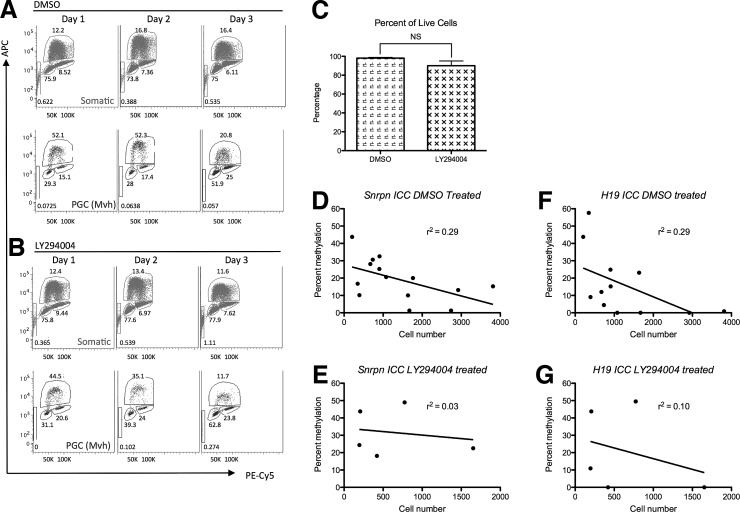
To determine whether loss of cytosine methylation from the Spnrn or H19 ICC correlates with proliferation, we cultured E10.5 AGMs in the presence of either DMSO or the PI3-kinase inhibitor, LY294004 (Fig. 5A, B). PI3-kinase regulates PGC proliferation through the cKIT receptor downstream of kit ligand signaling [34]. Addition of 10 μM LY294004, a potent inhibitor of PI3-kinase, effectively blocks PGC proliferation in vitro [34]. Therefore, we hypothesized that the culture of E10.5 genital ridges in the presence of 10 μM LY294004 would cause PGCs to arrest in the cell cycle and provide a novel approach for examining the role of PI3-kinase-dependent PGC proliferation in ICC erasure. To test this, we first performed a cell cycle assay in the presence and absence of LY294004 (Fig. 5A, B). In this assay, the cell cycle dynamics of PGCs were distinguished from somatic cells in the same sample using intracellular staining for Mvh [28]. Similar to previous reports [19], our data show that the somatic cells of the organ culture are mostly in the G1 phase of the cell cycle (73%–75%). In contrast, the majority of Mvh-positive PGCs are in the S phase during days 1 and 2 of organ culture (Fig. 5A). In LY294004-treated cultures, the majority of PGCs are not in the S phase and instead are in G1 and G2/M of the cell cycle by day 2 of organ culture (Fig. 5B). To evaluate whether LY294004 causes toxicity, we determined cell viability by calculating the percentage of cells that are negative for 7aad; this result showed no significant difference between DMSO and LY294004-treated cultures, with viability being on average>90% (Fig. 5C).
FIG. 5.
Imprint erasure dynamics at the Snrpn and H19 ICCs in PGCs after treatment with PI3-kinase inhibitor. (A) Cell cycle analysis of Mvh-positive PGCs and Mvh-negative somatic cells using Edu incorporation of in vitro cultured AGMs from days 1–3 of culture with DMSO or (B) 10 μM LY294004. Somatic cells (red), Mvh-positive PGCs (blue), Edu incorportation (APC), DNA content (PE-Cy5). (C) Percentage of viable cells between DMSO (vehicle) and LY294004-treated samples. (D) Percent cytosine methylation at the Snrpn ICC plotted as a function of the cell number during days 1–3 of AGM cell culture in the presence of DMSO or (E) LY294004. (F) Percent cytosine methylation at the H19 ICC plotted as a function of the cell number during days 1–3 of AGM cell culture in the presence of DMSO or (G) LY294004.
To determine whether addition of 10 μM LY294004 blocks demethylation, we added either DMSO or LY294004 to the AGM organ culture at the time of plating and used linear regression analysis, where CpG methylation in individual samples treated with either DMSO or LY294004 was plotted relative to the PGC number (Fig. 5D–G). Our results demonstrate that CpG methylation at the Snrpn ICC in control (DMSO-treated) conditions is correlated (r2=0.29) with increasing PGC number (Fig. 5D). In contrast, culture of AGMs with 10 μM LY294004 abolished this correlation (r2=0.03), with retention of CpG methylation at the Snrpn ICC in the presence of LY294004 (Fig. 5E). Therefore, blocking PI3K in the AGM organ culture model blocks replication-coupled demethylation of the Snrpn ICC. Similarly, analysis of the H19 ICC under control (DMSO-treated) conditions also revealed a correlation (r2=0.29) between cell number and the loss of CpG methylation (Fig. 5F). However, unlike the results at the Snrpn ICC, culture of AGMs with 10 μM of LY294004 had a modest effect, but did not prevent demethylation at the H19 ICC (r2=0.10). Taken together, this suggests that PI3-kinase-dependent proliferation does not play a major role in loss of CpG methylation at the H19 ICC (Fig. 5G).
Discussion
In the present study, our results reveal that PI3-kinase-dependent PGC proliferation plays an important role in the process of imprint erasure at the Snrpn ICC in PGCs, but not a major role at the H19 ICC, indicating that different genomic loci undergo demethylation through different mechanisms. We have also shown that a 3-day ex vivo AGM organ culture recapitulates the classic epigenetic reprogramming events in murine PGCs during embryo development from E11.5 to E13.5, including the formation of 5hmC foci; the loss of cytosine methylation from the Snrpn and H19 ICCs with retention of some methylation at IAP. Therefore, our results show that the AGM organ culture constitutes a faithful ex vivo model to study different modes of PGC methylation reprogramming.
Loss of imprinting at the Snrpn locus in humans is associated with the Prader-willi or Angelman Syndrome [29]. Conversely, altered imprinting at the H19 locus is associated with the Beckwith–Wiedemann Syndrome [35]. The majority of cases are due to genetic mutations or rearrangements that result in permanent changes to the mechanism of allele-specific gene expression. However, given that allele-specific gene expression is regulated primarily by DNA methylation, the role of aberrant epigenetic mechanisms (epimutations) in causing imprinting syndromes in children is a concern. There are currently two mechanistic models for ICC erasure in mammalian PGCs. These include, replication-independent active demethylation involving base excision repair and a second mechanism that involves replication-coupled demethylation [19,21–23].
In previous studies, evidence for replication-coupled demethylation of ICCs was proposed due to an undeniable correlation of the PGC cell number with the kinetics of CpG demethylation [19]. In the current study, our data support a major role for PI3-kinase-dependent replication-coupled loss of CpG methylation at the Snrpn ICC, but a minor to negligible role in demethylation of the H19 ICC. Evidence for active demethylation was hypothesized to occur downstream of poly(ADP-ribosyl)ation by experiments that involved inhibiting PARP in the E10.5 AGM organ culture model [23] as well as injection of PARP inhibitors into pregnant mice [22]. In both cases of experimental contexts, it was proposed that demethylation of the H19/IGF2 ICC was sensitive to PARP inhibition. In the current study, our data provide independent support that demethylation of the H19 ICC is active because, unlike the Snrpn ICC, this locus continues to demethylate even when cell proliferation is blocked. Taken together, our data support the theory that multiple independent mechanisms act to promote demethylation of ICCs in the germ line.
The first step in demethylation at the majority of ICCs involves conversion of 5mC to 5hmC by Tet1 [14,16]. In the current study, we show that the E10.5 AGM organ culture model recapitulates the unique rearrangement of 5hmC in PGCs [17], with the conversion of 5hmC territories into 5hmC-enriched foci. Therefore, use of the AGM organ culture model to evaluate oxidation of 5mC and potential roles as an independent epigenetic mark is warranted. It is clear in both the embryo and AGM organ culture model that 5hmC does not disappear completely from the germ line genome and instead remains as discrete foci. The purpose of residual 5hmC staining in the germ line at the conclusion of the PGC period is currently unknown.
In conclusion, our data support the idea that mechanisms of demethylation in PGCs are locus specific such that a PI3-kinase-dependent replication-coupled mechanism supports the removal of CpG methylation from the Snrpn ICC, whereas removal of CpG methylation from the H19 ICC is largely independent of PI3-kinase. PI3-kinase-dependent PGC proliferation is hypothesized to occur downstream of estrogen signaling on the somatic cells of the gonad [34]. This finding is critical given that maternal exposure of bisphenol A (an endocrine disruptor) causes epigenetic changes at ICCs in exposed embryos [36]. Our study demonstrates that the E10.5 AGM organ culture model provides a new opportunity to evaluate modifiers of locus-specific cytosine demethylation during a highly reproducible nuclear reprogramming event in mammalian cells.
Acknowledgments
The authors would like to thank the UCLA BSCRC flow cytometry core for flow and FACS assistance. This work was supported by an R01 grant from the NIH (NIH/NICHD R01 HD058047) awarded to ATC as well as the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research. The authors would also like to acknowledge the support of the California Institute for Regenerative Medicine (CIRM) Predoctoral training grant (TG2-01169) for funding J.H.C.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Anway MD, Cupp AS, Uzumcu M. and Skinner MK. (2005). Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. and B PTRS. (2013). Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci 368:20110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skinner MK, Haque CG-BM, Nilsson E, Bhandari R. and McCarrey JR. (2013). Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and the subsequent germ line. PLoS One 8:e66318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Waal E, Yamazaki Y, Ingale P, Bartolomei M, Yanagimachi R. and McCarrey JR. (2012). Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci U S A 109:4163–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M. and Matsui Y. (2005). Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278:440–458 [DOI] [PubMed] [Google Scholar]
- 6.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J. and Surani MA. (2002). Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117:15–23 [DOI] [PubMed] [Google Scholar]
- 7.Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J. and Reik W. (2003). Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35:88–93 [DOI] [PubMed] [Google Scholar]
- 8.Maatouk DM, Kellam LD, Mann MRW, Lei H, Li E, Bartolomei MS. and Resnick JL. (2006). DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 133:3411–3418 [DOI] [PubMed] [Google Scholar]
- 9.Ng J, Kumar V, Muratani M, Kraus P, Yeo J, Yaw L. and Xue K. (2013). In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell 24:324–333 [DOI] [PubMed] [Google Scholar]
- 10.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W. and Reik W. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48:849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mochizuki K, Tachibana M, Saitou M, Tokitake Y. and Matsui Y. (2012). Implication of DNA demethylation and bivalent histone modification for selective gene regulation in mouse primordial germ cells. PLoS One 7:e46036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachs M, Onodera C, Blaschke K, Ebata KT, Song JS. and Ramalho-Santos M. (2013). Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep 3:1777–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP. and Surani MA. (2010). Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329:78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi S, Shen L, Liu Y, Sendler D. and Zhang Y. (2013). Role of Tet1 in erasure of genomic imprinting. Nature 504:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24:310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackett J a, Sengupta R, Zylicz JJ, Murakami K, Lee C, a Down T. and Surani MA. (2013). Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339:448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K. and Zhang Y. (2013). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res 23:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K. and Zhang Y. (2012). Tet1 controls meiosis by regulating meiotic gene expression. Nature 5:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagiwada S, Kurimoto K, Hirota T, Yamaji M. and Saitou M. (2013). Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32:340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X. and Cheng X. (2012). Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40:4841–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki Y, Lee J, Matsuzawa A, Kohda T, Kaneko-Ishino T. and Ishino F. (2014). Active DNA demethylation is required for complete imprint erasure in primordial germ cells. Sci Rep 4:3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciccarone F, Klinger FG, Catizone A, Calabrese R, Zampieri M, Bacalini MG, De Felici M. and Caiafa P. (2012). Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One 7:e46927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H. and Saitou M. (2012). Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science 338:971–975 [DOI] [PubMed] [Google Scholar]
- 25.Hayashi K, Ohta H, Kurimoto K, Aramaki S. and Saitou M. (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146:519–532 [DOI] [PubMed] [Google Scholar]
- 26.Vincent JJ, Huang Y, Chen P-Y, Feng S, Calvopiña JH, Nee K, Lee SA, Le T, Yoon AJ, et al. (2013). Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell 12:470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengner CJ, Camargo FD, Hochedlinger K, Grant G, Zaidi S, Gokhale S, Scholer HR. and Tomilin A. (2007). Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 1:403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakeling SI, Miles DC. and Western PS. (2013). Identifying disruptors of male germ cell development by small molecule screening in ex vivo gonad cultures. BMC Res Notes 6:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, et al. (2001). Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet 27:341–344 [DOI] [PubMed] [Google Scholar]
- 30.Koopman P, Munsterberg A, Capel B, Vivian N. and Lovell-Badge R. (1990). Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348:450–452 [DOI] [PubMed] [Google Scholar]
- 31.Sekido R. and Lovell-Badge R. (2008). Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453:930–934 [DOI] [PubMed] [Google Scholar]
- 32.McLaren ABM. (1990). Development of mouse germ cells in cultures of fetal gonads. Cell Differ Dev 31:185–195 [DOI] [PubMed] [Google Scholar]
- 33.Munsterberg A. and Lovell-badge R. (1991). Expression of the mouse anti-Mullerian hormone gene suggests a role in both male and female sexual differentiation. Development 624:613–624 [DOI] [PubMed] [Google Scholar]
- 34.Moe-Behrens GH, Klinger FG, Esklid W, Grotmol T, Haugen TB. and De Felici M. (2003). Akt/PTEN signaling mediates estrogen-dependent proliferation of primordial germ cells in vitro. Mol Endocrinol 17:2630–2638 [DOI] [PubMed] [Google Scholar]
- 35.Netchine I, Rossignol S, Azzi S, Brioude F. and Le Bouc Y. (2012). Imprinted anomalies in fetal and childhood growth disorders: the model of Russell-Silver and Beckwith-Wiedemann syndromes. Endocr Dev 23:60–70 [DOI] [PubMed] [Google Scholar]
- 36.Susiarjo M, Sasson I, Mesaros C. and Bartolomei MS. (2013). Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet 9:e1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]