Abstract
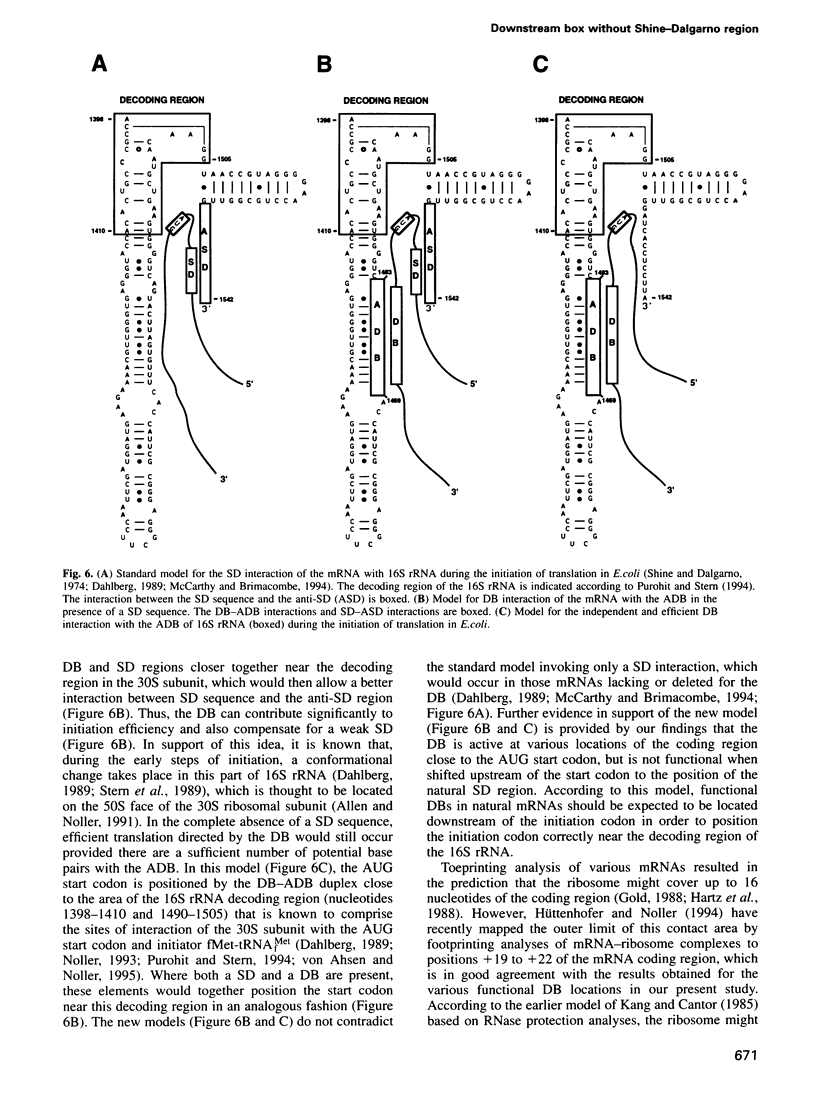
The downstream box (DB) was originally described as a translational enhancer of several Escherichia coli and bacteriophage mRNAs located just downstream of the initiation codon. Here, we introduced nucleotide substitutions into the DB and Shine-Dalgarno (SD) region of the highly active bacteriophage T7 gene 10 ribosome binding site (RBS) to examine the possibility that the DB has an independent and functionally important role. Eradication of the SD sequence in the absence of a DB abolished the translational activity of RBS fragments that were fused to a dihydrofolate reductase reporter gene. In contrast, an optimized DB at various positions downstream of the initiation codon promoted highly efficient protein synthesis despite the lack of a SD region. The DB was not functional when shifted upstream of the initiation codon to the position of the SD sequence. Nucleotides 1469-1483 of 16S rRNA ('anti-downstream box') are complementary to the DB, and optimizing this complementarity strongly enhanced translation in the absence and presence of a SD region. We propose that the stimulatory interaction between the DB and the anti-DB places the start codon in close contact with the decoding region of 16S rRNA, thereby mediating independent and efficient initiation of translation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen P. N., Noller H. F. A single base substitution in 16S ribosomal RNA suppresses streptomycin dependence and increases the frequency of translational errors. Cell. 1991 Jul 12;66(1):141–148. doi: 10.1016/0092-8674(91)90146-p. [DOI] [PubMed] [Google Scholar]
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., White J., Ward J. M., Janssen G. R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol Microbiol. 1994 Nov;14(3):533–545. doi: 10.1111/j.1365-2958.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- Bujard H., Gentz R., Lanzer M., Stueber D., Mueller M., Ibrahimi I., Haeuptle M. T., Dobberstein B. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- Calogero R. A., Pon C. L., Canonaco M. A., Gualerzi C. O. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier-Miller M., Jacques N., Raibaud O., Dreyfus M. Transcription of single-copy hybrid lacZ genes by T7 RNA polymerase in Escherichia coli: mRNA synthesis and degradation can be uncoupled from translation. Nucleic Acids Res. 1990 Oct 11;18(19):5787–5792. doi: 10.1093/nar/18.19.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G. E., Calendar R. Bacteriophage P2 late promoters. II. Comparison of the four late promoter sequences. J Mol Biol. 1985 Feb 5;181(3):373–382. doi: 10.1016/0022-2836(85)90226-8. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989 May 19;57(4):525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988 Nov 5;204(1):79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Bulmer M. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 1993 Sep 25;21(19):4599–4603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatscher H. P., Geisen R. M., Fuchs E. Only one out of the three strong ribosomal binding sites of the early region of bacteriophage T7 exhibits high translational efficiency in fragments of about 30 base pairs. Eur J Biochem. 1988 Aug 15;175(3):461–465. doi: 10.1111/j.1432-1033.1988.tb14217.x. [DOI] [PubMed] [Google Scholar]
- Faxén M., Plumbridge J., Isaksson L. A. Codon choice and potential complementarity between mRNA downstream of the initiation codon and bases 1471-1480 in 16S ribosomal RNA affects expression of glnS. Nucleic Acids Res. 1991 Oct 11;19(19):5247–5251. doi: 10.1093/nar/19.19.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R., Kado C. I. A translational enhancer derived from tobacco mosaic virus is functionally equivalent to a Shine-Dalgarno sequence. Proc Natl Acad Sci U S A. 1989 Jan;86(1):129–132. doi: 10.1073/pnas.86.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisen R. M., Fatscher H. P., Fuchs E. More than 150 nucleotides flanking the initiation codon contribute to the efficiency of the ribosomal binding site from bacteriophage T7 gene 1. Nucleic Acids Res. 1987 Jun 25;15(12):4931–4943. doi: 10.1093/nar/15.12.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Helke A., Geisen R. M., Vollmer M., Sprengart M. L., Fuchs E. An unstructured mRNA region and a 5' hairpin represent important elements of the E. coli translation initiation signal determined by using the bacteriophage T7 gene 1 translation start site. Nucleic Acids Res. 1993 Dec 11;21(24):5705–5711. doi: 10.1093/nar/21.24.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A., de Boer H. A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Noller H. F. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994 Aug 15;13(16):3892–3901. doi: 10.1002/j.1460-2075.1994.tb06700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Kawakami K., Nakamura Y. Multiple control of Escherichia coli lysyl-tRNA synthetase expression involves a transcriptional repressor and a translational enhancer element. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):302–306. doi: 10.1073/pnas.90.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey-Hoyle M., Steege D. A. Mutational analysis of an inherently defective translation initiation site. J Mol Biol. 1992 Apr 20;224(4):1039–1054. doi: 10.1016/0022-2836(92)90468-y. [DOI] [PubMed] [Google Scholar]
- Jacob W. F., Santer M., Dahlberg A. E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. W., Cantor C. R. Structure of ribosome-bound messenger RNA as revealed by enzymatic accessibility studies. J Mol Biol. 1985 Jan 20;181(2):241–251. doi: 10.1016/0022-2836(85)90088-9. [DOI] [PubMed] [Google Scholar]
- Klock G., Hillen W. Expression, purification and operator binding of the transposon Tn1721-encoded Tet repressor. J Mol Biol. 1986 Jun 20;189(4):633–641. doi: 10.1016/0022-2836(86)90493-6. [DOI] [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994 Jul 1;8(13):1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loechel S., Inamine J. M., Hu P. C. A novel translation initiation region from Mycoplasma genitalium that functions in Escherichia coli. Nucleic Acids Res. 1991 Dec 25;19(24):6905–6911. doi: 10.1093/nar/19.24.6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C., von Ossowski I., Switala J., Mulvey M. R. KatF (sigma S) synthesis in Escherichia coli is subject to posttranscriptional regulation. J Bacteriol. 1993 Apr;175(7):2150–2153. doi: 10.1128/jb.175.7.2150-2153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. E., Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994 Nov;10(11):402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990 Mar;6(3):78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Schairer H. U., Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985 Feb;4(2):519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melançon P., Leclerc D., Destroismaisons N., Brakier-Gingras L. The anti-Shine-Dalgarno region in Escherichia coli 16S ribosomal RNA is not essential for the correct selection of translational starts. Biochemistry. 1990 Apr 3;29(13):3402–3407. doi: 10.1021/bi00465a037. [DOI] [PubMed] [Google Scholar]
- Nagai H., Yuzawa H., Yura T. Interplay of two cis-acting mRNA regions in translational control of sigma 32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins P. O., Rangwala S. H. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. J Biol Chem. 1989 Oct 15;264(29):16973–16976. [PubMed] [Google Scholar]
- Petersen C. Multiple determinants of functional mRNA stability: sequence alterations at either end of the lacZ gene affect the rate of mRNA inactivation. J Bacteriol. 1991 Apr;173(7):2167–2172. doi: 10.1128/jb.173.7.2167-2172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G. B., Stockwell P. A., Hill D. F. Messenger RNA recognition in Escherichia coli: a possible second site of interaction with 16S ribosomal RNA. EMBO J. 1988 Dec 1;7(12):3957–3962. doi: 10.1002/j.1460-2075.1988.tb03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner J., Krämer J., Meyer T. F. A plasmid system for high-level expression and in vitro processing of recombinant proteins. Gene. 1993 Aug 16;130(1):121–126. doi: 10.1016/0378-1119(93)90354-6. [DOI] [PubMed] [Google Scholar]
- Porter A. G., Hindley J. Characterization of extended sequences around the coat and replicase cistron ribosome binding sites in phage Q RNA. FEBS Lett. 1972 Oct 1;26(1):139–144. doi: 10.1016/0014-5793(72)80560-x. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Purohit P., Stern S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994 Aug 25;370(6491):659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- Ringquist S., Shinedling S., Barrick D., Green L., Binkley J., Stormo G. D., Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992 May;6(9):1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Vachon G., Laalami S., Morel-Deville F., Cenatiempo Y., Grunberg-Manago M. Both forms of translational initiation factor IF2 (alpha and beta) are required for maximal growth of Escherichia coli. Evidence for two translational initiation codons for IF2 beta. J Mol Biol. 1992 May 5;225(1):67–80. doi: 10.1016/0022-2836(92)91026-l. [DOI] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Shean C. S., Gottesman M. E. Translation of the prophage lambda cl transcript. Cell. 1992 Aug 7;70(3):513–522. doi: 10.1016/0092-8674(92)90175-c. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley B. L., Lupski J. R., Svec P. S., McMacken R., Godson G. N. Sequences of the Escherichia coli dnaG primase gene and regulation of its expression. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4550–4554. doi: 10.1073/pnas.79.15.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengart M. L., Fatscher H. P., Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990 Apr 11;18(7):1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl W. R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992 Mar 11;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj T. A., Pandit M. W. An additional ribosome-binding site on mRNA of highly expressed genes and a bifunctional site on the colicin fragment of 16S rRNA from Escherichia coli: important determinants of the efficiency of translation-initiation. Nucleic Acids Res. 1989 Apr 25;17(8):2973–2985. doi: 10.1093/nar/17.8.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz A., Pirrotta V., Ineichen K. Lambda repressor regulates the switch between PR and Prm promoters. Nature. 1976 Aug 19;262(5570):665–669. doi: 10.1038/262665a0. [DOI] [PubMed] [Google Scholar]
- Warburton N., Boseley P. G., Porter A. G. Increased expression of a cloned gene by local mutagenesis of its promoter and ribosome binding site. Nucleic Acids Res. 1983 Sep 10;11(17):5837–5854. doi: 10.1093/nar/11.17.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchuk O., Jacques N., Guillerez J., Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J Mol Biol. 1992 Aug 5;226(3):581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J Mol Biol. 1994 Jan 7;235(1):173–184. doi: 10.1016/s0022-2836(05)80024-5. [DOI] [PubMed] [Google Scholar]
- von Ahsen U., Noller H. F. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995 Jan 13;267(5195):234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]