Abstract
Electronic and vibrational circular dichroism are often used to determine the secondary structure of proteins, because each secondary structure has a unique spectrum. In order to determine these spectral features, polypeptides that are known to adopt a particular conformation along their entire length are studied ideally. Little is known about the vibrational circular dichroic spectroscopic features of the β-hairpin. In this study, the VCD spectral features of a decapeptide, YYDPETGTWY (CLN025), which forms a stable β-hairpin that is stabilized by intramolecular weakly polar interactions and hydrogen bonds were determined. Molecular Dynamics simulations and ECD spectropolarimetry were used to confirm that CLN025 adopts a β-hairpin in water, TFE, MeOH and DMSO and to examine differences in the secondary structure, hydrogen bonds and weakly polar interactions. CLN025 was synthesized by microwave-assisted solid phase peptide synthesis with Nα-Fmoc protected amino acids. The VCD spectra displayed a (−,+,−) pattern with bands at 1640 to 1656 cm−1, 1667 to 1687 cm−1 and 1679 to 1686 cm−1 formed by the overlap of a lower frequency negative couplet and a higher frequency positive couplet. A maximum IR absorbance was observed at 1647 to 1663 cm−1 with component bands at 1630 cm−1, 1646 cm−1, 1658 cm−1 and 1675 to 1680 cm−1 that are indicative of the β-sheet, random meander, either random meander or loop and turn, respectively. These results are similar to the results of others, who examined the VCD spectra of β-hairpins formed by DPro-Xxx turns and indicate that observed pattern is typical of β-hairpins.
Keywords: CLN025, Chignolin, vibrational circular dichroism, electronic circular dichroism, beta-hairpins, miniprotein, molecular dynamics
Introduction
Model β-hairpin miniproteins are frequently studied because they are the simplest form of anti-parallel β-sheet and often display characteristics indicative of proteins such as two-state folding.1 β-hairpins are stabilized by cross-strand hydrogen bonds, disulfide bonds and weakly polar interactions.2–4 Weakly polar interactions play an important role due to their strength and ubiquitousness in peptides. Weakly polar interactions can be as strong as hydrogen bonds.5–12 They play an important role in the stabilization of secondary structure such as α-helices,5 and the tertiary structure of peptides such as the TC5b8 and APP10, 12 miniproteins. Numerous β-hairpins have been designed to take advantage the aromatic-aromatic (Ar-Ar) interactions,13–17 cation-π interactions,16–18 and CH-π interactions.16, 17
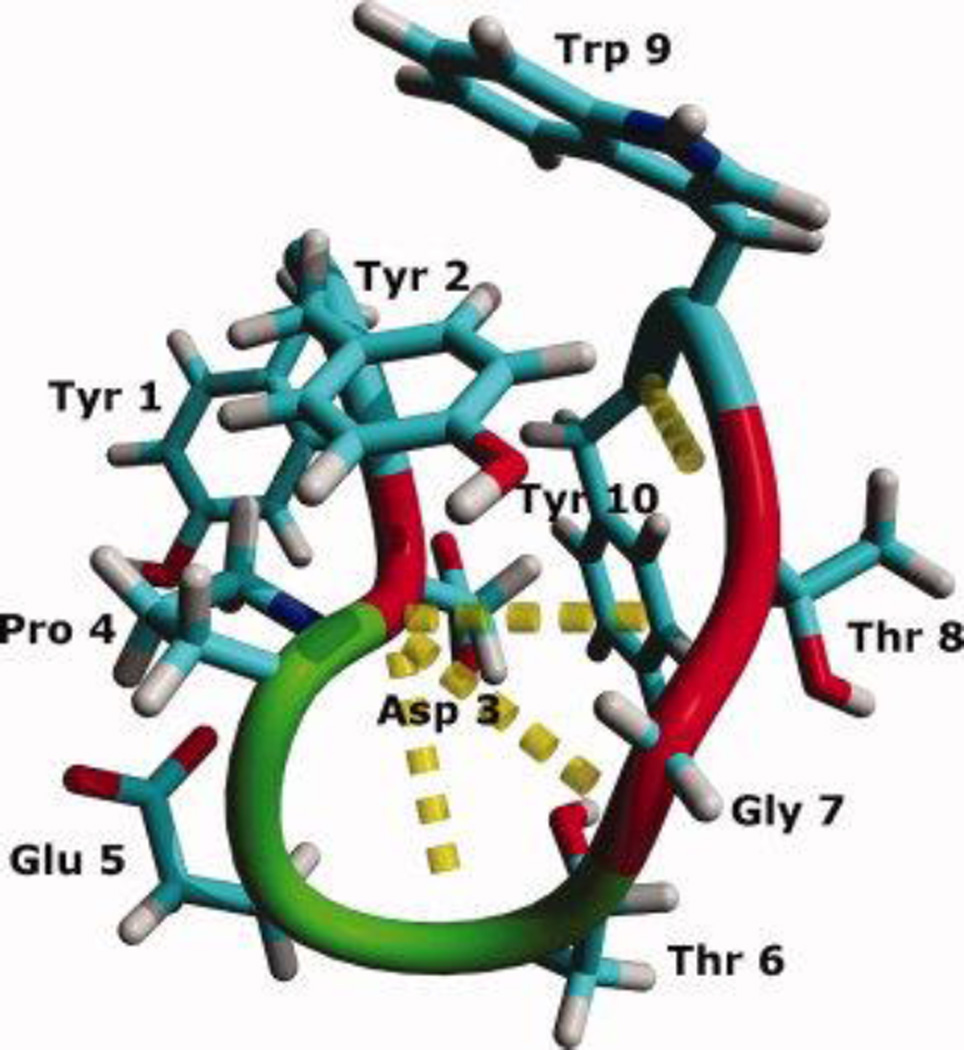
CLN025, Tyr-Tyr-Asp-Pro-Glu-Thr-Gly-Thr-Trp-Tyr, (Figure 1), is a miniprotein that forms a stable β-hairpin with a Tm of 70 °C, which undergoes a reversible cooperative transition upon thermal denaturation.19 CLN025 is a chignolin variant in which the terminal Gly residues are replaced with Tyr residues. The β-hairpin has a turn-initiating Asp-Pro sequence,20 and is stabilized by an Ar-Ar interaction between the side chains of Tyr2 and Trp9 and hydrogen bonds between Asp3 and Thr6, Gly7 and Thr8.19
Figure 1.
The backbone and side chains of the crystal structure of CLN025. Random meander is cyan, β-sheet is red, turn is green and the H-bonds are represented by yellow dotted lines.
The vibrational circular dichroism spectroscopic properties of helices,21–24 turns,6, 25–27 random meander23, 28 and β-sheets23, 27, 29–31 have been studied extensively but the properties of the β-hairpin have not. Previous studies of the VCD spectroscopic properties of β-hairpins were of peptides with DPro-Xxx turns or head-to-tail cyclization to stabilize the structure in chloroform or aqueous solvents rather than solvents such as TFE, MeOH or DMSO.32–34 Many of the designed β-hairpins aggregate at higher concentrations in water. This hampers experiments to examine VCD spectra of β-hairpins containing only L-amino acids. In this study, the VCD spectroscopic properties of CLN025, which has been shown to form a stable β-hairpin containing only naturally occurring amino acids, in aqueous solution and in different organic solvents (TFE, MeOH and DMSO) were determined. Molecular Dynamics simulations and ECD were used to confirm the β-hairpin conformation of CLN025 in the solvents.
Methods
CLN025 synthesis and purification
CLN025 was synthesized with a CEM Liberty microwave peptide synthesizer (CEM, Matthews, NC, USA) at a 0.1 mmole scale using a H-Tyr(OtBu) HMPB ChemMatrix resin and 5 molar excess of Nα-Fmoc protected amino acids. t-butyl protection was used to protect the reactive side chains of Asp, Glu, Thr and Tyr while t-Boc protection was used for Trp residues. Couplings were performed using HATU and DIEA at 75 °C for 10 minutes with 25 W radiation energy. Deprotections were performed using 20% (v/v) piperidine and 0.1 M HOBt at 75 °C for 3 minutes with 35 W radiation energy.
The peptide was cleaved from the resin by stirring in a 95/2.5/2.5 (v/v/v) TFA/TIS/water mixture for 10 minutes at 0 °C followed by 110 minutes at room temperature. The peptide was precipitated with cold diethyl ether, filtered, dissolved in TFA and filtered. The filtrate was evaporated to 1 mL and the peptide was precipitated with cold diethyl ether and collected by filtration. The peptide was dissolved in glacial acetic acid which was diluted to 5% acetic acid with nanopure water and the solution was lyophilized.
Purification was performed by reverse-phase high pressure liquid chromatography using a Gilson dual pump apparatus (Gilson, Inc, Middleton, WI, USA). The peptide was purified on a Phenomenex Jupiter C18 column (5 µm, 200 mm × 10 mm; Phenomenex, Torrance, CA, USA) using an aqueous solvent of 0.1% TFA in water and an organic solvent of 0.09% TFA in acetonitrile. The peptide was purified at 30 °C using a gradient of 10% to 50% organic solvent over 60 minutes with a flow rate of 4 mL min−1. Pure fractions were pooled and lyophilized. The peptide was identified using a API150EX mass spectrometer (PE SCIEX, Foster City, CA, USA)and characterized by RP-HPLC with a Jupiter C18 column (5 µm, 200 mm × 4.6 mm; Phenomenex, Torrance, CA, USA) using a 3% to 60% organic solvent gradient over 40 minutes at 1 mL min−1.
ECD
ECD spectra were recorded using a Jasco J-810 Spectropolarimeter (JASCO, Inc., Easton, MD, USA) by performing 20 scans from 185 to 250 nm at 100 nm min−1 scan speed in a 0.05 cm path length quartz cell. The background spectra of the solvents were subtracted and the mean residue molar ellipticities were calculated using peptide concentrations determined by quantitative RP-HPLC35 with a Jupiter C18 column (5 µm, 200 mm × 4.6 mm; Phenomenex, Torrance, CA, USA). CLN025 was dissolved in either 20 mM KH2PO4-K2HPO4 buffer (pH 7.0), TFE or MeOH at a final concentration of 100 µM. CDSSTR36–38 analysis was performed with data set 6 using the DichroWeb website.39–43 The concentration of 100 µM CLN025 was used because at concentrations above 1 mM CLN025 aggregates.
VCD
CLN025 was dissolved in 0.1 M DCl in D2O and lyophilized three times prior to VCD measurements in order to remove all trifluoroacetic acid salts and perform a H/D exchange on the backbone of the peptide. All samples for VCD measurement were prepared to have a concentration of 20 mg mL−1 CLN025. Buffer solutions were prepared by dissolving 4 mg of CLN025 in 10 µL DMSO-d6 and gradually diluting this solution to 5% DMSO-d6 with 20 mM KD2PO4-K2DPO4 buffer (pD 7.4). All other solutions were prepared by dissolving 2 mg CLN025 in either TFE-d, MeOH-d4 or DMSO-d6.
VCD spectra were recorded using a dual PEM BOMEM Biotools Chiralir Fourier Transform VCD Spectropolarimeter (BioTools, Inc., Jupiter, FL, USA). Either 4 or 8 block measurements consisting of 4500 scans for VCD and 225 scans for IR absorption at 8 cm−1 resolution were performed in a 75 µm pathlength CaF2 cell. 8 blocks were recorded for VCD measurements of CLN025 in buffer, TFE-d, MeOH-d4 and DMSO-d6 at 20 °C, while 4 blocks were recorded for VCD measurements in buffer at 0 °C, 20 °C and 60 °C. Spectra of the blocks were averaged and the background spectra of the solvent and water vapor were subtracted. Spectra were set to 0 at 1800 cm−1 and normalized44 by dividing each data point by the maximum IR absorbance in the amide I' region. The spectra were normalized in order to remove the effect of small differences in the sample concentration on the intensity of the spectra.
Maximum-entropy deconvolution and peak fitting analysis of the FT-IR spectra were performed with the PROTA v 3.0 software package (BioTools, Inc., Jupiter, FL, USA) using the tyrosyl C-C ring vibration band at 1515 cm−1 as the representative peak shape, which has been found to be a good marker due to its sharpness, narrow width, and discrete location.45 Peak-fitting analysis is not conclusive, because it produces no unique band assignment, molar absorptivities of all conformations are assumed to be equal and conclusions can be subjective.46 The analysis performed by the PROTA software package, however is less subjective, by using Bayesian derivatives and maximum likelihood functions to generate a list of peaks that meet acceptable signal to noise criteria and determines the areas of the individual peaks with the Levenberg-Marquardt maximum likelihood algorithm.
Molecular Dynamics Simulations
Molecular Dynamics simulations were performed using the crystal structure of CLN02519 with the GROMACS 3.2.1 software package47, 48 using the GROMOS 96 force field with the 53a6 parameter set.49, 50 CLN025 was solvated in either 1438 SPC51 water, 302 TFE, 1866 MeOH or 286 DMSO molecules in a 45.61 nm3 dodecahedral box with one chloride and three sodium ions to neutralize the system. The MeOH, TFE and DMSO models used were optimized for the 53a6 parameter set52 and the experimentally determined compressibilities (4.92 × 10−5 bar−1, 4.50 × 10−5 bar−1, 4.45 × 10−5 bar−1, 1.22 × 10−4 bar−1, 1.29 × 10−4 bar−1, 4.50 × 10−5 bar−1) and dielectric constants (86, 78, 68, 27, 32, 47) were used for the simulations of CLN025 in SPC water at 278 K, 300 K and 333 K and TFE, MeOH and DMSO at 300 K, respectively.53 The solvated structures were energy-minimized using steepest descent.54 An NVT simulation of the positionally restrained peptide was performed for 100 ps at 300 K (27 °C) followed by a 200.1 ns NPT simulation at 300 K and 1 bar using Berendsen coupling.55 All bonds were constrained56 with Shake57 in the TFE simulation and Lincs58 in the other simulations. In addition, MD simulations were performed in SPC water at 278 K (5 °C) and 333 K (60 °C).
MD simulations were analyzed by examining the middle structure of the largest cluster determined by the backbone RMSD of structures sampled every 10 ps along the trajectory. Secondary structure of CLN025 was analyzed by the DSSP method.59
Results and Discussion
ECD spectroscopy
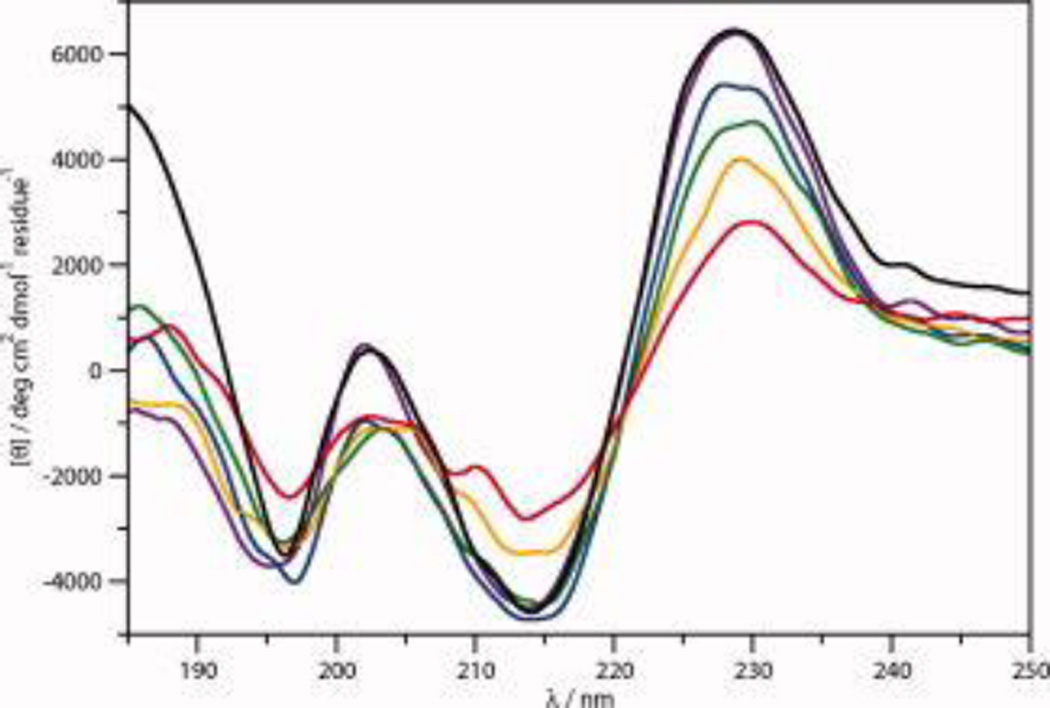
The ECD spectra of 100 µM CLN025 in 20 mM potassium phosphate buffer (pH 7.0) from 0 °C to 80 °C and cooled from 80 °C to 0 °C are shown in Figure 2. These spectra are similar to those measured by Honda and coworkers,19 except their spectra showed a more intense negative band at 197 nm. The spectra indicate that the peptide has β-sheet, type II β-turn and random meander, as well as an aromatic couplet caused by the Ar-Ar interactions.
Figure 2.
The ECD spectra of 100 µM CLN025 in 20 mM potassium phosphate buffer at 0 °C (violet), 20 °C (blue), 40 °C (green), 60 °C (orange) and 80 °C (red) and cooled from 80 °C to 0 °C (black).
As the temperature increases, the intensity and position of the positive band of the aromatic couplet shifts from 225 to 230 nm and at some temperatures more than one couplet is apparent most likely due to contributions from the Tyr2-Trp9 and Tyr1-Tyr10 Ar-Ar interactions as well as the Tyr1-Tyr9 interaction at higher temperatures. This provides further evidence that using the aromatic couplet as a measure of folding is not accurate since, in peptides containing more than two aromatic rings, the interactions could shift causing a shift in the aromatic couplet that is not indicative of the overall fold.
As the temperature increases above 60 °C a positive band at 212 nm becomes visible. This is indicative of random meander, indicating that the β-hairpin is destabilized. The increase in random meander content most likely results from a loss of β-sheet which would explain why the intensity of the 195 nm band does not change. The spectra, however, still suggests β-hairpin conformation and the aromatic couplet indicates that the ends of the peptide remain in close proximity. The ECD spectra of the peptide at 0 °C and the peptide cooled from 80 °C to 0 °C are similar, indicating reversible unfolding.
The ECD spectra of CLN025 in buffer, TFE and MeOH are shown in Figure 3. The spectra have similar characteristics, but the peaks in the spectrum in TFE are blue-shifted 2 to 3 nm while in MeOH they are red shifted 1 to 2 nm from the positions in the spectrum of the peptide in buffer. This is most likely due to solvent effects including differing degrees of hydration of individual amide groups46, 60–62 and differences in the dipolar character of the solvent.63–66 The aromatic couplet in TFE is less intense due to the tendency of high concentrations of TFE to weaken the interactions between hydrophobic side chains.67, 68 The higher intensity of the aromatic couplet in MeOH suggests a stronger Ar-Ar interaction. Due to the overlap of the aromatic couplet with signals conveying information about the secondary structure,69, 70 it is difficult to determine if small changes in the conformation of the peptide occur in the different solvents. CDSSTR analysis (Table 1) however, suggests that the peptide is 46% sheet, 23% turn and 31% random meander in buffer. In TFE and MeOH, the sheet component increases to 56% and 59%, respectively, while the random meander component decreases to 20% in TFE and the turn component decreases to 13% in MeOH.
Figure 3.
The ECD spectra of 100 µM CLN025 in 20 mM potassium phosphate buffer (black), TFE (red) and MeOH (blue) at 20 °C.
Table 1.
Structural content (%) of CLN025 in different solvents as determined by the analysis of ECD spectra using the CDSSTR method.
| Solvent* | Strand | Turn | Random meander |
|---|---|---|---|
| Buffer | 46 | 23 | 31 |
| TFE | 56 | 24 | 20 |
| MeOH | 59 | 13 | 28 |
See methods
The ECD spectra of CLN025 are similar at all temperatures and in the different solvents indicating no significant conformational change in the structure of CLN025 upon heating and in either TFE or MeOH. This demonstrates the stability of the β-hairpin in CLN025. The small changes in the ECD spectra of CLN025 indicate that at high temperatures the amount of random meander increases and the Ar-Ar interactions decrease. In TFE, CLN025 displays a higher β-sheet content and a decrease in the amount of random meander. In MeOH, the β-sheet and random meander content of CLN025 increase and the turn content decreases.
VCD spectroscopy
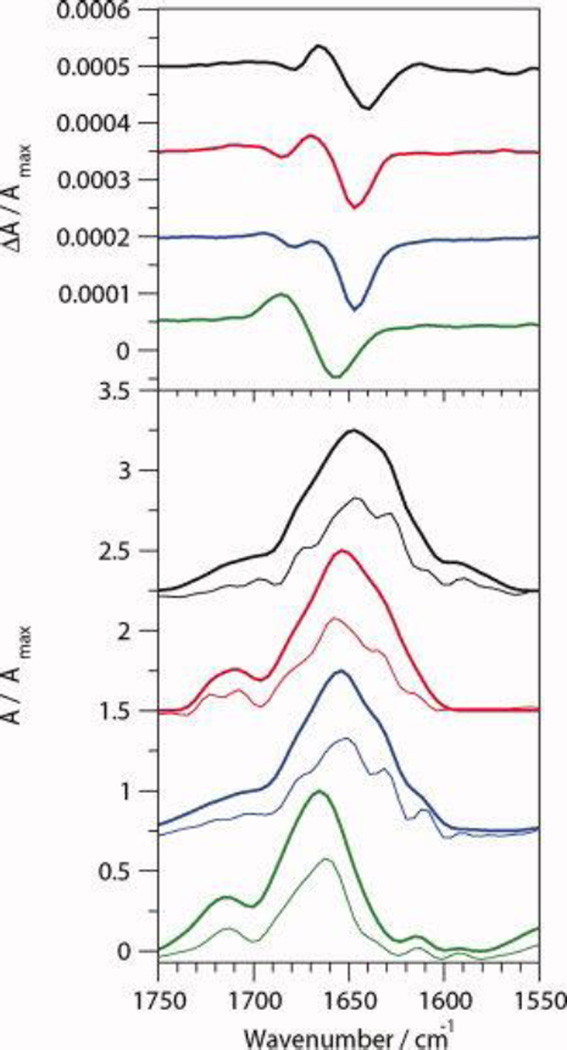
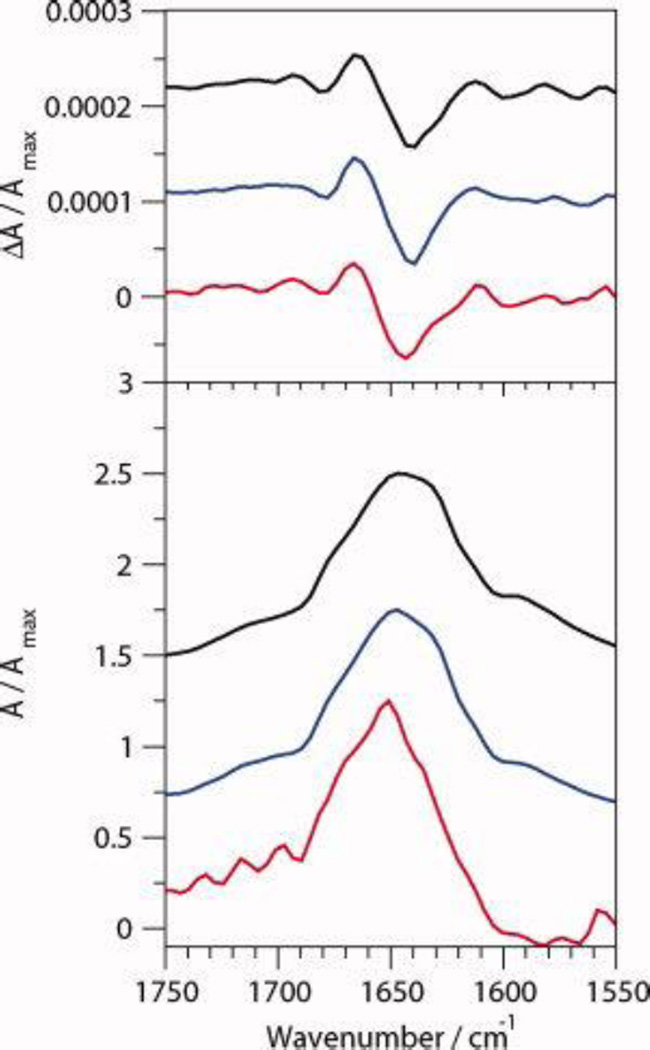
The VCD and FT-IR spectra of CLN025 in various solvents are shown in Figure 4 and the position of the VCD bands are listed in Table 2. The IR spectrum of CLN025 in buffer has a maximum absorbance at 1647 cm−1 with shoulders at 1675 cm−1, 1646 cm−1, 1630 cm−1 and 1612 cm−1 in the deconvoluted spectra, which are indicative of turn, random meander, β-sheet and tertiary amide or tyrosine side-chain vibration, respectively.15, 23, 27, 45, 71 The VCD spectra show a (−,+,−) pattern, formed by the overlap of the positive bands of a negative and positive couplet, with bands at 1640 cm−1, 1667 cm−1 and 1679 cm−1, respectively. The first couplet, centered at 1653 cm−1, is indicative of random meander and may indicate a local left handed twist28 while the second couplet, centered at 1673 cm−1, is indicative of a type II' β-turn.25, 72
Figure 4.
Normalized VCD (top) and IR absorption (bottom) spectra of 20 mg mL−1 CLN025 in 5% (v/v) DMSO-d6 in 20 mM deuterated potassium phosphate buffer (black), TFE-d (red), MeOH-d4 (blue) and DMSO-d6 (green). The deconvoluted absorption spectra are shown by the thin curves.
Table 2.
Position of bands (cm−1) in the VCD spectra of CLN025
| Solvent | − band | + band | − band |
|---|---|---|---|
| Buffer | 1679 | 1667 | 1640 |
| TFE | 1686 | 1671 | 1647 |
| MeOH | 1679 | 1671 | 1647 |
| DMSO | 1687 | 1656 |
The IR absorbance spectra of the peptide in TFE and MeOH are similar to the IR spectrum of the peptide in buffer, except the maximum absorbance is shifted to 1655 cm−1. The peaks of the VCD spectra of the peptide in TFE and MeOH are also shifted from positions in buffer with the negative band shifted to 1647 cm−1 and the positive band to 1671 cm−1. In MeOH the second negative VCD band is at the same location as the second negative VCD band of the peptide in buffer, while in TFE this band is shifted up to 1686 cm−1 which indicates a change in the turn structure of CLN025 in TFE.
In DMSO, the maximum IR absorbance is shifted to 1663 cm−1 and the 1612 cm−1 absorbance peak is resolved due to a reduced shoulder around 1630 cm−1, indicating a loss of β-sheet. The peaks at 1706 cm−1 and 1712 cm−1 are indicative of the Glu and Asp side chains in deuterated solvents, respectively,45 or a change in the turn-like structure of the peptide. The VCD of CLN025 in DMSO shows only a negative couplet with a negative component at 1656 cm−1 and positive component at 1687 cm−1, attributed to a greater relaxation of the turn that causes the second couplet to be obscured by the first couplet.
Peak-fitting analysis of the FT-IR spectra, shown in Table 3, indicates that the 1612 cm−1 signal contributes equally in the different solvents, which indicates that the environment around the prolyl residue does not change significantly. The 1630 cm−1 signal indicates that CLN025 is approximately 16%, 24% and 11% β-sheet in buffer, TFE and MeOH, respectively, while a signal accounting for 3% of the amide I' area in DMSO is due either to β-sheet or the low frequency absorbance of turn structures. The signals at 1646 cm−1 and 1658 cm−1 are due either to random meander of loop conformations. The major signal shifts from 1646 cm−1 to 1658 cm−1 when the peptide is in non-aqueous solutions, which causes the shift of the negative VCD couplet in these solvents. The signal at 1675 cm−1 is due either to the turn or the high frequency absorbance of β-sheet23, 45 although turn is more likely due to the high signal of this peak in DMSO. The shift of the 1675 cm−1 band to 1680 cm−1 in TFE is the likely cause of the shift in the positive couplet in the VCD spectrum. The 1705 cm−1 peak in TFE and MeOH is most likely to be due to the glutamic acid side chain, indicating that this side chain is not shielded. The 1712 cm−1 band seen in buffer and especially in DMSO, where it accounts for 25% of the amide I' signal, is due either to the aspartic acid side chain or a change in structure. The aspartic acid side chain provides a better explanation due to the lack of a VCD signal in this region and because of the shift in the orientation of the side chain in the structure of CLN025 in DMSO (Figure 6d).
Table 3.
The areas of the component peaks in the deconvoluted FT-IR spectrum at the amide I' region of CLN025 as a percentage of the total area of the amide I' peak.
| Wavenumber / cm−1 | Buffer | TFE | MeOH | DMSO | Assignment |
|---|---|---|---|---|---|
| 1722 | 5 | Turn | |||
| 1712 | 5 | 24 | Turn or Asp | ||
| 1705 | 6 | 2 | Turn or Glu | ||
| 1696 | 2 | Turn | |||
| 1680 | 8 | Turn | |||
| 1675 | 5 | 5 | 25 | Turn or β-sheet | |
| 1658 | 27 | 53 | 57 | 45 | Random meander or loop |
| 1646 | 40 | 22 | Random meander | ||
| 1630 | 16 | 24 | 11 | 3* | β-sheet |
| 1612 | 4 | 4 | 3 | 3 | Pro |
May be due to the low frequency absorbance of turn structure rather than β-sheet.
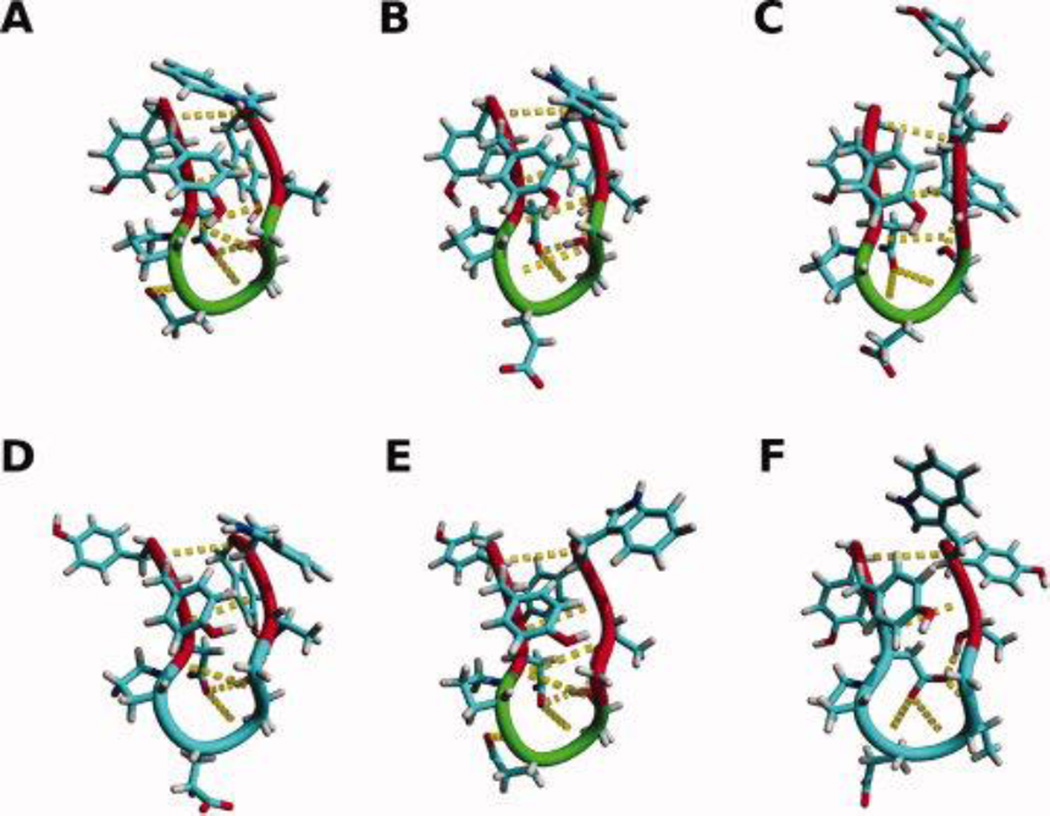
Figure 6.
The backbone and side chains of the middle structure of the most populated cluster of trajectories of MD simulations of CLN025 in A, H2O 5 °C; B, H2O 27 °C; C, H2O 60 °C; D, TFE; E, MeOH and F, DMSO. Random meander is cyan, β-sheet is red, turn is green and the H-bonds are yellow dotted lines.
The CDSSTR analysis of the ECD spectra (Table 1) and the peak fitting analysis of the FT-IR spectra (Table 3) show similarities in the differences of β-sheet, turn and random meander content in different solvents. Each analysis indicates that in TFE the β-sheet content of CLN025 increases and the random meander content decreases. In MeOH, both analyses indicate a loss of turn content but the peak-fitting analysis indicates a slight loss of β-sheet and an increase in the random meander content of CLN025. This discrepancy between the results of the two analyses is most probably due to the interference of the signal from the aromatic couplet in the ECD spectra. Even though the amount of turn in the peptide in MeOH decreases, the position of the second couplet and the relative contributions of the amide bands at 1646 cm−1 and 1658 cm−1 suggest that the turn in the peptide in MeOH is more similar to that in buffer than the turn in TFE is to that in buffer. While the CDSSTR analysis of the ECD spectra of CLN025 in buffer indicates that the β-hairpin is 46% sheet, 23% turn and 31% random meander, the peak fitting analysis of the VCD indicates that the β-hairpin is 16% sheet, 12% turn and 67% random meander. The major differences between the overall amounts of the β-sheet, turn and random meander assigned by the two analyses could be due to the interference of the aromatic couplet in the ECD spectra and the faulty assumption in peak fitting analysis that the molar absorbtivities of all conformations are equal.46
The VCD spectra of CLN025 in buffer at 5 °C, 20 °C and 60 °C, shown in Figure 5, are similar indicating that the β-hairpin is stable. The less pronounced shoulder at 1630 cm−1 in the IR absorption spectrum indicates that the amount of β-sheet content decreases at 60 °C.
Figure 5.
Normalized VCD (top) and IR absorption (bottom) spectra of 20 mg mL−1 CLN025 in 5% (v/v) DMSO-d6 in 20 mM deuterated potassium phosphate buffer at 5 °C (black), 20 °C (blue) and 60 °C (red).
The analysis indicates that the VCD spectra of CLN025 and β-hairpins in general consist of overlapping negative and positive couplets. The negative couplet is due to random meander or loop structure, while the higher frequency positive couplet is due to the turn. This produces a (−,+,−) pattern with bands at 1640 to 1656 cm−1, 1667 to 1687 cm−1 and 1679 to 1686 cm−1. The β-sheet component of the VCD spectra is not visible due to its low intensity29 and overlap with the first negative band (1640 to 1656 cm−1). The β-sheet, however, might contribute to the greater width of the first negative band. The IR absorption spectra show a broad amide I' band with a maximum at 1647 to 1663 cm−1 and bands at 1630 cm−1, 1646 cm−1, 1658 cm−1 and 1675 to 1680 cm−1 which are indicative of the β-sheet, random meander, random meander or loop and turn, respectively. The shifts in the peaks of the VCD and IR absorption spectra of the peptides in the different solvents is likely to be due to solvent effects, which have been shown to alter the spectral shapes of β-hairpins due to differing degrees of hydration of the individual amide groups46, 60–62 and differences in the dipolar character of the solvent.63–66
The results are similar to the results found by other groups for β-hairpins that have DPro-Xxx turns. Zhao and coworkers32 examined the spectra of numerous octapeptides with a DPro-Xxx turn in chloroform and found a (−,+,−,+) pattern of VCD peaks at 1643 to 1659 cm−1, 1655 to 1670 cm−1, 1670 to 1678 cm−1 and 1690 to 1697 cm−1, respectively with a maximum IR absorbance at 1690 to 1697 cm−1. Hilario and associates33 examined the VCD spectra of a dodecapeptide with a DPro-Gly turn in aqueous solution and found a negative couplet with negative band at 1638 cm−1, positive band at 1660 to 1680 cm−1 in the VCD spectra and a maximum IR absorbance at 1638 cm−1. These results are similar to those here, except that the maximum IR absorbance was 9 cm−1 lower. This is most likely to be due to a greater amount of β-sheet structure in dodecapeptide. Hilario and coworkers34 examined spectra of 4 to 16 residue peptides with a DPro-Gly turn in aqueous solution and found a negative couplet with negative band at 1635 to 1645 cm−1 and positive band at 1660 to 1670 cm−1 in the VCD spectra and a maximum IR absorbance at 1635 to 1645 cm−1.
MD simulations
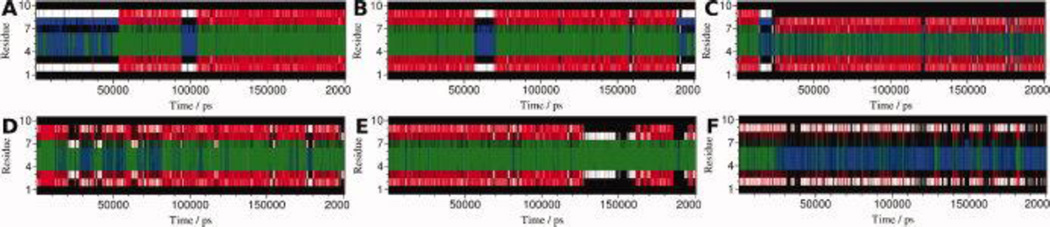
Cluster analysis, based on the backbone RMSD of the peptide, generated 8, 15, 76, 3, 19 and 6 clusters for CLN025 in H2O at 5 °C, H2O at 27 °C, H2O at 60 °C, TFE at 27 °C, MeOH at 27 °C and DMSO at 27 °C, respectively. The middle structures of the largest clusters are seen in Figure 9 and account for 92.3%, 94.4%, 72%, 99.8%, 83.9% and 98.2%, respectively, of the 20,000 structures sampled. The small number of clusters and the large contribution of the largest cluster indicates the CLN025 forms a stable structure in all environments studied. The middle structures indicate that the β-hairpin conformation is maintained in all environments but the turn is relaxed in TFE and DMSO showing a random meander from Pro4 to Gly7 in TFE and from Asp3 to Gly7 in DMSO. In DMSO the Asp3 side chain moves closer to the opposite strand and rotates so that the oxygen atoms are pointing towards the turn and strand. This allows the side chain to participate in more hydrogen bonds. In MeOH, the peptide maintains a tight turn though shortened and the Gly7 residue is assigned to the β-sheet region. In H2O at 60 °C, the turn is flattened causing the Gly7 residue to rotate and the C-terminal strand changes conformation by elongation. This causes the hydrogen bond pattern to change and the Trp9 residue to move further away from the Tyr2 residue breaking the Ar-Ar interaction and forming a new Ar-Ar interaction with the Tyr1 residue.
DSSP analysis (Figure 7) further confirms that the β-hairpin structure of CLN025 in all environments studied has a Pro-Glu-Thr-Gly turn flanked by two residues on each side in a β-sheet or β-bridge conformation. At 60 °C, Trp9 changes from being in a β-sheet conformation to a random meander conformation. In DMSO, the β-sheet structure of the peptide is diminished significantly. Tyr2 and Trp9 form a β-bridge, the turn is relaxed and the Asp3, Gly7 and Thr8 residues are in a random meander conformation.
Figure 7.
DSSP analysis of the trajectories of simulations of CLN025 in A, H2O 5 °C; B, H2O 27 °C; C, H2O 60 °C; D, TFE; E, MeOH and F, DMSO. Random meander is black, β-sheet is red, β-bridge is white, bend is blue and turn is green.
The MD simulations confirm the results of the ECD and VCD spectral analyses. CLN025 maintained a β-hairpin conformation in all environments studied. In TFE, the turn is relaxed as shown by the shift in the second couplet of the VCD spectra and its corresponding amide I' bands in the IR spectra as well as by the middle structures of the most populated cluster from the MD simulations and the DSSP analysis. In MeOH, the turn in CLN025 is shortened, as seen by the loss of turn content from the CDSSTR analysis of the ECD spectra, the results of the peak-fitting analysis of the VCD spectra and the middle structure of the peptide in MD analyses. In MeOH, however, the peptide maintains the same tight conformation as the peptide in water as indicated by the position of the second couplet in the VCD spectra and the assignment of the turn region in the middle structures of the peptide. In DMSO, the turn is relaxed and is mostly random meander as seen by the lack of an evident second couplet in the VCD spectra as well as by the DSSP analysis and the middle MD structures of the peptide.
Conclusions
The VCD spectral properties of a 10 residue miniprotein, CLN025, that forms a stable β-hairpin, with only naturally occurring amino acids and without D-amino acids or cyclization to stabilize the β-hairpin, are reported. ECD, VCD and Molecular Dynamics simulations confirm that CLN025 adopts a stable β-hairpin conformation in the environments studied. The VCD spectra exhibit a (−,+,−) pattern with bands at 1640 to 1656 cm−1, 1667 to 1687 cm−1 and 1679 to 1686 cm−1. A maximum IR absorbance was observed at 1647 to 1663 cm−1 with bands at 1630 cm−1, 1646 cm−1, 1658 cm−1 and 1675 to 1680 cm−1 in the deconvoluted spectra, indicating β-sheet, random meander, random meander or loop and turn, respectively. These results are similar to those reported for peptides in which DPro was incorporated to nucleate the β-turn in the hairpin and suggest that the VCD and IR absorbance spectra reported here are typical of β-hairpins.
Acknowledgments
This work was supported by the NIH-INBRE grant P20 RR016469.
References
- 1.Ptitsyn OB. FEBS Lett. 1991;131:197. doi: 10.1016/0014-5793(91)80799-9. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez-Alvarado M, Kortemme T, Blanco FJ, Serrano L. Bioorg & Med Chem. 1999;7:93–103. doi: 10.1016/s0968-0896(98)00215-6. [DOI] [PubMed] [Google Scholar]
- 3.Maynard AJ, Sharman GJ, Searle MS. J Am Chem Soc. 1998;120:1996–2007. [Google Scholar]
- 4.Andersen NH, Dyer RB, Fesinmeyer RM, Gai F, Liu Z, Neidigh JW, Tong H. J Am Chem Soc. 1999;121:9879–9880. [Google Scholar]
- 5.Palermo NY, Csontos J, Owen MC, Murphy RF, Lovas S. J Comput Chem. 2007;28:1208–1214. doi: 10.1002/jcc.20578. [DOI] [PubMed] [Google Scholar]
- 6.Borics A, Murphy RF, Lovas S. Protein Pept Lett. 2007;14:353–359. doi: 10.2174/092986607780363907. [DOI] [PubMed] [Google Scholar]
- 7.Csontos J, Palermo NY, Murphy RF, Lovas S. J Comput Chem. 2008;29:1344–1352. doi: 10.1002/jcc.20898. [DOI] [PubMed] [Google Scholar]
- 8.Hatfield MPD, Palermo NY, Csontos J, Murphy RF, Lovas S. J Phys Chem B. 2008;112:3503–3508. doi: 10.1021/jp077674h. [DOI] [PubMed] [Google Scholar]
- 9.Csontos J, Murphy RF, Lovas S. Biopolymers. 2008;89:1002–1011. doi: 10.1002/bip.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palermo NY, Csontos J, Murphy RF, Lovas S. Int J Quantum Chem. 2008;108:814–819. doi: 10.1002/qua.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csontos J, Murphy RF, Lovas S. Adv Exp Med Biol. 2009;611:79–80. doi: 10.1007/978-0-387-73657-0_35. [DOI] [PubMed] [Google Scholar]
- 12.Palermo NY, Csontos J, Murphy RF, Lovas S. Adv Exp Med Biol. 2009;611:89–90. doi: 10.1007/978-0-387-73657-0_40. [DOI] [PubMed] [Google Scholar]
- 13.Cochran AG, Skelton NJ, Starovasnik MA. Proc Natl Acad Sci USA. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Oyola R, Gai F. J Am Chem Soc. 2003;125:15388–15394. doi: 10.1021/ja037053b. [DOI] [PubMed] [Google Scholar]
- 15.Takekiyo T, Wu L, Yoshimura Y, Shimizu A, Keiderling TA. Biochemistry. 2009;48:1543–1552. doi: 10.1021/bi8019838. [DOI] [PubMed] [Google Scholar]
- 16.Eidenschink LA, Kier BL, Huggins KNL, Andersen NH. Proteins. 2009;75:308–322. doi: 10.1002/prot.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen NA, Olsen KA, Fesinmeyer Rm, Tan X, Hudson FM, Eidenschink LA, Farazi SR. J Am Chem Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riemen AJ, Waters ML. Biochemistry. 2009;48:1525–1531. doi: 10.1021/bi801706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda S, Akiba T, Kato YS, Sawada Y, Sekijima M, Ishimura M, Ooishi A, Watanabe H, Odahara T, Harata K. J Am Chem Soc. 2008;130:15327–15331. doi: 10.1021/ja8030533. [DOI] [PubMed] [Google Scholar]
- 20.Terada T, Satoh D, Mikawa T, Ito Y, Shimizu K. Proteins. 2008;73:621–631. doi: 10.1002/prot.22100. [DOI] [PubMed] [Google Scholar]
- 21.Borics A, Murphy RF, Lovas S. Biopolymers. 2003;72:21–24. doi: 10.1002/bip.10247. [DOI] [PubMed] [Google Scholar]
- 22.Copps J, Murphy RF, Lovas S. Peptide Science. 2007;88:427–437. doi: 10.1002/bip.20709. [DOI] [PubMed] [Google Scholar]
- 23.Krimm S, Bandekar J. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 24.Prestrelski SJ, Byler DM, Thompson MP. Int J Pept Protein Res. 1991;37:508–512. [PubMed] [Google Scholar]
- 25.Borics A, Murphy RF, Lovas S. Biopolymers. 2006;85:1–11. doi: 10.1002/bip.20593. [DOI] [PubMed] [Google Scholar]
- 26.Wyssbrod HR, Diem M. Biopolymers. 1992;32:1237–1242. doi: 10.1002/bip.360320912. [DOI] [PubMed] [Google Scholar]
- 27.Shanmugam G, Polararapu PL. Biophysical Journal. 2004;87:622–630. doi: 10.1529/biophysj.104.040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keiderling TA, Silva RAGD, Yoder G, Dukor RK. Bioorganic & Medicinal Chemistry. 1999;7:133–141. doi: 10.1016/s0968-0896(98)00217-x. [DOI] [PubMed] [Google Scholar]
- 29.Kubelka J, Keiderling TA. J Am Chem Soc. 2001;123:12048–12058. doi: 10.1021/ja0116627. [DOI] [PubMed] [Google Scholar]
- 30.Pancoska P, Yasui SC, Keiderling TA. Biochemistry. 1989;28:5917. doi: 10.1021/bi00440a031. [DOI] [PubMed] [Google Scholar]
- 31.Baumruk V, Keiderling TA. J Am Chem Soc. 1993;115:6939. [Google Scholar]
- 32.Zhao C, Polavarapu PL, Das C, Balaram P. J Am Chem Soc. 2000;122:8228–8231. [Google Scholar]
- 33.Hilario J, Kubelka J, Syud FA, Gellman SH, Keiderling TA. Biopolymers. 2002;67:233–236. doi: 10.1002/bip.10094. [DOI] [PubMed] [Google Scholar]
- 34.Hilario J, Kubelka J, Keiderling TA. J Am Chem Soc. 2003;125:7562–7574. doi: 10.1021/ja030039e. [DOI] [PubMed] [Google Scholar]
- 35.Szendrei GI, Fabian H, Mantsch HH, Lovas S, Nyeki O, Schon I, Otvos L., Jr Eur J Biochem. 1994;226:917–924. doi: 10.1111/j.1432-1033.1994.t01-1-00917.x. [DOI] [PubMed] [Google Scholar]
- 36.Compton LA, Johnson WC., Jr Anal Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 37.Manavalan P, Johnson WC., Jr Anal Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 38.Sreerama N, Woody RW. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 39.Whitmore L, Wallace BA. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 40.Whitmore L, Wallace BA. Nucleic Acids Research. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobley A, Whitmore L, Wallace BA. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 42.Sreerama N, Woody RW. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 43.Sreerama N, Venyaminov SY, Woody RW. Anal Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 44.Baumruk V, Huo D, Dukor RK, Keiderling TA, Lelieure D, Brack A. Biopolymers. 1994;34:1115–1121. doi: 10.1002/bip.360340815. [DOI] [PubMed] [Google Scholar]
- 45.Jung C. J Mol Recognit. 2000;13:325–351. doi: 10.1002/1099-1352(200011/12)13:6<325::AID-JMR507>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 46.Surewicz WK, Mantsch HH, Chapman D. Biochemistry. 1993;32:389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- 47.Lindahl E, Hess B, Van der Spoel D. J Mol Mod. 2001;7:306–317. [Google Scholar]
- 48.Berendsen HJC, Van der Spoel D, Van Drunen R. Comp Phys Comm. 1995;91:43–56. [Google Scholar]
- 49.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. J Comput Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 50.Oostenbrink C, Soares TA, van der Vegt NF, van Gunsteren WF. Eur Biophys J. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 51.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. Dordrecht: D. Reidel Publishing Company; 1981. pp. 331–342. [Google Scholar]
- 52.Oostenbrink C, Villa A, Mark AE, Gunsteren WFV. J Comp Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 53.Weast RC. CRC Handbook of Chemistry and Physics 1st Student Edition. Boca Raton, Florida: CRC Pres, Inc; 1988. [Google Scholar]
- 54.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comp Chem. 1997;18:1463–1472. [Google Scholar]
- 55.Berendsen HJC, Postma JPM, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 56.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 57.Miyamoto S, Kollman PA. J Comp Chem. 1992;13:952–962. [Google Scholar]
- 58.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comp Chem. 1997;18:1463–1472. [Google Scholar]
- 59.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 60.Bour P, Keiderling TA. J Phys Chem B. 2005;109:23687–23697. doi: 10.1021/jp054107q. [DOI] [PubMed] [Google Scholar]
- 61.Pancoska P, Wang L, Keiderling TA. Protein Sci. 1993;2:411. doi: 10.1002/pro.5560020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dukor RK, Pancoska P, Keiderling TA, Prestrelski SJ, Arakawa T. Arch Bioch Biophys. 1992;298:678. doi: 10.1016/0003-9861(92)90465-9. [DOI] [PubMed] [Google Scholar]
- 63.Lee H, Kim SS, Choi JH, Cho M. J Phys Chem B. 2005;109:5331. doi: 10.1021/jp0461302. [DOI] [PubMed] [Google Scholar]
- 64.Choi JH, Cho M. J Chem Phys. 2004;120:4383. doi: 10.1063/1.1644100. [DOI] [PubMed] [Google Scholar]
- 65.Bour P, Keiderling TA. J Chem Phys. 2003;119:11253. [Google Scholar]
- 66.Ham S, Kim JH, Kochan H, Cho M. J Chem Phys. 2003;118:3491. [Google Scholar]
- 67.Pantoja-Uceda D, Rico M, Jimenez MA. Biopolymers. 2005;79:150–162. doi: 10.1002/bip.20345. [DOI] [PubMed] [Google Scholar]
- 68.Buck M. Q Rev Biophys. 1998;31:297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- 69.Grishina IB, Woody RW. Faraday discuss. 1994;99:245–262. doi: 10.1039/fd9949900245. [DOI] [PubMed] [Google Scholar]
- 70.Cochran AG, Skelton NJ, Starovasnik MA. Proc Natl Acad Sci USA. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Susi H, Byler DM. Arch Biochem Biophys. 1987;258:465–469. doi: 10.1016/0003-9861(87)90367-5. [DOI] [PubMed] [Google Scholar]
- 72.Shanmugam G, Polavarapu PL, Gopinath D, Jayakumar R. Biopolymers. 2005;80:636–642. doi: 10.1002/bip.20132. [DOI] [PubMed] [Google Scholar]