Abstract
The drug-loading properties of nanocarriers depend on the chemical structures and properties of their building blocks. Here, we customize telodendrimers (linear-dendritic copolymer) to design a nanocarrier with improved in vivo drug delivery characteristics. We do a virtual screen of a library of small molecules to identify the optimal building blocks for precise telodendrimer synthesis using peptide chemistry. With rationally designed telodendrimer architectures, we then optimize the drug binding affinity of a nanocarrier by introducing an optimal drug-binding molecule (DBM) without sacrificing the stability of the nanocarrier. To validate the computational predictions, we synthesize a series of nanocarriers and evaluate systematically for doxorubicin delivery. Rhein-containing nanocarriers have sustained drug release, prolonged circulation, increased tolerated dose, reduced toxicity, effective tumor targeting and superior anticancer effects owing to favourable doxorubicin-binding affinity and improved nanoparticle stability. This study demonstrates the feasibility and versatility of the de novo design of telodendrimer nanocarriers for specific drug molecules, which is a promising approach to transform nanocarrier development for drug delivery.
Introduction
Nanoparticle-based drug encapsulation increases drug solubility and stability, minimizes toxic side effects,1, 2 and more importantly, delivers drug molecules specifically to tumors through the enhanced permeability and retention (EPR) effect.3, 4 Several nanodrugs have been approved by US Food and Drug Administration.1, 5 For example, Doxil®, a stealth liposomal nanoformulation of doxorubicin (DOX), has significantly reduced cardiotoxicity.6 However, Doxil shows only marginal improvement in efficacy over free DOX in clinical practice, especially for solid tumor treatment.6, 7, 8 It is due to the poor intratumoral diffusion (~100 nm) 7 and unfavorable drug release profile reducing drug availability, despite of more drug delivered to tumor sites by EPR effects.6, 7, 8 This indicates that the balance between drug retention and drug release is critical in determining the in vivo fate and efficacy of a nanoformulation in cancer treatment. In the literatures, numerous DOX delivery systems have been developed including liposomes,9 dendrimers,10, 11 polymer nanoparticles12, polymer-DOX conjugations13, 14, polymer micelles,17–18 and inorganic nanoparticles.15 Of these, polymer micelles (10–100 nm in size) are one of the most versatile nanocarriers for the delivery of DOX and other chemotherapeutic drugs due to the abundant chemical diversity, functionality and tunable physical properties.16
“Like dissolves like” is a principle rule that is applicable to mixture systems. A docetaxel-conjugated polyethylene glycol-poly(ε-caprolactone) (PEG-PCL) polymer showed higher docetaxel loading capacity and stability than the parent polymer PEG-PCL.17 Polymer-drug conjugations via labile bonds are considered to be an effective prodrug strategy to increase the solubility and reduce the toxicity of the hydrophobic drug molecules.14, 18 Despite some polymeric prodrugs can self-assemble into micelles for further drug loading,19 this approach may be hindered by the availability of functional groups on a drug molecule and the high cost of production. Instead, a molecule with structural similarity and a complimentary conformation to the drug molecule is promising to be an efficient host after being conjugated onto a polymeric nanocarrier to improve drug delivery. However, it is still challenging to introduce these molecules freely into polymers with the precise control of location and density.
The growth of the polymer field has benefited from new developments in synthetic and catalytic chemistry. The biocompatible polymers for drug delivery are still limited to a few, which hinders the development and optimization of nanocarriers to deliver the compounds/drugs in preclinical and clinical development. In addition, the uncertain relationship between the structure and property of polymer nanoparticles for drug delivery is a problem for pharmaceutical companies, whose expertise are to probe the well-defined drug-biologic interactions using systematic and computer-aided approaches.20 Computational chemistries, such as theoretical methods and molecular simulations, have been applied in nanoparticle system to understand drug-loading properties.21 Unlike proteins, nanoparticle systems have no defined conformations and are too big in size for computation chemistry to build an affordable and reliable model for drug loading predictions. Up-to-date, the structure-based de novo design and optimization of nanocarriers for a given drug delivery has not been documented, due to the lack of both reliable theoretical models and precise polymer synthesis for the systematic validation and evaluation.21
Here we developed a novel well-defined telodendrimer nanoplatform to leverage the synergism between computational design and combinatorial chemistry for drug-specific nanocarrier development. We found that the optimized telodendrimer nanoformulations of DOX significantly improved the treatment of lymphoma in animal models, in comparison with free DOX and Doxil®.
Results
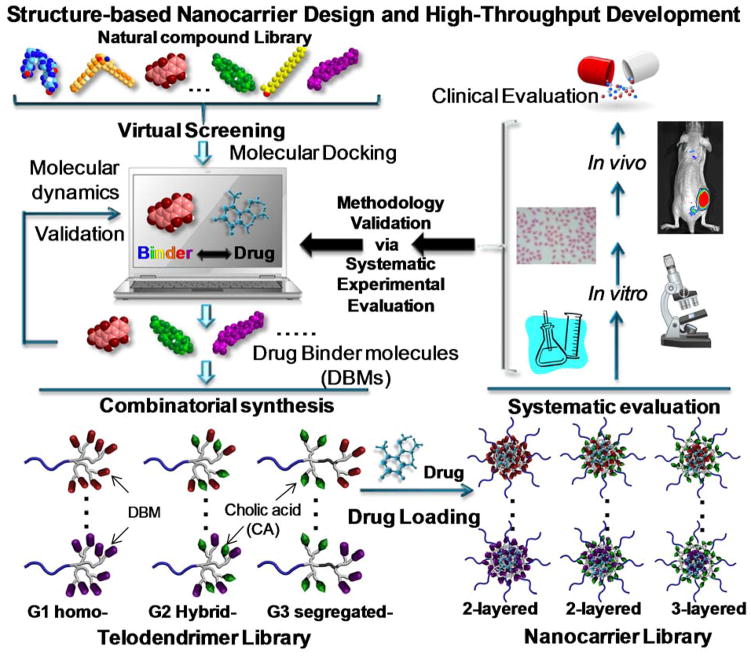
We have developed a hybrid polymer system, a telodendrimer, composed of linear polyethylene glycol (PEG)-blocking–dendritic polylysine and the capping peripheral building blocks, e.g. cholic acid (CA). 22, 23 It self-assembles into micelles in aqueous solution for efficient delivery of anticancer drugs.22, 23, 24, 25, 26, 27, 28 The efficient peptide chemistry used in telodendrimer synthesis allows for free and precise control over the architecture and the functionality of the telodendrimer. The peripheral groups on the dendritic polylysine have more flexibility in interacting with drug molecules when compared to the main chains or pendant groups in linear polymers. This interaction greatly influences drug-loading properties of nanocarriers. We hypothesize that the optimal drug-binding molecules (DBMs) could be identified by molecular docking and introduced into telodendrimer in parallel to make a library of nanocarriers for systematic evaluation and optimization (Figure 1). Herein, the telodendrimer system provides a blueprint for the customized nanocarrier design in the delivery of a given drug.
Figure 1. Structure-based nanocarrier design and development.
Schematic illustration of the rational design and combinatorial synthesis of telodendrimers for systematic evaluation and optimization of nanocarriers.
Structure-based telodendrimer design
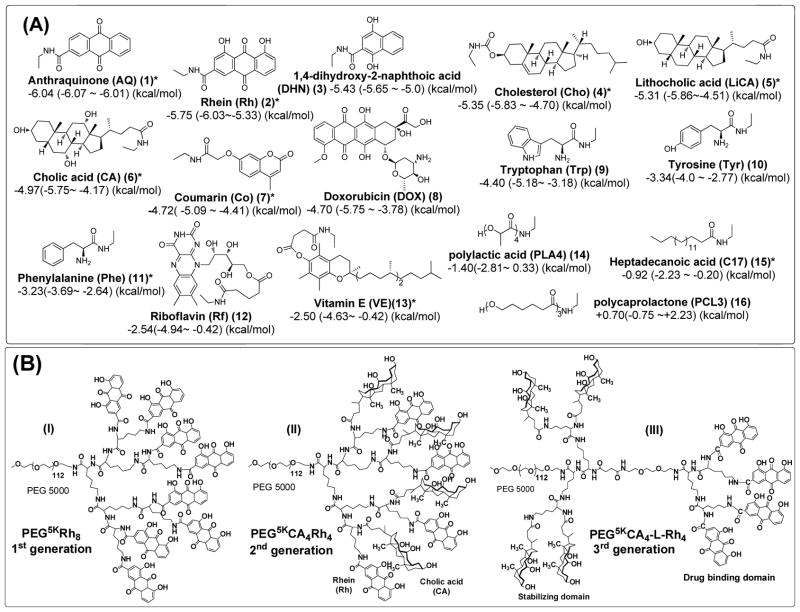
To prove the concept, a number of lipophilic vitamins and natural molecules were collected into a model library for virtual screening against DOX via molecular docking. As shown in Figure 2A, the docking energies were ranked based on the mean of 100 docking assays (Supplementary Table 1). DOX was not shown as the most favorable binder to itself in the docking study due to steric hindrance. As expected, the aromatic molecules, such as anthraquinone (Aq), Rhein (Rh) and 1,4-dihydroxy-2-naphthoic acid (DHN), and the polycyclic steroid molecules, e.g., cholesterol (Cho), lithocholic acid (LiCA) and CA, could bind to DOX strongly with minimum docking energy of −5.65 ~ −6.07 kcal/mol via pi-pi and pi-polycyclic hydrophobic interactions and hydrogen bonding, respectively. In contrast, the segments of typical synthetic polymers, e.g., poly(ε-caprolactone) (PCL) and poly(lactic acid) (PLA) and a fatty acid of heptadecanoic acid (C17) showed less negative docking energies with DOX (−0.75 ~ −2.81 kcal/mol).
Figure 2. Chemical structures of building blocks and telodendrimers.
(A) Representative DBMs ranked by docking energies against DOX-binding in molecular docking. The docking energies are presented as the average and the range of docking energy. *Building blocks selected for telodendrimer synthesis. (B) Three generations of telodendrimers with distinct architectures synthesized by peptide chemistry: the 1st generation of telodendrimers PEG5kRh8 (I), 2nd generation of telodendrimer PEG5kCA4Rh4 (II) and the 3rd generation of the functional segregated telodendrimer PEG5kCA4-L-Rh4 (III). As an example, Rh is presented as one of the DBMs together with an amphiphilic CA on the periphery.
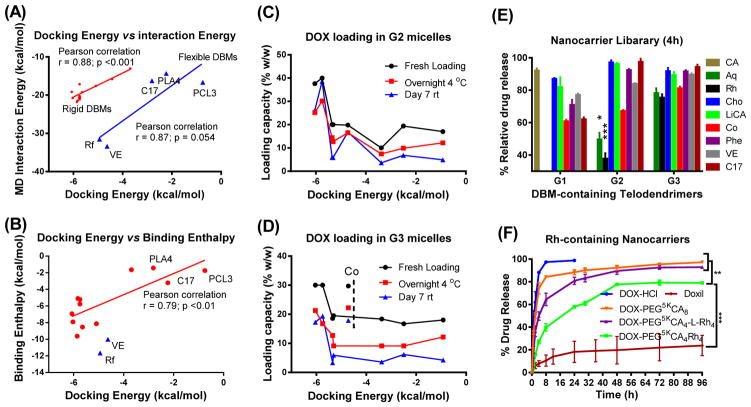
It has been a concern that the lack of water in molecular docking decreases its accuracy in predicting binding events in an aqueous environment, e.g. drug loading process. To address this issue, we conducted molecular dynamic simulations on the DBM-DOX complexes in the presence of explicit water. The DBM-DOX complexes with the lowest docking energies were taken as the initial conformations for molecular dynamics (MD) simulation for 5 ns. The mean interaction energies between DBMs and DOX over the simulation were extracted (Supplementary Table 1) and compared with docking energy (Figure 3A). The docking energies were observed to be less negative than MD interaction energy, due to the consideration of desolvation and torsion energies in Autodock 4 energy evaluation.29 Importantly, a good monotonic correlation (Pearson’s r: 0.88) was observed for the DBMs with the relatively rigid structures, e.g., aromatic or steroidal compounds with high dock rankings. However, the docking energy of flexible molecules, such as fragments of typical synthetic polymers of PCL, PLA and a fatty acid of C17 were underestimated by docking, which exhibited more favorable interactions in MD simulations, although still higher than those of rigid aromatic and steroidal compounds. Riboflavin (Rf) and vitamin E (VE, α-tocopherol) have both aromatic and flexible components in their structures, and exhibited the most favorable interactions with DOX in MD simulation via adapting hairpin binding models (Supplementary Figure 2A). Interestingly, the binding energy for DOX-Rf underwent sudden changes with time, which was also observed for DOX-DOX binding (Supplementary Figures 2F&2G), may due to their hydrophilic side chains. In contrast, a converging energy profile was observed for VE binding with DOX via hydrophobic interactions.
Figure 3. Methodology validation and systematic evaluation.
(A, B) Most favorite docking conformations between DBM-DOX were simulated by molecular dynamics for 5 ns in the present of explicit water. (A) Unbound interaction energy computed from MD simulations plotted against the minimum docking energies. (B) The binding enthalpies analyzed by a solvent-balance method were plotted against the lowest docking energies. Linear regression model was fit via Ordinary Least Square to calculate the Pearson correlation coefficient and the associated p values. (C, D) Drug loading capacities of G2 (C) and G3 (D) telodendrimer micelles at fresh loading and after storage were plotted against docking energies of the DBMs. (E) Percentage of DOX release from G1, G2 and G3 telodendrimer micelle library relative to the release rate of free DOX and PEG5kCA8 at 4 h. (F) In vitro drug release profiles were studied using a dialysis method under sink condition for free DOX, Doxil and DOX loaded in various telodendrimer nanocarriers. Data were presented as mean ± SEM (n=3). Student’s t-test: *P<0.05; **P<0.01; ***P<0.001.
Further, the binding enthalpy of DBMs and DOX were calculated by a solvent-balance method30 from three MD simulations for the complex, and individual molecules with the same number of water molecules before and after molecular binding (Supplementary Figure 3A). The average potential energies (U) of the initial systems, i.e. (DBM + nH2O) and (DOX + mH2O) and final (DBM-DOX + (m+n)H2O) system were analyzed, as shown in Supplementary Table 2. The enthalpy changes (ΔH) were calculated as:
| (1) |
It showed that the enthalpy changes were at a similar level and correlated monotonically with docking energies (Figure 3B) and the MD interaction energies (Supplementary Figure 3C). In addition, enthalpy calculations can differentiate the DBMs with the similar docking energies, such as aromatic molecules (Rh, AQ and DHN) and steroid compounds (CA, LiCA and Cho). These studies demonstrated that MD simulation provides a valuable validation for DBMs docking results, especially for flexible molecules.
Indicated by both molecular docking and MD simulations, Rh exhibited consistent, strong DOX binding affinities. It can be conjugated onto telodendrimer through its readily available carboxylic acid functional group. In addition, Rh is an attractive bioactive molecule in traditional Chinese medicine.31, 32 Rh binds to DOX with much negative docking energies (−6.03 vs −4.9 kcal/mol) and much narrower energy distributions (−0.76 vs 1.77 kcal/mol) than DOX-DOX binding. Experimentally, 1H NMR and NOESY spectra of a DOX-Rh solution mixture in DMSO-d6 indicated the correlations of both polar hydrogen and aromatic protons between DOX and Rh (Supplementary Figures 4A–C), suggesting the formation of intermolecular H-bonding and pi-pi stacking. In addition, we found that DOX stabilizes Rh from crystallization in DMSO at a concentration of approximately 10 mM during NMR study. In contrast, only hydrogen bonding was indicated by NMR studies and no hydrophobic interactions were detected between CA and DOX in DMSO (Supplementary Figures 4D&4E), which correlates with a slightly higher docking energy (−5.75 kcal/mol) between CA-DOX with broader energy distributions than Rh-DOX (2.18 vs 0.76 kcal/mol). This may be the reason for the fair stability of DOX-loading in the typical CA-contacting telodendrimer PEG5kCA8 micelles.25
As shown in Figure 2B, PEG5kRh8 was first prepared. It is almost insoluble in aqueous solution due to the overwhelming intermolecular pi-pi stacking and hydrogen bonding between Rh moieties. To develop an efficient nanocarrier, the strong adherent forces need to be restricted within the core of the nanocarrier for drug loading, which is achieved mostly by the PEGylation of nanoparticles. However, it is not sufficient sometimes to use the conventional PEG with chain length from 2 to 5 kDa in molecular weight to stabilize the cluster of hydrophobic components introduced in telodendrimer for optimization, such as PEG5kRh8. In our previous studies, the facial amphiphilic CA was found to be important for telodendrimers to form stable micelles, in comparison with its hydrophobic analogues.23 We have demonstrated that the hydrophilic hydroxyl-rich surface of CA faces towards the aqueous PEG layer on the core-shell interface of PEG5kCA8 micelles to minimize the surface tension, therefore stabilizing the nanocarriers.33 With this in mind, we developed another two versions of telodendrimers with the rational architecture design: i.e. the second generation (G2) of hybrid telodendrimers and the third generation (G3) of functional-segregated telodendrimers (Figure 2B) using CA as co-building blocks. The proximal positions of CA within both G2 and G3 telodendrimers are expected to stabilize nanocarriers and payloads. As a result, G2 PEG5kCA4Rh4 and G3 PEG5kCA4-L-Rh4 were observed to be easily dispersed into aqueous solution and readily for DOX loading.
Combinatorial nanocarrier synthesis and characterization
In order to systematically validate the computational prediction in guiding telodendrimer design, eight representative DBMs with different docking energy levels (Figure 2A) were selected for G1, G2 and G3 telodendrimer synthesis, respectively. Table 1 shows twenty-four telodendrimers synthesized in a combinatorial manner by conjugating DBMs on the peripheral of polylysine scaffolds (Supplementary Figure 1). All the telodendrimers were fully characterized by 1H NMR and MALDI-TOF MS (Supplementary Figure 10). As expected, most of the G1 telodendrimers have poor solubility with polydispersed particle sizes, especially for DBMs with high partition coefficient (LogP), e.g., Cho, LiCA, C17 and VE, which are commonly used as building blocks in amphiphilic polymer micelle preparation. Alike PEG5kRh8, an anthraquinone (AQ)-containing telodendrimer PEG5kAQ8 has very poor solubility in aqueous solution, due to its overwhelming pi-pi stacking. In contrast, G1 telodendrimers derivatives from smaller DBMs, e.g., coumarin (Co) and phenylalanine (Phe) self-assemble into micelles with uniform and small particle sizes of approximately 35 and 42 nm, respectively. In addition, these two telodendrimers exhibited relatively good DOX loading capacities with about 20–30% drug loading. G2 and G3 telodendrimers mostly formed well-dispersed micelles with sub-40 nm size before and after DOX loading at a 20% feed content with good to excellent loading efficiency. Throughout the library of the 24 nanocarriers, Rh- and AQ-containing G2 and G3 telodendrimers represent the best nanocarriers with about 30~40% (w/w) of DOX loading capacity and ~100% initial loading efficiency (Figures 3C&3D).
Table 1. Drug loading properties of nanocarrier library.
The drug loading capacity, efficiency, CMC and particle sizes of the telodendrimer nanocarrier library made from eight selected DBMs.
| DBM (rank-ing) | DOX | PEG5KDBM8 (G1) | PEG5KCA4DBM4 (G2) | PEG5KCA4-L-DBM4 (G3) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CMC (μM) | Size (nm) | DOX LE (%)a LE0 - LE1 - LE7 | CMC (μM) | Size (nm) | DOX LE (%)a LE0 - LE1 - LE7 | CMC (μM) | Size (nm) | DOX LE (%)a LE0 - LE1 - LE7 | |||
| AQ(1) | 0 | 2.84 | PPT | N/A | 2.1 | MP | N/A | 2.36 | MP | N/A | |
| 10% | - | PPT | 49- - | - | 24±7 | 100-100-91 | - | 27±7 | 100-94-100 | ||
| 20% | - | - | - | - | 25±6 | 100-87-93 | - | 29±9 | 100-96-100 | ||
| 30% | - | - | - | - | 20 ±5 | 100-84-61 | - | 30±8 | 100-71-58 | ||
| 40% | - | - | - | - | MP | 94-54-66 | - | - | - | ||
| Rh(2) | 0 | ND | PPT | N/A | 0.93 | 17±5 | N/A | 2.7 | 32±8 | N/A | |
| 10% | - | PPT | - | 0.89 | 20±5 | 100-100-100 | 1.1 | 30±8 | 100- 100-100 | ||
| 20% | - | - | - | - | 24±7 | 100-98-100 | - | 28±7 | 100-78-86 | ||
| 30% | - | - | - | - | 19±4 | 100-85-95 | - | 21±5 | 100-56-64 | ||
| 40% | - | - | - | - | 15±4 | 100-75-97 | - | - | - | ||
| LiCA(4) | 0 | 1.18 | 36±11 | N/A | 1.45 | 17±4 | N/A | 1.2 | 17±4 | N/A | |
| 10% | - | 350/49 b | 100-100-87 | - | 14±3 | 100-100-57 | 15±3 | 100- 93-59 | |||
| 20% | - | 59/33 b | 99-70-52 | - | 16±4 | 100-64-29 | Mp | 98-46-19 | |||
| Cho(5) | 0 | 0.89 | PPT | N/A | 0.75 | 17±4 | N/A | 1.2 | 18±4 | N/A | |
| 10% | - | MP | 100-100-87 | - | 39±12 | 100-100-42 | - | 23±7 | 100-84-33 | ||
| 20% | - | - | - | - | 41±16 | 100-72-30 | - | 24±8 | 93-63-18 | ||
| Co(8) | 0 | 2.59 | 35 ± 12 | N/A | 1.99 | 14 ± 4 | N/A | 1.7 | 36 ± 13 | N/A | |
| 10% | - | 28 ± 9 | 100-100-92 | - | 13 ± 2 | 100-97-97 | - | 36 ± 13 | 100-100-85 | ||
| 20% | - | 27 ± 8 | 100-94-73 | - | 385/133 b | 99-82-84 | - | 38 ± 13 | 100-68-80 | ||
| 30% | - | 32 ± 10 | 100-75-65 | - | - | - | - | 342/32 b | 99-60-74 | ||
| Phe (11) | 0 | 1.77 | 42±19 | N/A | 2.86 | 13±3 | N/A | 2.9 | 103±39 | N/A | |
| 10% | - | 34±9 | 100-100-92 | - | 185/14 | 100-75-36 | - | 240/22 b | 100-91-36 | ||
| 20% | - | 34±14 | 100- 95-100 | - | - | - | - | 17±4 | 92-45-13 | ||
| 30% | - | 29 ±8 | 100-53-40 | - | - | - | - | - | - | ||
| VE(12) | 0 | 2.5 | MP | N/A | 1.6 | MP | N/A | 1.4 | 25±6 | N/A | |
| 10% | MP | 78-64-55 | - | 38±15 | 100-90-65 | - | 23±6 | 100-85-61 | |||
| 20% | N/A | N/A | - | 36±12 | 96-50-34 | - | 24±6 | 84-46-25 | |||
| C17 (13) | 0 | 1.66 | PPT | N/A | 1.33 | 13±4 | N/A | 2.7 | MP | N/A | |
| 10% | - | 408/61 | 86- - | - | 14±2 | 100-92-32 | - | 196±99 | 100-92-43 | ||
| 20% | - | - | - | - | 14±3 | 85-61-24 | - | MP | 90-44-17 | ||
Note:
DOX loading efficiency (LE) were measured by fluorescence (λex=470 nm, λem=590 nm) measurements of samples before and after spin at the speed of 13,000g for 5 min; The drug loading efficiencies were measured right after drug loading process (LE0) and after a storage at 4 °C overnight (LE1) or after a storage at room temperature for one week (LE7). Therefore drug loading efficiencies were presented as LE0 - LE1 - LE7.
Two peaks were detected in DLS particle size analysis; N/A: not applicable; ND: not detected; MP: multiple peaks; PPT: precipitation; -: not tested
As shown in Table 1, critical micelle concentrations (CMCs) of the telodendrimer micelles in the library are mostly less than 3 μM, indicating stable micelle construction. The DOX release profiles from all micelle formulations were tested by dialysis methods. The G1 telodendrimer nanocarriers released the drug slower than G2 and G3 telodendrimers with the same building blocks, likely due to the significantly reduced drug contents after removal of drug precipitations as well as the enhanced entanglement and larger particle sizes. Of the DBMs tested, Rh- and AQ-containing G2 telodendrimers sustained DOX better from release (Figure 3E) and exhibited the best stabilities (Table 1) than all other formulations. In contrast, a significant amount of drug precipitations were observed upon storage for all other formulations, including G1-Ph, G1-Co and G3-Co nanocarriers. Overall, the drug loading capacity and stability of DOX-loaded telodendrimer correlated with the DOX-binding affinities of DBMs as evaluated computationally. Given the rationally designed architectures, the drug binding affinity of a nanocarrier can be increased without sacrificing the stability and dispersibility of nanocarriers.
Characterization of Rh-containing telodendrimer nanocarriers
DOX release profiles from G2 and G3 Rh-containing telodendrimer micelles were then compared with free DOX, Doxil and DOX-PEG5kCA8 (Figure 3F). As expected, a burst release and a very slow drug release of DOX were observed, respectively, for free DOX and stealth liposomal Doxil. DOX loaded PEG5kCA8 formulation was observed similarly to the free DOX release profile. DOX release was significantly slower from G2 telodendrimer PEG5kCA4Rh4 than G3-Rh telodendrimer, DOX-PEG5kCA8 and free DOX. After DOX loading, the CMCs of the G2-Rh and G3-Rh micelles were slightly decreased to 0.83 and 1.1 μM, respectively (Table 1), using Nile Red (NR) as a fluorescent probe.34 Interestingly, NR, as well as DOX were fluorescently quenched after being loaded in Rh-containing G1 and G2 telodendrimer micelles, indicating the close pi-pi interactions between DOX or NR with Rh moieties in telodendrimers (Supplementary Figures 5A&5B). In contrast, the fluorescent signal of DOX was enhanced after being loaded in non-aromatic PEG5kCA8 micelles (Supplementary Figure 5E). Meanwhile, pronounced fluorescent signals were observed after DOX or NR was loaded in the G3-Rh telodendrimer nanocarriers (Supplementary Figures 5C&5E). This suggests that DOX or NR can be encapsulated partially in the CA-rich proximal layer in the domain-segregated G3-Rh micelles (Supplementary Figure 5F) and remain fluorescent. This also explains the relatively faster DOX release from G3-Rh micelles than that from G2-Rh micelles due to the lower DOX-CA binding affinity.
The reproducibility and quality control of nanocarrier synthesis is critical for clinical development of nanomedicine. To test the reproducibility, G2 and G3 Rh/CA-containing telodendrimers were re-synthesized following the procedure shown in Supplementary Figure 1. Both NMR and MALDI-TOF MS analysis revealed the precise telodendrimers synthesis via peptide chemistry (Supplementary Figures 6A–D). The G2 and G3 Rh-telodendrimers self-assemble into uniform micelles with sizes of 17 nm and 32 nm, respectively, determined by DLS analysis (Supplementary Figure 6E). Transmission electron microscopy (TEM) images revealed spherical micelles formed by G2 hybrid PEG5kCA4Rh4 telodendrimer before and after DOX loading. In contrast, the G3 telodendrimers self-assemble into short rod-shaped micelles (~10 nm in width) before and after DOX loading. Segregated oligo-Rh in the G3 telodendrimer has a good chance to form the long range pi-pi stacking in one dimension, whereas Rh is separated by CA in the G2 hybrid telodendrimer. TEM indicated the increased thickness of G3 worm-like micelles from 10 to 16 nm with increasing DOX loading contents from 10–30% by weight (Supplementary Figures 6F&6G). Proton NMR spectra of the fresh DOX-loaded micelle solutions in D2O were recorded with no recognizable free DOX signals detected, indicating the completion of drug encapsulation (data not shown). The DOX-loaded G2 and G3 micelle solutions at 20% drug content maintained stable particle size and drug loading content upon storage at 4 °C for months monitored by DLS and UV-vis spectroscopy, respectively, while precipitation usually appears for DOX-loaded PEG5kCA8 micelles at 4 °C overnight.
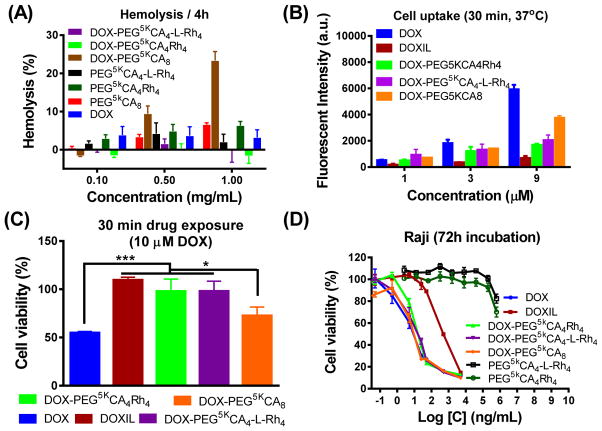
Given the enhanced stability, the hemolytic activities of both G2 and G3 Rh-containing telodendrimers were almost undetectable before and after DOX loading up to 1 mg/mL concentration after 4 h and overnight incubation with red blood cells in vitro (Figure 4A). DOX-PEG5kCA8 was observed to have moderate hemolysis (~20%) at 1 mg/mL, despite very low hemolysis for the blank ones (<5%). Reduced drug uptake in Raji lymphoma cells was observed in the cell lysate after incubation with both G2 and G3 DOX nanoformulations when compared to free DOX and DOX-PEG5kCA8 (Figure 4B & Supplementary Figures 7A–C). Doxil had even lower cell uptake after 30 min incubation, reflecting the stealth property and superior drug encapsulation stability. The cell uptake of G2 and G3 telodendrimer micelles and Doxil were significantly enhanced at 37 °C when compared to 4 °C in a concentration-dependent manner, indicating nanoparticle uptake through the endocytosis pathway. As expected, reduced cytotoxicity of Doxil, DOX-G2-Rh and DOX-G3-Rh formulations on Raji lymphoma cells was observed after a drug exposure of 30 min (Figure 4C) or 2 h (Supplementary Figure 7D), which was correlated with the reduced cell uptake. This may indicate reduced off-target toxicity of payload drug in blood circulation. Blank G2-Rh and G3-Rh telodendrimer micelles didn’t show toxicity at a concentration of 500 μg/mL in cell culture. As shown in the in vitro cell viability assays after a 72 h drug incubation in three lymphoma cell lines (Figure 4D & Supplementary Figures 7E–H), the telodendrimer nanoformulations of DOX exhibited similar half-maximum growth inhibitory concentrations (IC50s) compared to free DOX due to the efficient drug release as shown in Figure 3F. While Doxil exhibited about 40–100 fold reduction in IC50 compared to the other formulations, which was due to reduced cell uptake (Figure 4B) and slow drug release (Figure 3F). It demonstrated that the balanced drug sustainability and drug release are important to reduce side effects and maximize drug efficacy.
Figure 4. Biocompatibility and cytotoxicity of optimized nanoformulations.
(A) Reduced hemolytic properties of Rh-containing optimized telodendrimers with/without DOX loading after incubation with red blood cell at 37 °C for 4 h. (B) Slower cell uptake of optimized nanoformulations measured by the fluorescent signals of the cell lysate of Raji lymphoma cells after the incubation with free DOX, Doxil and three DOX nanoformulations with concentrations of 1, 3 and 9 μM at 37 °C for 30 min. (C) Reduced cytotoxicity of nanoformulations to Raji cells after being incubated with various DOX formulations for 30 min, followed by the incubation with refreshed cell culture medium for 72 h. (D) Maintained efficacy of optimized nanoformulations on Raji lymphoma cells after continuous 72 h incubation with blank micelles, free DOX, Doxil and DOX nanoformulations. Data were presented as mean ± SEM (n =3). Student’s t-test: * p <0.05; ** P<0.01; *** P<0.001.
Reduced toxicity and increased tumor targeting
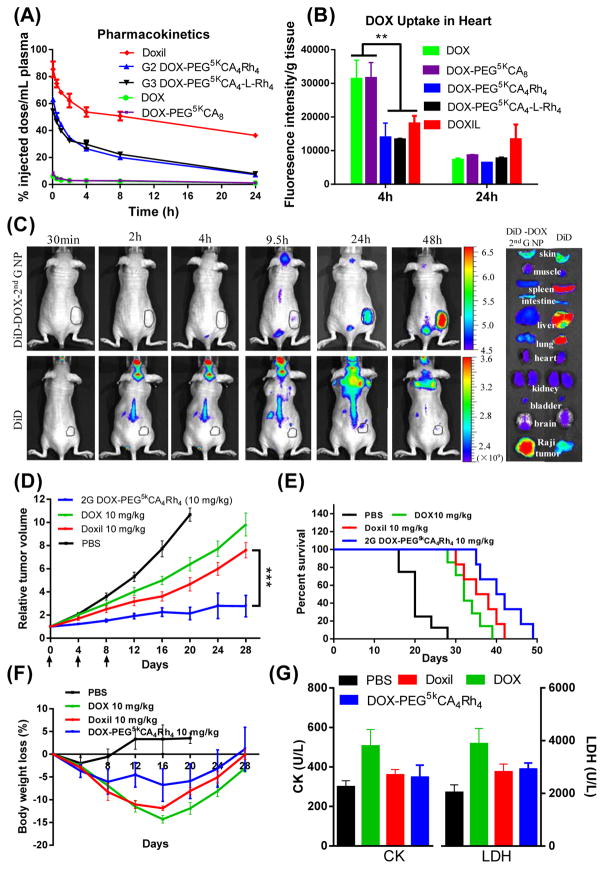
The maximum tolerated doses (MTD) of different DOX-formulations were tested in BALB/c mice (Supplementary Figures 8A&8B). A single dose MTD of a G2 DOX-PEG5kCA4Rh4 was determined to be between 20 mg/kg and 25 mg/kg, which is about 2 to 2.5 fold higher than the MTD dose of DOX and Doxil (10 mg/kg).25 The MTD for G3 of DOX-PEG5kCA4-L-Rh4 was observed to be between 15 mg/kg to 20 mg/kg. The blank G2-Rh and G3-Rh telodendrimer micelles were tested in animals: no toxicity and no body weight loss were observed after a single injection at 125 mg/kg. In order to identify proper doses for in vivo cancer treatment, repeated dosages were given at a dose of 15 mg/kg on days 0 and 4. The G2 DOX-PEG5kCA4Rh4 nanoformulation was tolerated well in the animals. One animal receiving G3 DOX-PEG5kCA4-L-Rh4 was euthanized on day 9 due to body weight loss over 30%. All other animals in this treatment group had a weight loss of less than 15%. The higher toxicity of G3 formulation may be related to the faster drug release profile.
The total DOX concentrations in the plasma of mice were determined by fluorescent measurements after intravenous administration of free DOX and DOX nanoformulations (Figure 5A). Free DOX was eliminated rapidly from circulation. Consistent with the previous report,25 Doxil had an extremely long circulation time with about 85% and 35% of drug detected at 5 min and 24 h post-injection, respectively. Similar to free DOX, only 8% of DOX-PEG5kCA8 was detected at 5 min, indicating its poor in vivo stability for drug encapsulation. In contrast, Rh-containing G2 and G3 telodendrimer nanocarriers showed remarkably prolonged blood circulation, with more than 63% and 54% of the injected dosage retained in blood after 5 minutes, and 7.3% of G2 and 7.9% of G3 were detected at 24 hours post-injection. A pharmacokinetics (PK) analysis using a three-compartment model was performed for both free drug and nanoformulations (Supplementary Table 3). Compared with free DOX administration, Doxil has a about 37-fold slower elimination constant (K01) and 36-fold increase of area under curve (AUC) of the plasma drug concentration. The stealth PEG layer and the stable crystallized DOX in Doxil enabled the extremely prolonged drug circulation. DOX-PEG5kCA8 was observed to have a similar PK profile with free drug, which correlates with its low drug binding affinity and poor in vitro stability. In contrast, Rh-containing G2 and G3 nanoformulations of DOX showed significantly improved PK profiles with greater than a 6-fold decrease in K01 and a 6-fold increase of plasma AUC than free DOX and DOX-PEG5kCA8. Similar to the reported studies, 8 Doxil had a terminal half-life (t1/2) of 32.8 h, the longest one among all the formulations. In comparison, G2 and G3 DOX-nanoformulations consistently prolonged t1/2 to about 11 hours, which was increased 72-fold when compared to that of free DOX. Consistently, the steady-state volume of distribution (Vss) of Doxil is very small, 0.076 L/kg, which approximates the blood volume of animal, indicating the dominant blood pool residency. Similarly, both G2 and G3 Rh-containing formulations have a small Vss of 0.12 L/kg. However, free DOX and DOX-PEG5kCA8 have about more than a 10-fold increase in Vss, 1.35 L/kg and 1.26 L/kg, respectively, indicative of rapid distribution/dispersion of free drug and unstable nanoformulations into the tissues in mice.
Figure 5. In vivo performance of nanoformulations.
(A) PK profiles of DOX concentration in plasma and (B) drug uptake in heart analyzed at different time points after tail vein injection of free DOX, Doxil and DOX-loaded micelles at a dose of 10 mg/kg in BALB/C mice (5–6 weeks, both sexes). (C) In vivo and ex vivo near infrared fluorescence (NIRF) optical images of Raji lymphoma bearing nude mice (~10 weeks, female) injected intravenously with free DiD dye and DiD-DOX-co-loaded G2 PEG5kCA4Rh4 micelles, respectively. Tumors and major organs were excised for ex vivo imaging at 48 h post-injection. (D) In vivo tumor growth inhibition, (E) Kaplan-Meier survival curve and (F) body weight changes of Raji lymphoma bearing nude mice (9–11 weeks, both sexes) treated with different DOX formulations at 10 mg/kg for three doses on day 0, 4, 8 (n = 5–8) on nude mice bearing Raji lymphoma tumors with the initial volume of 100–150 mm3: (G) The serum CK and LDH levels were analyzed 7 days after the last treatment. Data were presented as mean ± SEM. Student’s t-test: * p <0.05; ** P<0.01; *** P<0.001.
About a 50% reduction in heart uptake of drug for Doxil and G2-Rh, G3-Rh DOX-nanoformulations were observed relative to free DOX and DOX-PEG5kCA8 at 4 h post administrations. Despite the prolonged blood circulations, there was no significant difference in heart uptake at 24 h post-injection for all formulations. The overall reduced AUC in the heart and the reduced peak values of free drug in circulation via Doxil, G2-Rh and G3-Rh DOX-nanoformulations correlated with the reduced cardiotoxicity (Figure 5B). The near infrared (NIR) in vivo and ex vivo imaging showed that G2-Rh DOX-nanoformulation could deliver fluorescent payload to tumor site preferentially as shown in Figure 5C. Three-fold greater tumor uptake and four-fold less uptake in liver, lung and spleen were observed in the animals treated with G2-Rh DOX-nanoformulation in comparison to the free payload administration (Supplementary Figure 8).
Enhanced anticancer effects
Raji lymphoma xenograft models were treated with DOX nanoformulations, free DOX, and Doxil intravenously on days 0, 4, and 8 (n = 5–8) at the equivalent DOX dose of 10 mg/kg, which is the MTD level for free DOX and Doxil. Initial tumor volumes were 150~200 mm3. As shown in Figure 5D, DOX-G2-Rh was the most efficient one in inhibiting tumor growth (P < 0.05) with the relative tumor volume of 2.1 on day 20. The DOX-G2-Rh treatment group also had a significantly prolonged median survival time (41 days) compared to groups treated with PBS (20 days), DOX (32 days) and Doxil (36.5 days) (Figure 5E). The body weights of animals were monitored with a maximum loss of <15% seen in all groups, which were recovered by the week four (Figure 5F). Similar red blood cell and platelet counts were observed for all the groups on day 7 after the last administration. There was slightly reduced white blood cell (WBC) numbers observed in the animals treated with free DOX (Supplementary Table 4), indicating myelosupression. No liver and kidney damage was observed as indicated by stable levels of ALT, AST, and BUN in all groups (Supplementary Table 5). Of note, the biomarkers for the cardiotoxicity, e.g., serum creatine kinase (CK) and lactate dehydrogenase (LDH) were elevated significantly in animals treated with free DOX (Figure 5G). G2-Rh DOX-nanoformulation and Doxil did not show significant CK and LDH elevation when compared to free DOX, indicating reduced cardiotoxicity.
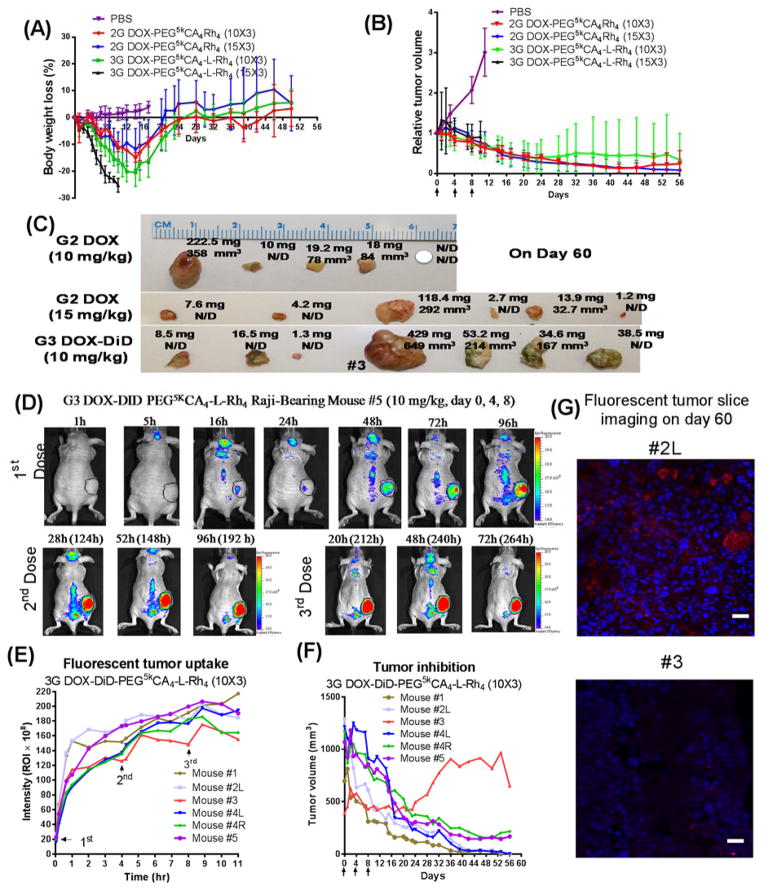
Given the reduced toxic side effects and the increased MTD for G2-Rh and G3-Rh DOX-nanoformulations, the anticancer effects could be further enhanced with higher tolerated dosages. Advanced tumors with large tumor volumes pose challenges for disease control. A separate in vivo cancer treatment study was designed to apply a higher dose (15 mg/kg) of telodendrimer nanoformulations to treat animals with an average initial lymphoma tumor volume of 700 mm3. Two dose levels of G2-Rh and G3-Rh DOX-nanoformulations, 10 and 15 mg/kg, were used to treat lymphoma cancers in nude mice bearing xenografted tumors for efficacy and toxicity studies. As shown in Figure 6A, the mice tolerated three-dosage treatments of G2-Rh DOX-nanoformulations at both 10 mg/kg and 15 mg/kg with maximum body weight loss of less than 15%. All mice in the group treated with G3-Rh DOX-nanoformulation died at 15 mg/kg after three treatments. Mice in DOX-G3-10 mg/kg group survived from three dosages with maximum body weight loss less than 20% at day 14, which gradually recovered over the next ten days. When compared to the PBS control group, none of the groups showed a decrease in the number of WBCs, red blood cells (RBCs), platelets or hemoglobin (Supplementary Table 4). In the animals treated with G3-Rh DOX-nanoformulation at 10 mg/kg, a slight increase of WBCs was observed (11.6±3.0 K/ul, normal range: 1.8–10.7).
Figure 6. Enhanced anticancer effects.
In vivo cancer treatment and theranostic tumor imaging in nude mice (9–11 weeks, both sexes) bearing Raji lymphoma tumors with the initial volume of 600–800 mm3: (A) Tumor inhibition curves and (B) body weight changes of mice after intravenous treatment with G2 and G3 nanoformulations of DOX at 10 and 15 mg/kg, respectively, for three doses on day 0, 4, 8 (n = 5–7). PBS injection was used in the control group. (C) In vivo volume and ex vivo weight of tumors in the G2 and G3 treatment groups. (D) In vivo NIRF optical images of Raji lymphoma bearing mice #5 treated with the intravenous injection of DiD-DOX-coloaded G3 PEG5kCA4-L-Rh4 micelles (10 mg/kg) for three times on days 0, 4, 8. (E) Fluorescent quantitative tumor uptake analysis and (F) tumor volume measurements of individual mice in the G3 DiD-DOX treatment group. (G) Lymphoma tumor tissue #2L and #3 stained with DAPI and observed under fluorescent confocal microscope: Red fluorescent is the signal from DiD and nuclei are in blue. Scale bar: 20 μm.
After four weeks, the majority of tumors shrunk dramatically in size to approximately 30% of the initial tumor volume in all three continuous treatment groups, e.g., DOX-G2-Rh at 10 and 15 mg/kg and DOX-G3-Rh at 10 mg/kg. The initial decrease was followed by the stable tumor size or further decreases to undetectable on day 60 (Figure 6B). In the control group treated with PBS, tumors progressed quickly with tumor volumes over 1500 mm3 on day 11 post-administration. The DOX-G2-Rh formulation decreased tumor size more efficiently at the elevated dose of 15 mg/kg. In this treatment group, four of the six tumors were not detectable by the end of treatment (Figure 6C). Even 10 mg/kg of DOX-G2-Rh nanoformulation exhibited a superior efficacy in shrinking tumor volume until day 46, whereas one tumor recurred. The DOX-G3-Rh nanoformulation also exhibited antitumor effects in the first three weeks of the treatment with significant decrease in tumor volume (Figure 6C). The enhanced anticancer effects of nanoformulations in large tumors might be explained by the advanced angiogenesis increasing EPR effects.
To monitor tumor-targeted drug delivery, a NIR dye, DiD, was co-loaded with DOX in the G3-Rh telodendrimer micelles for the treatment group at 10 mg/kg. Continuous accumulation of fluorescent signals was observed on the xenografted tumors (Figure 6D & Supplementary Figure 9). Interestingly, a unique zigzag profile of a tumor uptake in animal #3 was observed during the treatment (Figure 6E). A fast efflux indicated pronounced washout effects within this tumor, which correlated with less effective treatment in animal #3 (Figure 6F). A lower fluorescent signal was observed for tumor #3 on pathological studies when compared to tumor #2L (Figure 6G). This demonstrated the theranostic value of the nanoformulations in combination of NIR fluorescent imaging. The replacement of NIR probe with radio isotopes will allow for the theranostic imaging in human patients via positron emission tomography (PET) or single-photon emission computed tomography (SPECT) techniques.
Discussion
Rational nanocarrier design and structural optimization are great challenges to nanocarrier-based drug delivery. It mostly relies on the empirical approaches21, such as compatibility/solubility analysis to predict drug loading and release properties,51, 52, 53 which omit the detailed molecular interactions in drug loading. MD and Monte Carlo (MC) simulations have been applied to simulate drug-excipient interactions54, 55 and nanoparticle formation.56, 57, 58 A computational approach to dock drug molecules into a polymer matrix provides molecular level information on drug-matrix interaction in the random polymer aggregates57, 59 or dendrimer systems.60, 61 In fact, there is no such virtually empty pocket pre-existing in polymer aggregates for the drug molecule to diffuse in. Instead, drug loading mostly is a dynamic process of the instant co-assembly of drug-polymer complex into the core of micelle, which is too expensive for atomic molecular simulation. Overall, the capability and liability of the computational approaches in simulating polymer-drug aggregates is a bottleneck for structure-based nanocarrier design. Experimentally, combinatorial chemistry has been applied in material discovery to search for positively-charged polymers62, 63 and lipidoids64 for gene delivery. However, typical amphiphilic polymers have a very limited freedom and functionality for further optimization due to the monotonic nature of polymer chemistry. It is still not practical to use polymerization approaches to synthesize a large number of well-defined polymers with diverse structures for drug-loading test or for the validation of the theoretical design. In summary, significant technical difficulties lie in both theoretical nanocarrier design and precise high-throughput nanocarrier synthesis. In addition, the synergistic combination of these two approaches in nanocarrier development is still lacking.21
The majority mass (~75%) of the core of telodendrimer micelle is composed of DBMs on the periphery of the dendron, which has sufficient flexibility to interact with drug molecules spatially. Therefore, drug-DBM interactions determine the drug binding affinities within nanocarrier. Such small molecular interactions between drug-DBM can be efficiently calculated and ranked through computational approaches, thus bypassing the bottleneck of computational chemistry in studying polymer aggregates. Stepwise synthetic route via peptide chemistry affords telodendrimers with well-defined and easily diversifiable chemical structures and architectures. All of these features enable the telodendrimer system to serve as a blueprint for both computational design and combinatorial synthesis of nanocarriers for systematical evaluation.
The stability of a nanoformulation determines its fate and in vivo efficacy. The polymeric micelle system is a dynamic self-assembled system in nature, which dissociates upon dilution and interactions with hydrophobic components in vivo. As results, some micelle formulations in clinic development have the similar PK profiles with free drug,65, 66 which reduces the EPR effects for tumor-targeted drug delivery. Reversible crosslinking strategies have been applied to improve the stability of nanocarriers, e.g., telodendrimer micelles27, 67 and other delivering systems68 for on-demand drug delivery. However, most of the crosslinking strategies are facing significant hurdles for clinical translation, such as the crosslinking efficiency, toxicity of crosslinking treatments/reagents and tedious purification process.68 In reality, the simple system is preferred in terms of reproducibility, quality control, and regulations for clinical development. As shown in this study, the stability and drug sustainability of the non-crosslinked micelles could be enhanced by engineering a nanocarrier with specific drug-binding molecules. In concept, large numbers of biocompatible molecules could be virtually screened via molecular docking to identify DBMs, which can be verified by MD simulation. MD simulation of the drug-telodendrimer interactions at atomic level further provides hints for drug loading properties. The resulting telodendrimer library provides numerous candidates to optimize the drug loading capacities and stabilities for in vitro and in vivo evaluations.
Targeted drug delivery by a nanoparticle formulation is based on both elongated circulation time of nanoparticle and leaky blood vessels present in solid tumors. Doxil has a superior half-life about 20–35 hours in animals and 50–80 hours in human adult8 with the majority of DOX still encapsulated when detected in plasma, due to the limited drug release properties of the salt form of DOX-HCl within liposomes.8 Doxil is able to efficiently target tumors via EPR effects and can efficiently reduce the cardiotoxicity of DOX. In clinical practice, Doxil only produced a marginally improved therapeutic benefit over free DOX.69 Due to the large particle size of Doxil and very slow drug release, the intratumoral penetration is limited and the drug availability within tumor is reduced. Even reduced anticancer effects for cisplatin were reported after encapsulation into the stealthy lipsomes.70 The combination of the stable drug encapsulation in blood stream and efficient drug release at tumor sites is believed to be critical in determining the in vivo efficacy of nanotherapeutics. The smaller sizes of polymer micelles represent intrinsic merit for intratumoral drug delivery. Both G2 and G3 DOX nanoformulations exhibited a better anticancer effect than free DOX, Doxil and the less stable telodendrimer formulations.25 This is due to the enhanced stability and prolonged blood circulation for passive tumor targeting and the smaller sizes with the steady drug release for efficient intratumoral drug delivery and cancer killing.
In summary, we have demonstrated for the first time the application of molecular docking/MD simulation techniques in screening building blocks to synthesize nanocarriers for the delivery of a given drug. The computational predictions have been validated by combinatorial synthesis and systematic evaluation of a telodendrimer nanocarrier library. Rh-containing telodendrimers were identified to have the most favorable DOX-binding affinity, as well as biocompatibility and bioactivity of a building block. Novel two-layered and three-layered telodendrimers have been synthesized using amphiphilic CA as co-core forming building blocks with other DBMs, which efficiently stabilize nanocarriers into small particle sizes and prevent further aggregation. The Rh-containing G2 and G3 telodendrimer nanocarriers exhibited superior DOX loading capacity, stability and reduced toxic side effects when compared to the typical CA-only telodendrimers. The G2 telodendrimer could sustain DOX better with a slow release profile than the G3 telodendrimer nanocarrier. The enhanced stability of the G2 and G3 nanoformulations leads to the prolonged circulation, increased MTD, reduced cardiotoxicity, and enhanced tumor targeted drug delivery. The balanced stability and drug release profiles of G2 and G3 nanoformulations yielded the significantly improved anticancer effects when compared to both free DOX and Doxil formulations.
Methods
Materials and nomenclature
Doxorubicin hydrochloride (DOX·HCl) (Novaplus) and Doxil® (Ben Venue Laboratories, Inc., Bedford, OH) were obtained from the Regional Oncology Center Pharmacy, SUNY Upstate Medical University. Monomethyl-terminated poly(ethylene glycol) monoamine (MeO-PEG-NH2, Mw 5 kDa) were purchased from JenKem Technology, USA Inc. (Fmoc)lys(Boc)-OH, (Fmoc)Lys(Dde)-OH, (Fmoc)Lys(Fmoc)-OH were obtained from AnaSpec Inc. (San Jose, CA). Tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, MTS] and phenazine methosulfate (PMS) were purchased from Promega (Madison, Wisconsin). Rhein, cholic acid, triethylamine (TEA), and all other chemicals were purchased from Sigma-Aldrich (St. Louis). The preparations of cholic acid derivatives (Cholic acid NHS ester) were described in the previous publication.23 A bisamino-triethylene glycol mono-succinimide linker molecule was synthesized following a reported procedure.35
The nomenclature of the telodendrimers followed the system used in the previous publications.23 For example, telodendrimer PEG5kCA8 indicates that the molecular weight of PEG is 5 kDa and there are eight cholic acids (CAs) conjugated the periphery of polylysine; PEG5kCA4Rh4 indicates that four CAs are conjugated on the α-amino of polylysine and four rheins (Rhs) are conjugated in the ε-amino groups of polylysine; PEG5kCA4-L-Rh4 indicates that PEG is 5 kDa and four CAs are conjugated in the intermediate layer of telodendrimer and four Rhs are conjugated in the interior layer of telodendrimer and there is a triethylene glycol linker molecule between two layers.
Molecular mechanics
The 3D structure of doxorubicin and the 3D SDF files of small molecules of interest were collected from PubChem compound website and input in Meastro software package from Schodinger.36 The carboxylic acids in these molecules were pre-masked with N-ethyl amide structure to mimic the structures after conjugation in the telodendrimer. By default, MacroModel (Schrodinger) energy minimization are performed using MMFF94 force field37 in the present of implicit water with the normal cutoff distances in place for unbounded interactions, for example 7 Å for van der Waals interactions and 12 Å for electrostatics and 4 Å for H-bond were applied. The minima was searched via PRCG with the maximum iterations of 2500 and the gradient converge threshold was set as 0.05.
Molecular docking
AutoDock 4 program was used to measure docking energies between DOX and selected small molecules. The minima conformations of DOX obtained from Macromodel were used as a receptor in docking studies. The cube size has been optimized to be 4 nm in side length. AutoDock Tools (ADT) was used to add polar hydrogen and Kollman charge to receptor molecules (doxorubicin). The number of points in x-, y-, and z-dimension was 60 with the spacing of 0.375 angstrom of each grid points. Lamarckian Genetic Algorithm was used. The population size was set as 150 individuals; the maximum number of evaluations was 2.5 million; the maximum number of generations was 27,000. The rate of gene mutation was set as 0.02 and the rate of crossover was 0.8. The other docking parameter options such as random number generator, energy parameters, step size parameters and output format parameters were set as default values. Over 50 small molecular candidates were docked with the doxorubicin for 100 runs for each docking. The binding energies with the unit of kcal/mol were provided in the docking result. The conformations with the same binding energy are grouped in the same cluster.
Molecular dynamic simulation
A series of docking complexes with high to low docking energies were further examined by MD simulation with the explicit waters using Desmond software (Desmond/Maestro academic version 2014.2)38, 39 and analyzed using Maestro’s trajectory visualizer. DBM-DOX complex with the lowest docking energy was input as the initial conformation for MD simulation using OPLS-2005 force field. The water box type was orthorhombic with the boundary distance of 10 Å and the water molecules were modeled with TIP3P model,40 which was demonstrated to be a propitiate model for studying small molecular interactions.30 No ion was added into the system. The solvated systems were relaxed with the default multi-stage protocol in Desmond, followed by a series of short NVT and NPT Berendsen41 simulations at T = 10 K with varying constraints on solute and solvent atoms. MD production runs were carried out in the NPT ensemble for 10 ns for each DBM-DOX and for 5 ns for individual DBM. The temperature was regulated to 300 K with the Nose–Hoover chain thermostat42, 43 with a relaxation time of 1.0 ps. Pressure was regulated to 1 bar with the Martyna–Tobias–Klein barostat using isotropic coupling and a relaxation time of 2.0 ps.44 The RESPA integrator45 was used to integrate equations of motions with a 2.0 fs time step for bonded and near interactions and a 6.0 fs time step for far interactions. A cutoff of 9 Å was applied to non-bonded interactions. The smooth particle mesh Ewald method46 was used to treat long-range electrostatics with a tolerance of 10−9. The OPLS_2005 force field47, 48 was applied to the system.
The interaction energies between DOX-DBMs were analyzed via the “Simulation event analysis” function in Desmond/Maestro and the mean and SD/SEM of interaction energy was calculated. In addition, the average and standard deviation (Mean, SD) of total potential energies (U) of the DOX-DBM binding system in water, e.g., U(DBM-DOX + (m+n)H2O) was calculated via “simulation quality analysis” function in Desmond/Maestro using the 10 ps block length for averaging. Standard Error of Mean (SEM) was calculated based on the formula of SEM = SD/(square root of sample size). MD simulations of individual DBMs in the same condition were performed and the corresponding U(DBM + nH2O) was analyzed. A series of MD runs of DOX were performed with the increasing boundary distance from 10 to 11, 12, 13 and 15 Å to increase water molecules included in the system. It is hypothesized that the additional water molecules with the increased box boundary could be treated as bulky water because it is beyond the 9 Å cut off for non-bounded interactions. The potential energies of the DOX-water systems were calculated and plotted against numbers of water molecules included and the standard curve was obtained with the R2 of 1 (Supplementary Figure 3B). This working plot was used to calculate the potential energy U(DOX + mH2O) of a DOX-mH2O system that is required to balance water molecules (Supplementary Figure 3A) in the enthalpy calculation: ΔH = U(DBM-DOX + (m+n)H2O) − U(DBM + nH2O) −U(DOX + mH2O).49
Telodendrimer synthesis
General procedure: The telodendrimers were synthesized via solution-phase peptide amide bond condensation reactions starting from MeO-PEG-NH2. N-terminal protected lysine was used to synthesize the branched scaffold of telodendrimer using diisopropyl carbodimide (DIC, 3 equiv) and N-hydroxybenzotriazole (HOBt, 3 equiv) as coupling reagents in DMF. Upon the negative Kaiser test of the reaction, the ice-chilled ether was added to the reaction solution to precipitate polymer, which was further washed twice with chilled ether. Fmoc protecting groups were removed by the treatment with 20% piperidine in DMF for 30 min. Polymer was precipitated and washed with chilled ether. G1 homo telodendrimer PEG5k-DBM8 synthesis: A dendritic polylysine was synthesized via three repeated (Fmoc)Lys(Fmoc)-OH coupling as mentioned above. At the end, the polylysine was capped with NHS ester of DBM or DBM with free acid groups using HOBT/DIC as coupling reagents. G2 Hybrid telodendrimer PEG5k-CA4DBM4 synthesis: A dendritic polylysine with orthogonal protected α-(N-Fmoc) and ε-(N-Boc) amino groups was synthesized on MEO-PEG-NH2 via two repeated (Fmoc)Lys(Fmoc)-OH coupling followed by a (Fmoc)Lys(Boc)-OH coupling via HOBt/DIC chemistry. Then Fmoc group was removed by the treatment of 20% piperidine in DMF, followed by the coupling of cholic acid-OSu onto the α-position of lysine. Then Boc protecting groups were removed by the treatment with 50% TFA in DCM for 30 min. Then DBM-NHS ester reacted with the ε position of lysine to generate hybrid telodendrimers. G3 three-layered telodendrimer PEG5k-CA4-L-DBM4 synthesis: A two layered dendritic polylysine with Fmoc and Boc protections, respectively, was prepared on MEO-PEG-NH2 using orthogonal protected lysine, (Fmoc)Lys(Boc)-OH, via HOBt/DIC coupling chemistry. After removal of Fmoc groups, DBMs with free acid groups were coupled using HOBT/DIC as coupling reagents to the terminal end of interior dendritic polylysine. Then, cholic acid NHS ester was finally coupled to the adjacent amino groups of branched polylysine after removal of Boc group. Telodendrimer were precipitated and washed three times with cold ether, dialyzed for purification as mentioned above.
Drug loading and characterizations
DOX was encapsulated into telodendrimer micelles by a thin-film and hydration method. DOX·HCl was stirred with three molar equivalent of triethylamine in chloroform (CHCl3)/methanol (MeOH) (10:1, v/v) for 30 min to neutralize HCl. The telodendrimer was added into the above solution at certain polymer-drug ratio. Solvents were evaporated to dryness and a thin film of homogeneous drug-telodendrimer mixture was casted on the flask wall, which was further dried under high vacuum for 30 min. Then polymer film was reconstituted in 1 mL phosphate buffered solution (PBS), followed by a 30 min sonication. The particle size distributions of drug loaded micelles were characterized by dynamic light scattering (DLS, Microtrac) and transmission electron microscopy (TEM, a JEOL JEM-2100 HR instrument). The stability of DOX-loaded micelles upon storage was evaluated by monitoring the particle sizes of micelles using DLS.
Cell culture and animals
T-cell lymphoma cell lines (Jurkat and MOLT-4) and B-cell lymphoma cell lines (Raji and Ramos) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). All these cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin G, and 100 μg/mL streptomycin at 37 ºC using a humidified 5% CO2 incubator. Specific Pathogen Free (SPF) BALB/c mice, both sexes, 5–6 weeks age, were purchased from Charles River (Hollister, CA); athymic nude mice (Nu/Nu strain), 6–8 weeks age of both sexes, were purchased from Harlan (Livermore, CA). All animals were kept under pathogen-free conditions according to AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) guidelines and were allowed to acclimatize for at least 4 days prior to any experiments. All animal experiments were performed in compliance with institutional guidelines and according to protocol approved by the Committee for the Humane Use of Animals of State University of New York Upstate Medical University.
MTS assay
MTS assay was used to evaluate the effects of DOX-loaded micellar NPs on the cell viability against both T and B lymphoma cell lines. MOLT-4, Jurkat, Raji and Ramos cells were seeded in 96-well plate at the cell densities of 8 × 103 cells/well, respectively. After overnight incubation, the cells were treated with different concentrations of DOX·HCl, Doxil®, and DOX-loaded micelles, as well as the equivalent dose of blank micelles. After 72 h incubation, CellTiter 96® Aqueous Cell Proliferation Reagent, which is composed of MTS and an electron coupling reagent PMS, was added to each well according to the manufacturer’s instructions. The cell viability was determined by measuring the absorbance at 490 nm using a microplate reader (BioTek Synergy 2). Untreated cells served as a control. Results were shown as the average cell viability of triplicate wells via a formula of [(ODtreat − ODblank)/(ODcontrol − ODblank)×100%].
TB staining
Cytotoxicity can also be measured using trypan blue (TB) staining. Raji lymphoma cells were seeded in 96-well plate at the cell densities of 8 × 103 cells/well. And the cells were treated with different concentrations of DOX·HCl, Doxil®, and DOX-loaded micelles, as well as the untreated cells as controls. After 72 h incubation, mix the cell suspension with 0.4% trypan blue solution at a 1:1 v/v ratio. After 1–2 min incubation, carefully and continuously fill the hemocytometer chamber and count cells under the microscope in four 1 × 1 mm squares of one chamber and determine the average number of live (viable, unstained) and dead (blue) cells per square. The average cell viability of triplicate wells via a formula of No. of viable cells counted/total tells counted (viable and dead) × 100 = % viable cells.
Cell lysis and drug extraction
It may not be accurate to quantify cellular uptake of Doxil® by flow cytometry directly, because the fluorescence of DOX is quenched within the intact Doxil® lipsome even after cell uptake. So, we employ a cell lysis and drug extraction method to analyze quantitative cellular uptake of free DOX, Doxil® and DOX nano-formulations. Briefly, 1×105 Raji cells were incubated with free DOX, Doxil® and DOX-loaded micelles at different DOX concentrations (1, 3 and 9 μM) for 30 min or 2 h at 4°C or 37 °C, respectively. The cells were washed with PBS three times. Then, 100 μL of extraction buffer (10% Triton X-100, deionized water, and acidified isopropanol (0.75 N HCl) at a 1:2:15 volumetric ratio) were added to cells, and DOX was extracted overnight at −20 °C. The fluorescence of the supernatant was determined at excitation/emission of 470/590 nm using microplate reader (BioTek Synergy 2).
Hemolysis assays
One mL of fresh blood was collected from a BALB/c mouse by cardiac puncture under deep anesthesia into 10 mL of PBS solution in the presence of 20 mM EDTA. Red blood cells (RBCs) were then separated by centrifugation at 1000 g for 10 min. The RBCs were washed three times with 10 mL of PBS, and re-suspended in 20 mL PBS. 200 μL of diluted RBC suspension was incubated with polymers at a series of concentrations (100, 500 and 1000 μg/mL) with gentle shake at 37 °C for 0.5 hrs, 4 hrs and overnight, respectively. Free DOX was also tested at the same DOX concentration levels with the DOX-loaded nanoformulations, e.g. 10, 50 and 100 μg DOX/mL. The cell suspension was centrifuged at 1,000 g for 5 min. The free of hemoglobin in the supernatant was determined by measuring the absorbance at 540 nm using a UV-vis spectrometer. Incubations of RBCs with Triton-100 (2%) and PBS were used as the positive and negative controls, respectively. The percent hemolysis of RBCs was calculated using the following formula: RBC Hemolysis = 100% × (ODsample − ODPBS)/(ODtriton – ODPBS).
Maximum tolerated dose (MTD) studies
ealthy specific pathogen free BALB/c mice (5–6 weeks, both sexes) were administered intravenously with DOX·HCl, DOX-PEG5kCA4Rh4, DOX-PEG5kCA4-L-Rh4 (5 mg/ml DOX in 20 mg/ml telodendrimer) at the dose of 5, 10, 15 and 20 mg DOX/kg body weight, respectively (n = 4–5). Mice survival and body weight change were monitored daily for two weeks. At one week after the last injection, the blood was collected from each mouse to measure blood cell counts. The MTD was defined as the allowance of a median body weight loss of 15% and causes neither death due to toxic effects nor remarkable changes in the general signs within two weeks after administration.
Pharmacokinetic and biodistribution analysis
Healthy specific pathogen free BALB/c mice (5–6 weeks, both sexes) were administered intravenously with DOX·HCl, Doxil, DOX-PEG5kCA8, DOX-PEG5kCA4Rh4, DOX-PEG5kCA4-L-Rh4 (2 mg/ml DOX in 20 mg/ml telodendrimer) at the dose of 10 mg DOX/kg body weight, respectively (n = 3). At different time points, e.g., 5 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h, and 32 h post injection, 20 μL of blood samples were obtained by nicking the lateral tail vein using a sterile scalpel blade, respectively. Plasma was collected and diluted to 10 fold with DMSO for fluorescent measurements using a BioTek plate reader. After the last blood collection, mice were sacrificed and vital organs, e.g., heart and liver, lung, spleen and kidney were collected and homogenized and extract with extraction buffer (triton-100/water/acidified isopropanol with 0.05 N HCl 1:2.5:15 in volume) for fluorescence detection.
The pharmacokinetic parameters were calculated using a three-compartment model with PKsolver, an add-in program in Microsoft Excel.50 The following parameters were determined: area under the curve (AUC, 0 to infinity), maximum drug concentration (Cmax), total body clearance (Cl), terminal half-life of DOX (t1/2), steady-state volume of distribution (Vss), the apparent terminal elimination rate constant (Λ), and mean residence time (MRT).
Fluorescence animal imaging
Nude mice with subcutaneous lymphoma tumors of approximate 8 to 10 mm in diameter were subjected to in vivo near infrared dye (NIRF) optical imaging. Lymphoma xenograft mouse models were established by subcutaneously injecting 1×107 Raji lymphoma cells in a 100 μL of mixture of PBS and Matrigel (1:1 v/v) at the right flank in female nude mice (6–8 weeks). A hydrophobic NIRF dye (DiD) was encapsulated together with DOX into the micelles using the same method as described above. The DiD-DOX-co-loaded micelle formulation was filtered with a 0.22 μm filter to sterilize the sample. The Raji lymphoma tumor xenografts bearing nude mice were injected with free DiD and DiD-DOX-co-loaded micelles at 0.2:1:10 w/w/w ratio via the tail vein, respectively. Then the mice anesthetized and optically imaged by an IVIS 200 (PerkinElmer) with the excitation at 625 nm and the emission at 700 nm, at different time points (0.5 h, 1 h, 2 h, 4 h, 8 h, 24 h, 48 h and 72 h). At the end of the experiment, the animals were sacrificed and all the major organs and tumor were excised for ex vivo imaging. The associated fluorescence intensities were determined by Living Image software (Caliper Life Sciences) using operator-defined regions of interest (ROI) measurements.
Confocal fluorescence microscopic imaging
For the histological evaluation, the harvested tumors were frozen in O.C.T. (cryo-embedding medium) at 80 ºC. The corresponding slices (10 μm) were then prepared on a Minotomecryostat, air-dried for 30 min and fixed with 4% paraformaldehyde for 10 min. The nuclei were stained with DAPI, and the slides were mounted with cover slips and imaged with Nikon laser scanning confocal fluorescence microscopy.
In vivo anti-tumor efficacy studies
Subcutaneous Raji lymphoma xenograft mouse models were used to evaluate the therapeutic efficacy of different formulations of DOX. Lymphoma xenograft mouse models were established by subcutaneously injecting 1×107 Raji lymphoma cells in a 100 μL of mixture of PBS and Matrigel® (1:1 v/v) at the right flank in nude mice (6–8 weeks of both sexes). When tumor volume reached 150–300 mm3, mice were intravenously administrated with PBS, DOX·HCl, Doxil®, DOX- PEG5k-CA4 Rh4 at the dose of 10 mg/kg DOX equivalent (MTD of free DOX), respectively (n = 5–8). An alternative efficacy study, average tumor volume reached to about 500 mm3 before treatment to evaluate the effects of the angiogenesis. The treatments were given every four day on days 0, 4 and 8 for total three doses. Tumor sizes were measured with a digital caliper twice per week. Tumor volume was calculated by the formula (L×W2)/2, where L is the longest, and W is the shortest in tumor diameters (mm). To compare between groups, relative tumor volume (RTV) was calculated at each measurement time point (where RTV equals the tumor volume at given time point divided by the tumor volume prior to initial treatment). For humane reasons, animals were sacrificed when the implanted tumor volume reached 2000 mm3, which was considered as the end point of survival data. At day 7 after the last dosage, blood samples were obtained from all the mice for the measurement of blood cell counts, hepatic or renal function tests (ALT, AST, and BUN), and serum enzyme markers of cardiotoxicity including creatine kinase (CK) and lactate dehydrogenase (LDH). One mouse from each group was also sacrificed, and its heart was submitted for histopathology evaluation.
Statistical analysis
Data are presented as means ± standard error (SEM). Linear regression model was fit via ordinary least square in the correlation studies. The Pearson correlation coefficient and the associated p values were calculated to examine the correlation between molecular bindings obtained from molecular docking and molecular simulation. Cell viability curves were fit by a three-parameter model including top, bottom and IC50. The difference (p values) of IC50 between groups was tested by Wald z-test using the standard error from the curve fitting. The level of significance in all statistical analyses was set at a probability of P < 0.05. Statistical analysis was performed by Student’s t-test for comparison of two groups with specific variations, and one-way analysis of variance (ANOVA) for multiple groups, followed by Newman-Keuls test if overall P < 0.05.
Supplementary Material
Acknowledgments
We greatly appreciate Prof. Stephan Wilkinson for the help in TEM analysis. The authors thank Prof. Golam Mohi for the help in blood sample analysis. We appreciate Prof. Dongliang Wang for the helpful input for the statistic analysis. We thank Dr. Xiaobing Han for the helpful discussion on the computational studies. We thank Dr. Alexa Bodman for the proof reading of this manuscript. The financial supports from NIH/NCI R01CA140449 (Luo) and the institutional startup funds are greatly acknowledged.
Footnotes
Author contributions
C.S. performed in vitro and in vivo bioassays in characterized nanotherapeutics and wrote part of manuscript. G.D. performed the molecular docking experiments, synthesized, and characterized the telodendrimer nanocarrier library. K.X. contributed to the initial biology studies. X.W and L.W. facilitated the synthesis and characterization of nanocarriers. J.L. designed the study, initiated the project by conducting computational studies, initial telodendrimer synthesis and characterizations of nanocarriers, supervised the project and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZG. Small-molecule delivery by nanoparticles for anticancer therapy. Trends Mol Med. 2010;16:594–602. doi: 10.1016/j.molmed.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobbs SK, et al. Regulation of transport pathways in tumor vessels: role of tumor type and microenvironment. Proc Natl Acad Sci U S A. 1998;95:4607–4612. doi: 10.1073/pnas.95.8.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 5.Stern ST, et al. Translational considerations for cancer nanomedicine. J Control Release. 2010;146:164–174. doi: 10.1016/j.jconrel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Brien MER, et al. Reduced cardiotoxicity and comparable efficacy in a phase III trial of pegylated liposomal doxorubicin HCl (CAELYX™/Doxil®) versus conventional doxorubicin for first-line treatment of metastatic breast cancer. Ann Oncol. 2004;15:440–449. doi: 10.1093/annonc/mdh097. [DOI] [PubMed] [Google Scholar]
- 7.Unezaki S, et al. Direct measurement of the extravasation of polyethyleneglycol-coated liposomes into solid tumor tissue by in vivo fluorescence microscopy. InterJ Pharm. 1996;144:11–17. [Google Scholar]
- 8.Gabizon A, Shmeeda H, Barenholz Y. Pharmacokinetics of pegylated liposomal Doxorubicin: review of animal and human studies. Clin Pharmacokinet. 2003;42:419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 9.Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nature Reviews Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 10.Gillies ER, Frechet JMJ. Dendrimers and dendritic polymers in drug delivery. Drug Discovery Today. 2005;10:35–43. doi: 10.1016/S1359-6446(04)03276-3. [DOI] [PubMed] [Google Scholar]
- 11.Baker JR. Dendrimer-based nanoparticles for cancer therapy. ASH Education Program Book. 2009;2009:708–719. doi: 10.1182/asheducation-2009.1.708. [DOI] [PubMed] [Google Scholar]
- 12.Betancourt T, Brown B, Brannon-Peppas L. Doxorubicin-loaded PLGA nanoparticles by nanoprecipitation: preparation, characterization and in vitro evaluation. Nanomedicine (Lond) 2007;2:219–232. doi: 10.2217/17435889.2.2.219. [DOI] [PubMed] [Google Scholar]
- 13.Du J-Z, Du X-J, Mao C-Q, Wang J. Tailor-Made Dual pH-Sensitive Polymer–Doxorubicin Nanoparticles for Efficient Anticancer Drug Delivery. J Am Chem Soc. 2011;133:17560–17563. doi: 10.1021/ja207150n. [DOI] [PubMed] [Google Scholar]
- 14.Duncan R. Polymer conjugates as anticancer nanomedicines. Nat Rev Cancer. 2006;6:688–701. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Wang Y-C, Dou S, Xiong M-H, Sun T-M, Wang J. Doxorubicin-Tethered Responsive Gold Nanoparticles Facilitate Intracellular Drug Delivery for Overcoming Multidrug Resistance in Cancer Cells. ACS Nano. 2011;5:3679–3692. doi: 10.1021/nn200007z. [DOI] [PubMed] [Google Scholar]
- 16.Gaucher G, Dufresne MH, Sant VP, Kang N, Maysinger D, Leroux JC. Block copolymer micelles: preparation, characterization and application in drug delivery. J Control Release. 2005;109:169–188. doi: 10.1016/j.jconrel.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 17.Mikhail AS, Allen C. Poly(ethylene glycol)-b-poly(epsilon-caprolactone) micelles containing chemically conjugated and physically entrapped docetaxel: synthesis, characterization, and the influence of the drug on micelle morphology. Biomacromolecules. 2010;11:1273–1280. doi: 10.1021/bm100073s. [DOI] [PubMed] [Google Scholar]
- 18.Greenwald RB, Conover CD, Choe YH. Poly(ethylene glycol) conjugated drugs and prodrugs: a comprehensive review. Crit Rev Ther Drug Carrier Syst. 2000;17:101–161. [PubMed] [Google Scholar]
- 19.Veronese FM, et al. PEG-doxorubicin conjugates: influence of polymer structure on drug release, in vitro cytotoxicity, biodistribution, and antitumor activity. Bioconj Chem. 2005;16:775–784. doi: 10.1021/bc040241m. [DOI] [PubMed] [Google Scholar]
- 20.Zheng M, Liu X, Xu Y, Li H, Luo C, Jiang H. Computational methods for drug design and discovery: focus on China. Trends Pharmacol Sci. 2013;34:549–559. doi: 10.1016/j.tips.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh L, Neale C, Pomes R, Allen C. Computational approaches to the rational design of nanoemulsions, polymeric micelles, and dendrimers for drug delivery. Nanomedicine. 2012;8:20–36. doi: 10.1016/j.nano.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Xiao K, et al. A self-assembling nanoparticle for paclitaxel delivery in ovarian cancer. Biomaterials. 2009;30:6006–6016. doi: 10.1016/j.biomaterials.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo J, et al. Well-defined, size-tunable, multifunctional micelles for efficient paclitaxel delivery for cancer treatment. Bioconjug Chem. 2010;21:1216–1224. doi: 10.1021/bc1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao K, et al. The passive targeting of polymeric micelles in various types and sizes of tumor models. Nanoscience and Nanotechnology Letters. 2010;2:79–85. [Google Scholar]
- 25.Xiao K, Luo J, Li Y, Lee JS, Fung G, Lam KS. PEG-oligocholic acid telodendrimer micelles for the targeted delivery of doxorubicin to B-cell lymphoma. J Control Release. 2011;155:272–281. doi: 10.1016/j.jconrel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao K, et al. “OA02” Peptide Facilitates the Precise Targeting of Paclitaxel-Loaded Micellar Nanoparticles to Ovarian Cancer In Vivo. Cancer Res. 2012;72:2100–2010. doi: 10.1158/0008-5472.CAN-11-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. Well-Defined, Reversible Boronate Crosslinked Nanocarriers for Targeted Drug Delivery in Response to Acidic pH Values and cis-Diols. Angew Chem Int Ed Engl. 2012;51:2864–2869. doi: 10.1002/anie.201107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato J, et al. Disulfide cross-linked micelles for the targeted delivery of vincristine to B-cell lymphoma. Mol Pharm. 2012;9:1727–1735. doi: 10.1021/mp300128b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 30.Fenley AT, Henriksen NM, Muddana HS, Gilson MK. Bridging Calorimetry and Simulation through Precise Calculations of Cucurbituril–Guest Binding Enthalpies. J Chem Theo Comp. 2014;10:4069–4078. doi: 10.1021/ct5004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cong XD, Ding MJ, Dai DZ, Wu Y, Zhang Y, Dai Y. ER stress, p66shc, and p-Akt/Akt mediate adjuvant-induced inflammation, which is blunted by argirein, a supermolecule and rhein in rats. Inflammation. 2012;35:1031–1040. doi: 10.1007/s10753-011-9407-4. [DOI] [PubMed] [Google Scholar]
- 32.Huang Q, Lu G, Shen HM, Chung MC, Ong CN. Anti-cancer properties of anthraquinones from rhubarb. Med Res Rev. 2007;27:609–630. doi: 10.1002/med.20094. [DOI] [PubMed] [Google Scholar]
- 33.Jiang W, Luo J, Nangia S. Multiscale Approach to Investigate Self-Assembly of Telodendrimer Based Nanocarriers for Anticancer Drug Delivery. Langmuir. 2015 doi: 10.1021/la503949b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, et al. Novel SN38 conjugate-forming nanoparticles as anticancer prodrug: in vitro and in vivo studies. J Control Release. 2013;166:147–158. doi: 10.1016/j.jconrel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Song A, Wang X, Zhang J, Marik J, Lebrilla CB, Lam KS. Synthesis of hydrophilic and flexible linkers for peptide derivatization in solid phase. Bioorg Med Chem Lett. 2004;14:161–165. doi: 10.1016/j.bmcl.2003.09.067. [DOI] [PubMed] [Google Scholar]
- 36.Schrödinger Release 2014–1: Maestro v. Schrödinger, LLC; New York, NY: 2014. [Google Scholar]
- 37.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 38.Bowers KJ, et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. SC 2006 Conference, Proceedings of the ACM/IEEE; 2006. [Google Scholar]
- 39.Shan Y, Kim ET, Eastwood MP, Dror RO, Seeliger MA, Shaw DE. How Does a Drug Molecule Find Its Target Binding Site? J Am Chem Soc. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction Models for Water in Relation to Protein Hydration. In: Pullman B, editor. Intermolecular Forces. Springer Netherlands; 1981. [Google Scholar]
- 41.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 42.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys. 1984;81:511–519. [Google Scholar]
- 43.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985 doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 44.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 45.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 46.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 47.Tirado-Rives J, Jorgensen WL. Molecular dynamics of proteins with the OPLS potential functions. Simulation of the third domain of silver pheasant ovomucoid in water. J Am Chem Soc. 1990;112:2773–2781. [Google Scholar]
- 48.Banks JL, et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT) J Comput Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenley AT, Henriksen NM, Muddana HS, Gilson MK. Bridging Calorimetry and Simulation through Precise Calculations of Cucurbituril–Guest Binding Enthalpies. J Chem Theo Comput. 2014;10:4069–4078. doi: 10.1021/ct5004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Huo M, Zhou J, Xie S. PKSolver: An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput Meth Prog Biomed. 2010;99:306–314. doi: 10.1016/j.cmpb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Xiao Y, Allen C. Polymer-drug compatibility: a guide to the development of delivery systems for the anticancer agent, ellipticine. J Pharm Sci. 2004;93:132–143. doi: 10.1002/jps.10533. [DOI] [PubMed] [Google Scholar]
- 52.Huynh L, Grant J, Leroux JC, Delmas P, Allen C. Predicting the solubility of the anti-cancer agent docetaxel in small molecule excipients using computational methods. Pharm Res. 2008;25:147–157. doi: 10.1007/s11095-007-9412-3. [DOI] [PubMed] [Google Scholar]
- 53.Faller B, Ertl P. Computational approaches to determine drug solubility. Adv Drug Deliv Rev. 2007;59:533–545. doi: 10.1016/j.addr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Pajula K, Lehto VP, Ketolainen J, Korhonen O. Computational approach for fast screening of small molecular candidates to inhibit crystallization in amorphous drugs. Mol Pharm. 2012;9:2844–2855. doi: 10.1021/mp300135h. [DOI] [PubMed] [Google Scholar]
- 55.Li C, Wang JX, Le Y, Chen JF. Studies of bicalutamide-excipients interaction by combination of molecular docking and molecular dynamics simulation. Mol Pharm. 2013;10:2362–2369. doi: 10.1021/mp300727d. [DOI] [PubMed] [Google Scholar]
- 56.Cui Y. Using molecular simulations to probe pharmaceutical materials. J Pharm Sci. 2011;100:2000–2019. doi: 10.1002/jps.22392. [DOI] [PubMed] [Google Scholar]
- 57.Costache AD, Sheihet L, Zaveri K, Knight DD, Kohn J. Polymer-drug interactions in tyrosine-derived triblock copolymer nanospheres: a computational modeling approach. Mol Pharm. 2009;6:1620–1627. doi: 10.1021/mp900114w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivas G, Discher DE, Klein ML. Self-assembly and properties of diblock copolymers by coarse-grain molecular dynamics. Nat Mater. 2004;3:638–644. doi: 10.1038/nmat1185. [DOI] [PubMed] [Google Scholar]
- 59.Kuramochi H, Andoh Y, Yoshii N, Okazaki S. All-atom molecular dynamics study of a spherical micelle composed of N-acetylated poly(ethylene glycol)-poly(gamma-benzyl L-glutamate) block copolymers: a potential carrier of drug delivery systems for cancer. J Phys Chem B. 2009;113:15181–15188. doi: 10.1021/jp906155z. [DOI] [PubMed] [Google Scholar]
- 60.Tahir Ã, Guofeng W, Ryan M, Nicholas B, William AG., III Molecular modelling of dendrimers for nanoscale applications. Nanotechnology. 2000;11:77. [Google Scholar]
- 61.Jain V, Maingi V, Maiti PK, Bharatam PV. Molecular dynamics simulations of PPI dendrimer-drug complexes. Soft Matter. 2013;9:6482–6496. [Google Scholar]
- 62.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed Engl. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 63.Siegwart DJ, et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc Natl Acad Sci U S A. 2011;108:12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee KS, et al. Multicenter phase II trial of Genexol-PM, a Cremophor-free, polymeric micelle formulation of paclitaxel, in patients with metastatic breast cancer. Breast Cancer Res Treat. 2008;108:241–250. doi: 10.1007/s10549-007-9591-y. [DOI] [PubMed] [Google Scholar]
- 66.Shin HC, Alani AW, Rao DA, Rockich NC, Kwon GS. Multi-drug loaded polymeric micelles for simultaneous delivery of poorly soluble anticancer drugs. J Control Release. 2009;140:294–300. doi: 10.1016/j.jconrel.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, et al. Well-defined, reversible disulfide cross-linked micelles for on-demand paclitaxel delivery. Biomaterials. 2011;32:6633–6645. doi: 10.1016/j.biomaterials.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao Y, Huang W, Shi C, Atkinson ST, Luo J. Reversibly crosslinked nanocarriers for on-demand drug delivery in cancer treatment. Ther Deliv. 2012;3:1409–1427. doi: 10.4155/tde.12.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Juliano R. Nanomedicine: is the wave cresting? Nat Rev Drug Discov. 2013;12:171–172. doi: 10.1038/nrd3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bandak S, Goren D, Horowitz A, Tzemach D, Gabizon A. Pharmacological studies of cisplatin encapsulated in long-circulating liposomes in mouse tumor models. Anticancer Drugs. 1999;10:911–920. doi: 10.1097/00001813-199911000-00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.