Abstract
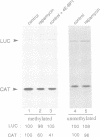
The immunosuppressant drug rapamycin blocks progression of the cell cycle at the G1 phase in mammalian cells and yeast. Here we show that rapamycin inhibits cap-dependent, but not cap-independent, translation in NIH 3T3 cells. Cap-dependent translation is also specifically reduced in extracts from rapamycin-treated cells, as determined by in vitro translation experiments. This inhibition is causally related to the dephosphorylation and consequent activation of 4E-BP1, a protein recently identified as a repressor of the cap-binding protein, eIF-4E, function. These effects of rapamycin are specific as FK506, a structural analogue of rapamycin, had no effect on either cap-dependent translation or 4E-BP1 phosphorylation. The rapamycin-FK506 binding protein complex is the effector of the inhibition of 4E-BP1 phosphorylation as excess of FK506 over rapamycin reversed the rapamycin-mediated inhibition of 4E-BP1 phosphorylation. Thus, inactivation of eIF-4E is, at least in part, responsible for inhibition of cap-dependent translation in rapamycin-treated cells. Furthermore, these results suggest that 4E-BP1 phosphorylation is mediated by the FRAP/TOR signalling pathway.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bierer B. E., Mattila P. S., Standaert R. F., Herzenberg L. A., Burakoff S. J., Crabtree G., Schreiber S. L. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C., Nakayama N., Goebl M., Tanaka K., Toh-e A., Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Aug;8(8):3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Albers M. W., Shin T. B., Ichikawa K., Keith C. T., Lane W. S., Schreiber S. L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994 Jun 30;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Chung J., Kuo C. J., Crabtree G. R., Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992 Jun 26;69(7):1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- De Benedetti A., Joshi-Barve S., Rinker-Schaeffer C., Rhoads R. E. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol. 1991 Nov;11(11):5435–5445. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont F. J., Melino M. R., Staruch M. J., Koprak S. L., Fischer P. A., Sigal N. H. The immunosuppressive macrolides FK-506 and rapamycin act as reciprocal antagonists in murine T cells. J Immunol. 1990 Feb 15;144(4):1418–1424. [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L. M., Bornfeldt K. E., Argast G. M., Krebs E. G., Kong X., Lin T. A., Lawrence J. C., Jr cAMP- and rapamycin-sensitive regulation of the association of eukaryotic initiation factor 4E and the translational regulator PHAS-I in aortic smooth muscle cells. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7222–7226. doi: 10.1073/pnas.92.16.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A., Mader S., Pause A., Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995 Nov 15;14(22):5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haystead T. A., Haystead C. M., Hu C., Lin T. A., Lawrence J. C., Jr Phosphorylation of PHAS-I by mitogen-activated protein (MAP) kinase. Identification of a site phosphorylated by MAP kinase in vitro and in response to insulin in rat adipocytes. J Biol Chem. 1994 Sep 16;269(37):23185–23191. [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hall M. N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991 Aug 23;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Hu C., Pang S., Kong X., Velleca M., Lawrence J. C., Jr Molecular cloning and tissue distribution of PHAS-I, an intracellular target for insulin and growth factors. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3730–3734. doi: 10.1073/pnas.91.9.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Howell M. T., Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990 Dec;15(12):477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- Jefferies H. B., Reinhard C., Kozma S. C., Thomas G. Rapamycin selectively represses translation of the "polypyrimidine tract" mRNA family. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. P., Warner J. R. Phosphorylation of the Saccharomyces cerevisiae equivalent of ribosomal protein S6 has no detectable effect on growth. Mol Cell Biol. 1987 Apr;7(4):1338–1345. doi: 10.1128/mcb.7.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas A. E., Lazaris-Karatzas A., Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992 Nov;11(11):4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., Hall M. N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993 May 7;73(3):585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamphear B. J., Kirchweger R., Skern T., Rhoads R. E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995 Sep 15;270(37):21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine K. S., Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990 Jun 7;345(6275):544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Lin T. A., Kong X., Haystead T. A., Pause A., Belsham G., Sonenberg N., Lawrence J. C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994 Oct 28;266(5185):653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- Lin T. A., Kong X., Saltiel A. R., Blackshear P. J., Lawrence J. C., Jr Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995 Aug 4;270(31):18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- Mader S., Lee H., Pause A., Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995 Sep;15(9):4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurides P. A., Akkaraju G. R., Jagus R. Evaluation of protein phosphorylation state by a combination of vertical slab gel isoelectric focusing and immunoblotting. Anal Biochem. 1989 Nov 15;183(1):144–151. doi: 10.1016/0003-2697(89)90182-6. [DOI] [PubMed] [Google Scholar]
- Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C., Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994 Oct 27;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Price D. J., Grove J. R., Calvo V., Avruch J., Bierer B. E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992 Aug 14;257(5072):973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- Rhoads R. E. Protein synthesis, cell growth and oncogenesis. Curr Opin Cell Biol. 1991 Dec;3(6):1019–1024. doi: 10.1016/0955-0674(91)90123-g. [DOI] [PubMed] [Google Scholar]
- Sabatini D. M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S. H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994 Jul 15;78(1):35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sabers C. J., Martin M. M., Brunn G. J., Williams J. M., Dumont F. J., Wiederrecht G., Abraham R. T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995 Jan 13;270(2):815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- Schreiber S. L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991 Jan 18;251(4991):283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985 Feb;40(2):223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- Stan R., McLaughlin M. M., Cafferkey R., Johnson R. K., Rosenberg M., Livi G. P. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem. 1994 Dec 23;269(51):32027–32030. [PubMed] [Google Scholar]
- Svitkin Y. V., Agol V. I. Complete translation of encephalomyocarditis virus RNA and faithful cleavage of virus-specific proteins in a cell-free system from Krebs-2 cells. FEBS Lett. 1978 Mar 1;87(1):7–11. doi: 10.1016/0014-5793(78)80121-5. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Lyapustin V. N., Lashkevich V. A., Agol V. I. Differences between translation products of tick-borne encephalitis virus RNA in cell-free systems from Krebs-2 cells and rabbit reticulocytes: involvement of membranes in the processing of nascent precursors of flavivirus structural proteins. Virology. 1984 Jun;135(2):536–541. doi: 10.1016/0042-6822(84)90207-1. [DOI] [PubMed] [Google Scholar]
- Terada N., Patel H. R., Takase K., Kohno K., Nairn A. C., Gelfand E. W. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. p70s6k/p85s6k: mechanism of activation, effects of rapamycin and role in mitogenesis. Biochem Soc Trans. 1993 Nov;21(4):901–904. doi: 10.1042/bst0210901. [DOI] [PubMed] [Google Scholar]