Abstract
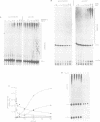
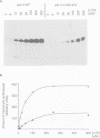
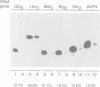
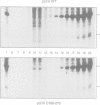
The alpha-amanitin domain or domain f of the largest subunit of RNA polymerases is one of the most conserved of these enzymes. We have found that the C-terminal part of domain f can be swapped between yeast RNA polymerase II and III. An extensive mutagenesis of domain f of C160, the largest subunit of RNA polymerase III, was carried out to better define its role and understand the mechanism through which C160 participates in transcription. One mutant enzyme, C160-270, showed much reduced transcription of a non-specific template at low DNA concentrations. Abortive synthesis of trinucleotides in a dinucleotide-primed reaction proceeded at roughly wild-type levels, indicating that the mutation did not affect the formation of the first phosphodiester bond, but rather the transition from abortive initiation to processive elongation. In specific transcription assays, on the SUP4 tRNA gene, pausing was extended but the rate of RNA elongation between pause sites was not affected. Finally, the rate of cleavage of nascent RNA transcripts by halted mutant RNA polymerase was increased approximately 10-fold. We propose that the domain f mutation affects the transition between two transcriptional modes, one being adopted during abortive transcription and at pause sites, the other during elongation between pause sites.
Full text
PDF


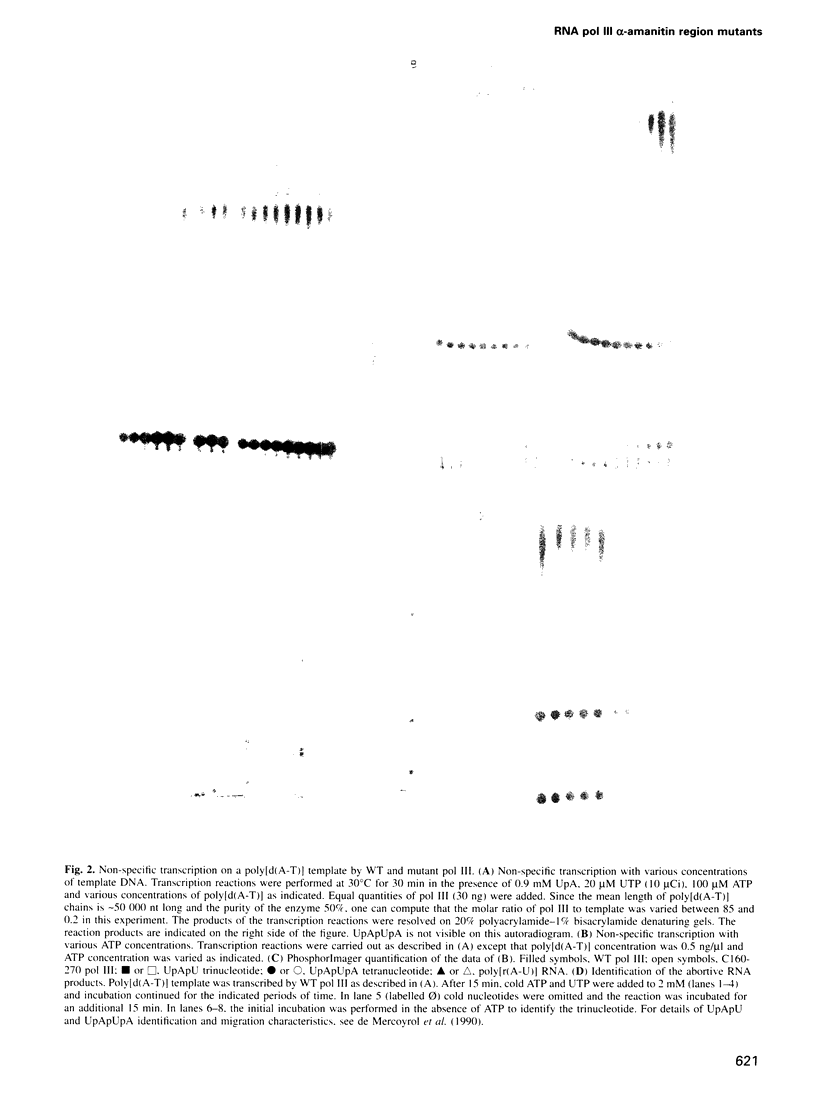








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmann C. R., Solow-Cordero D. E., Chamberlin M. J. RNA cleavage and chain elongation by Escherichia coli DNA-dependent RNA polymerase in a binary enzyme.RNA complex. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3784–3788. doi: 10.1073/pnas.91.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Lacroute F., Ruet A., Friesen J. D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992 Sep;12(9):4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Eigel A., Vögtel D., Feldmann H. Nucleotide sequences of yeast genes for tRNA(2), tRNA(2) and tRNA(1): homology blocks occur in the vicinity of different tRNA genes. EMBO J. 1982;1(3):291–295. doi: 10.1002/j.1460-2075.1982.tb01162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeleben C., Kassavetis G. A., Geiduschek E. P. Encounters of Saccharomyces cerevisiae RNA polymerase III with its transcription factors during RNA chain elongation. J Mol Biol. 1994 Jan 28;235(4):1193–1205. doi: 10.1006/jmbi.1994.1073. [DOI] [PubMed] [Google Scholar]
- Bartholomew B., Durkovich D., Kassavetis G. A., Geiduschek E. P. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993 Feb;13(2):942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Corden J. L. Clustered alpha-amanitin resistance mutations in mouse. Mol Gen Genet. 1995 Mar 20;246(6):778–782. doi: 10.1007/BF00290727. [DOI] [PubMed] [Google Scholar]
- Bartolomei M. S., Corden J. L. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987 Feb;7(2):586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berroteran R. W., Ware D. E., Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994 Jan;14(1):226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Lee J., Goldfarb A. Mapping of a contact for the RNA 3' terminus in the largest subunit of RNA polymerase. J Biol Chem. 1991 Dec 15;266(35):23932–23935. [PubMed] [Google Scholar]
- Borukhov S., Polyakov A., Nikiforov V., Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Sagitov V., Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993 Feb 12;72(3):459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Burton N., Cavallini B., Kanno M., Moncollin V., Egly J. M. Expression in Escherichia coli: purification and properties of the yeast general transcription factor TFIID. Protein Expr Purif. 1991 Oct-Dec;2(5-6):432–441. doi: 10.1016/1046-5928(91)90105-r. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992 1993;88:1–21. [PubMed] [Google Scholar]
- Chen Y., Weeks J., Mortin M. A., Greenleaf A. L. Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol Cell Biol. 1993 Jul;13(7):4214–4222. doi: 10.1128/mcb.13.7.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipres-Palacin G., Kane C. M. Cleavage of the nascent transcript induced by TFIIS is insufficient to promote read-through of intrinsic blocks to elongation by RNA polymerase II. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8087–8091. doi: 10.1073/pnas.91.17.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert T., Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992 Oct;6(10):1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985 Oct 25;260(24):13190–13198. [PubMed] [Google Scholar]
- Crerar M. M., Leather R., David E., Pearson M. L. Myogenic differentiation of L6 rat myoblasts: evidence for pleiotropic effects on myogenesis by RNA polymerase II mutations to alpha-amanitin resistance. Mol Cell Biol. 1983 May;3(5):946–955. doi: 10.1128/mcb.3.5.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Hermann-Le Denmat S., Lukhtanov E., Thuriaux P., Werner M., Sentenac A. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 1995 Aug 1;14(15):3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G. H., Lee D. N., Wang D., Chan C. L., Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994 Sep 2;269(35):22282–22294. [PubMed] [Google Scholar]
- Gabrielsen O. S., Hornes E., Korsnes L., Ruet A., Oyen T. B. Magnetic DNA affinity purification of yeast transcription factor tau--a new purification principle for the ultrarapid isolation of near homogeneous factor. Nucleic Acids Res. 1989 Aug 11;17(15):6253–6267. doi: 10.1093/nar/17.15.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen O. S., Oyen T. B. The requirement for the A block promoter element in tRNA gene transcription in vitro depends on the ionic environment. Nucleic Acids Res. 1987 Jul 24;15(14):5699–5713. doi: 10.1093/nar/15.14.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Zaychikov E. F., Abdukayumov M. N., Rabinov I. V., Richter V. I., Skoblov Y. S., Chistyakov P. G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989 Apr 1;180(3):577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- Hekmatpanah D. S., Young R. A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991 Nov;11(11):5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Riva M., Sentenac A., Fromageot P. Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985 Dec 5;260(28):15304–15310. [PubMed] [Google Scholar]
- Huet J., Sentenac A. The TATA-binding protein participates in TFIIIB assembly on tRNA genes. Nucleic Acids Res. 1992 Dec 25;20(24):6451–6454. doi: 10.1093/nar/20.24.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3'----5' direction in the presence of elongation factor SII. Genes Dev. 1992 Jul;6(7):1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Kashlev M., Lee J., Zalenskaya K., Nikiforov V., Goldfarb A. Blocking of the initiation-to-elongation transition by a transdominant RNA polymerase mutation. Science. 1990 May 25;248(4958):1006–1009. doi: 10.1126/science.1693014. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Joazeiro C. A., Pisano M., Geiduschek E. P., Colbert T., Hahn S., Blanco J. A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992 Dec 11;71(6):1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992 May 20;225(2):239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee D. N., Feng G., Landick R. GreA-induced transcript cleavage is accompanied by reverse translocation to a different transcription complex conformation. J Biol Chem. 1994 Sep 2;269(35):22295–22303. [PubMed] [Google Scholar]
- Lee J., Kashlev M., Borukhov S., Goldfarb A. A beta subunit mutation disrupting the catalytic function of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6018–6022. doi: 10.1073/pnas.88.14.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H., Kassavetis G. A., Geiduschek E. P. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994 Jan 28;235(4):1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- McDowell J. C., Roberts J. W., Jin D. J., Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994 Nov 4;266(5186):822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Moenne A., Camier S., Anderson G., Margottin F., Beggs J., Sentenac A. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 1990 Jan;9(1):271–277. doi: 10.1002/j.1460-2075.1990.tb08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Saurin W., Sentenac A. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. J Biol Chem. 1988 Jul 25;263(21):10048–10051. [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Nudler E., Kashlev M., Nikiforov V., Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995 May 5;81(3):351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- Olah J., Feldmann H. Structure of a yeast non-initiating methionine-tRNA gene. Nucleic Acids Res. 1980 May 10;8(9):1975–1986. doi: 10.1093/nar/8.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M., Newlands J., Das A., Goldfarb A., Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992 Feb 25;267(6):3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Rogalski T. M., Golomb M., Riddle D. L. Mutant Caenorhabditis elegans RNA polymerase II with a 20,000-fold reduced sensitivity to alpha-amanitin. Genetics. 1990 Dec;126(4):889–898. doi: 10.1093/genetics/126.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M. D., Izban M. G., Luse D. S. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagitov V., Nikiforov V., Goldfarb A. Dominant lethal mutations near the 5' substrate binding site affect RNA polymerase propagation. J Biol Chem. 1993 Jan 25;268(3):2195–2202. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Levin J. R., Agabian N. Molecular characterization of the Trypanosoma brucei RNA polymerase I and III largest subunit genes. J Biol Chem. 1989 Oct 25;264(30):18091–18099. [PubMed] [Google Scholar]
- Steinberg T. H., Burgess R. R. Tagetitoxin inhibition of RNA polymerase III transcription results from enhanced pausing at discrete sites and is template-dependent. J Biol Chem. 1992 Oct 5;267(28):20204–20211. [PubMed] [Google Scholar]
- Surratt C. K., Milan S. C., Chamberlin M. J. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier V., Stettler S., Sentenac A., Thuriaux P., Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995 Jan 16;14(2):351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher R., Hebron C., Feng G., Landick R. Termination-altering amino acid substitutions in the beta' subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994 Dec 1;8(23):2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- Werner M., Hermann-Le Denmat S., Treich I., Sentenac A., Thuriaux P. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol. 1992 Mar;12(3):1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman K. F., Drubin D. G., Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992 Oct;132(2):337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehall S. K., Bardeleben C., Kassavetis G. A. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J Biol Chem. 1994 Jan 21;269(3):2299–2306. [PubMed] [Google Scholar]
- Yon J., Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989 Jun 26;17(12):4895–4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mercoyrol L., Job C., Job D. Studies on the inhibition by alpha-amanitin of single-step addition reactions and productive RNA synthesis catalysed by wheat-germ RNA polymerase II. Biochem J. 1989 Feb 15;258(1):165–169. doi: 10.1042/bj2580165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mercoyrol L., Soulié J. M., Job C., Job D., Dussert C., Palmari J., Rasigni M., Rasigni G. Abortive intermediates in transcription by wheat-germ RNA polymerase II. Dynamic aspects of enzyme/template interactions in selection of the enzyme synthetic mode. Biochem J. 1990 Aug 1;269(3):651–658. doi: 10.1042/bj2690651. [DOI] [PMC free article] [PubMed] [Google Scholar]