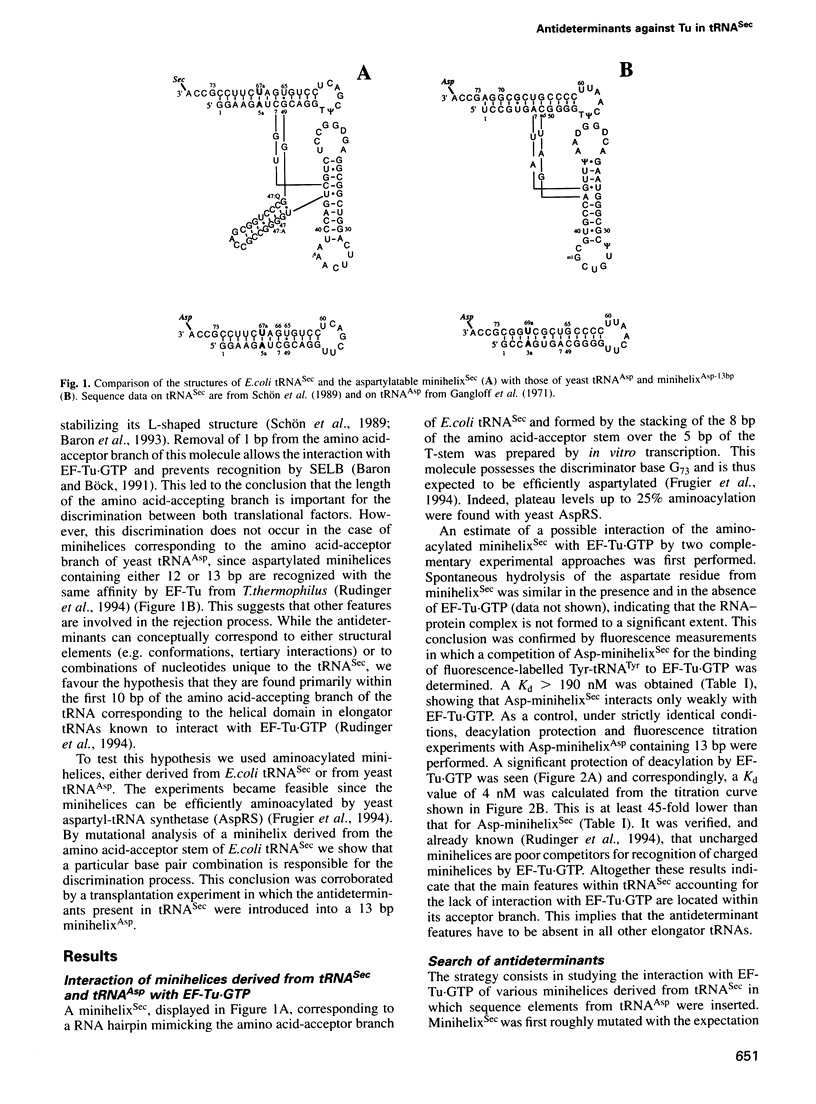
Abstract
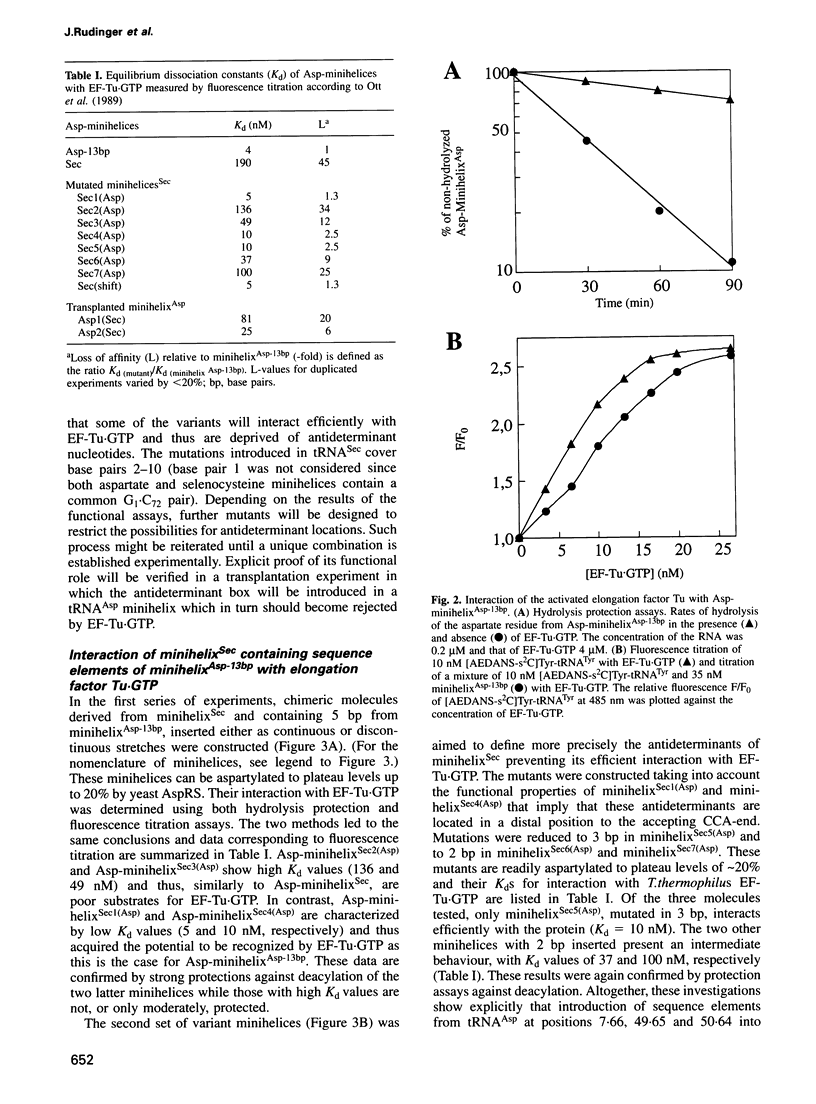
During protein biosynthesis, all aminoacylated elongator tRNAs except selenocysteine-inserting tRNA Sec form ternary complexes with activated elongation factor. tRNA Sec is bound by its own translation factor, an elongation factor analogue, e.g. the SELB factor in prokaryotes. An apparent reason for this discrimination could be related to the unusual length of tRNA Sec amino acid-acceptor branch formed by 13 bp. However, it has been recently shown that an aspartylated minihelix of 13 bp derived from yeast tRNA Asp is an efficient substrate for Thermus thermophilus EF-Tu-GTP, suggesting that features other than the length of tRNA Sec prevent its recognition by EF-Tu-GTP. A stepwise mutational analysis of a minihelix derived from tRNA Sec in which sequence elements of tRNA Asp were introduced showed that the sequence of the amino acid- acceptor branch of Escherichia coli tRNA Sec contains a specific structural element that hinders its binding to T.thermophilus EF-Tu-GTP. This antideterminant is located in the 8th, 9th and 10th bp in the acceptor branch of tRNA Sec, corresponding to the last base pair in the amino acid acceptor stem and the two first pairs in the T-stem. The function of this C7.G66/G49.U65/C50.G64 box was tested by its transplantation into a minihelix derived from tRNA Asp, abolishing its recognition by EF-Tu-GTP. The specific role of this nucleotide combination is further supported by its absence in all known prokaryotic elongator tRNAs.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain F. H., Varani G. Structure of the P1 helix from group I self-splicing introns. J Mol Biol. 1995 Jul 14;250(3):333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- Aström S. U., Byström A. S. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994 Nov 4;79(3):535–546. doi: 10.1016/0092-8674(94)90262-3. [DOI] [PubMed] [Google Scholar]
- Baron C., Böck A. The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNA(Sec) of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J Biol Chem. 1991 Oct 25;266(30):20375–20379. [PubMed] [Google Scholar]
- Baron C., Westhof E., Böck A., Giegé R. Solution structure of selenocysteine-inserting tRNA(Sec) from Escherichia coli. Comparison with canonical tRNA(Ser). J Mol Biol. 1993 May 20;231(2):274–292. doi: 10.1006/jmbi.1993.1282. [DOI] [PubMed] [Google Scholar]
- Been M. D. Cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem Sci. 1994 Jun;19(6):251–256. doi: 10.1016/0968-0004(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Boutorin A. S., Clark B. F., Ebel J. P., Kruse T. A., Petersen H. U., Remy P., Vassilenko S. A study of the interaction of Escherichia coli elongation factor-Tu with aminoacyl-tRNAs by partial digestion with cobra venom ribonuclease. J Mol Biol. 1981 Nov 5;152(3):593–608. doi: 10.1016/0022-2836(81)90271-0. [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Leinfelder W., Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989 Nov 23;342(6248):453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- Frugier M., Florentz C., Giegé R. Efficient aminoacylation of resected RNA helices by class II aspartyl-tRNA synthetase dependent on a single nucleotide. EMBO J. 1994 May 1;13(9):2218–2226. doi: 10.1002/j.1460-2075.1994.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster C., Chakraburtty K., Sprinzl M. Discrimination between initiation and elongation of protein biosynthesis in yeast: identity assured by a nucleotide modification in the initiator tRNA. Nucleic Acids Res. 1993 Dec 11;21(24):5679–5683. doi: 10.1093/nar/21.24.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster C., Ott G., Forchhammer K., Sprinzl M. Interaction of a selenocysteine-incorporating tRNA with elongation factor Tu from E.coli. Nucleic Acids Res. 1990 Feb 11;18(3):487–491. doi: 10.1093/nar/18.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff J., Keith G., Ebel J. P., Dirheimer G. Structure of aspartate-tRNA from brewer's yeast. Nat New Biol. 1971 Mar 24;230(12):125–126. doi: 10.1038/newbio230125a0. [DOI] [PubMed] [Google Scholar]
- Green R., Szostak J. W., Benner S. A., Rich A., Usman N. Synthesis of RNA containing inosine: analysis of the sequence requirements for the 5' splice site of the Tetrahymena group I intron. Nucleic Acids Res. 1991 Aug 11;19(15):4161–4166. doi: 10.1093/nar/19.15.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington K. M., Nazarenko I. A., Dix D. B., Thompson R. C., Uhlenbeck O. C. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry. 1993 Aug 3;32(30):7617–7622. doi: 10.1021/bi00081a003. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Cheong C., Tinoco I., Jr, Kim S. H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991 Oct 10;353(6344):579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Janiak F., Dell V. A., Abrahamson J. K., Watson B. S., Miller D. L., Johnson A. E. Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu.GTP: evidence for a new functional role for elongation factor Tu in protein biosynthesis. Biochemistry. 1990 May 8;29(18):4268–4277. doi: 10.1021/bi00470a002. [DOI] [PubMed] [Google Scholar]
- Joshi R. L., Faulhammer H., Chapeville F., Sprinzl M., Haenni A. L. Aminoacyl RNA domain of turnip yellow mosaic virus Val-RNA interacting with elongation factor Tu. Nucleic Acids Res. 1984 Oct 11;12(19):7467–7478. doi: 10.1093/nar/12.19.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesewetter S., Ott G., Sprinzl M. The role of modified purine 64 in initiator/elongator discrimination of tRNA(iMet) from yeast and wheat germ. Nucleic Acids Res. 1990 Aug 25;18(16):4677–4682. doi: 10.1093/nar/18.16.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Q., Yarus M. Bar to normal UGA translation by the selenocysteine tRNA. J Mol Biol. 1992 Jan 5;223(1):9–15. doi: 10.1016/0022-2836(92)90709-s. [DOI] [PubMed] [Google Scholar]
- Limmer S., Hofmann H. P., Ott G., Sprinzl M. The 3'-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6199–6202. doi: 10.1073/pnas.90.13.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer S., Reiser C. O., Schirmer N. K., Grillenbeck N. W., Sprinzl M. Nucleotide binding and GTP hydrolysis by elongation factor Tu from Thermus thermophilus as monitored by proton NMR. Biochemistry. 1992 Mar 24;31(11):2970–2977. doi: 10.1021/bi00126a018. [DOI] [PubMed] [Google Scholar]
- Lorber B., Kern D., Dietrich A., Gangloff J., Ebel J. P., Giegé R. Large scale purification and structural properties of yeast aspartyl-tRNA synthetase. Biochem Biophys Res Commun. 1983 Nov 30;117(1):259–267. doi: 10.1016/0006-291x(83)91569-3. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Chen Y. M., Foss K., Schneider J. Association of transfer RNA acceptor identity with a helical irregularity. Science. 1988 Dec 23;242(4886):1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- McClain W. H. Rules that govern tRNA identity in protein synthesis. J Mol Biol. 1993 Nov 20;234(2):257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991 Aug 16;253(5021):784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- Nazarenko I. A., Harrington K. M., Uhlenbeck O. C. Many of the conserved nucleotides of tRNA(Phe) are not essential for ternary complex formation and peptide elongation. EMBO J. 1994 May 15;13(10):2464–2471. doi: 10.1002/j.1460-2075.1994.tb06531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott G., Faulhammer H. G., Sprinzl M. Interaction of elongation factor Tu from Escherichia coli with aminoacyl-tRNA carrying a fluorescent reporter group on the 3' terminus. Eur J Biochem. 1989 Sep 15;184(2):345–352. doi: 10.1111/j.1432-1033.1989.tb15025.x. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990 Oct;72(10):735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Peter M. E., Reiser C. O., Schirmer N. K., Kiefhaber T., Ott G., Grillenbeck N. W., Sprinzl M. Interaction of the isolated domain II/III of Thermus thermophilus elongation factor Tu with the nucleotide exchange factor EF-Ts. Nucleic Acids Res. 1990 Dec 11;18(23):6889–6893. doi: 10.1093/nar/18.23.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A., Urbanke C. Aminoacyl transfer ribonucleic acid binding site of the bacterial elongation factor Tu. Biochemistry. 1980 May 13;19(10):2108–2112. doi: 10.1021/bi00551a017. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Additive, cooperative and anti-cooperative effects between identity nucleotides of a tRNA. EMBO J. 1993 Jul;12(7):2949–2957. doi: 10.1002/j.1460-2075.1993.tb05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. Initial stages of the thermal unfolding of yeast phenylalanine transfer RNA as studied by chemical modification: the effect of magnesium. Eur J Biochem. 1977 Nov 15;81(1):91–101. doi: 10.1111/j.1432-1033.1977.tb11930.x. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Dumas P., Ebel J. P., Giegé R. Comparison of the tertiary structure of yeast tRNA(Asp) and tRNA(Phe) in solution. Chemical modification study of the bases. J Mol Biol. 1987 May 5;195(1):193–204. doi: 10.1016/0022-2836(87)90336-6. [DOI] [PubMed] [Google Scholar]
- Rudinger J., Blechschmidt B., Ribeiro S., Sprinzl M. Minimalist aminoacylated RNAs as efficient substrates for elongation factor Tu. Biochemistry. 1994 May 17;33(19):5682–5688. doi: 10.1021/bi00185a003. [DOI] [PubMed] [Google Scholar]
- Schön A., Böck A., Ott G., Sprinzl M., Söll D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res. 1989 Sep 25;17(18):7159–7165. doi: 10.1093/nar/17.18.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong B. L., Lee C. P., RajBhandary U. L. Suppression of amber codons in vivo as evidence that mutants derived from Escherichia coli initiator tRNA can act at the step of elongation in protein synthesis. J Biol Chem. 1989 Apr 15;264(11):6504–6508. [PubMed] [Google Scholar]
- Seong B. L., RajBhandary U. L. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A., Cech T. R. Minor groove recognition of the conserved G.U pair at the Tetrahymena ribozyme reaction site. Science. 1995 Feb 3;267(5198):675–679. doi: 10.1126/science.7839142. [DOI] [PubMed] [Google Scholar]
- Sturchler-Pierrat C., Hubert N., Totsuka T., Mizutani T., Carbon P., Krol A. Selenocysteylation in eukaryotes necessitates the uniquely long aminoacyl acceptor stem of selenocysteine tRNA(Sec). J Biol Chem. 1995 Aug 4;270(31):18570–18574. doi: 10.1074/jbc.270.31.18570. [DOI] [PubMed] [Google Scholar]
- Tormay P., Wilting R., Heider J., Böck A. Genes coding for the selenocysteine-inserting tRNA species from Desulfomicrobium baculatum and Clostridium thermoaceticum: structural and evolutionary implications. J Bacteriol. 1994 Mar;176(5):1268–1274. doi: 10.1128/jb.176.5.1268-1274.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- White S. A., Nilges M., Huang A., Brünger A. T., Moore P. B. NMR analysis of helix I from the 5S RNA of Escherichia coli. Biochemistry. 1992 Feb 18;31(6):1610–1621. doi: 10.1021/bi00121a005. [DOI] [PubMed] [Google Scholar]
- Wikman F. P., Romby P., Metz M. H., Reinbolt J., Clark B. F., Ebel J. P., Ehresmann C., Ehresmann B. Crosslinking of elongation factor Tu to tRNA(Phe) by trans-diamminedichloroplatinum (II). Characterization of two crosslinking sites in the tRNA. Nucleic Acids Res. 1987 Jul 24;15(14):5787–5801. doi: 10.1093/nar/15.14.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikman F. P., Siboska G. E., Petersen H. U., Clark B. F. The site of interaction of aminoacyl-tRNA with elongation factor Tu. EMBO J. 1982;1(9):1095–1100. doi: 10.1002/j.1460-2075.1982.tb01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Q., Gross H. J. The length and the secondary structure of the D-stem of human selenocysteine tRNA are the major identity determinants for serine phosphorylation. EMBO J. 1994 Jan 1;13(1):241–248. doi: 10.1002/j.1460-2075.1994.tb06254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]