Abstract
RNAi is a major antiviral defense response in plant and animal model systems. RNA-dependent RNA polymerase 6 (RDR6) is an essential component of RNAi, which plays an important role in the resistance against viruses in the model plants. We found previously that rice RDR6 (OsRDR6) functioned in the defense against Rice stripe virus (RSV), and Rice Dwarf Phytoreovirus (RDV) infection resulted in down-regulation of expression of RDR6. Here we report our new findings on the function of OsRDR6 against RDV. Our result showed that down-regulation of OsRDR6 through the antisense (OsRDR6AS) strategy increased rice susceptibility to RDV infection while over-expression of OsRDR6 had no effect on RDV infection. The accumulation of RDV vsiRNAs was reduced in the OsRDR6AS plants. In the OsRDR6 over-expressed plants, the levels of OsRDR6 RNA transcript and protein were much higher than that in the control plants. Interestingly, the accumulation level of OsRDR6 protein became undetectable after RDV infection. This finding indicated that the translation and/or stability of OsRDR6 protein were negatively impacted upon RDV infection. This new finding provides a new light on the function of RDR6 in plant defense response and the cross-talking between factors encoded by host plant and double-stranded RNA viruses.
RNA silencing functions as a potent antiviral pathway in plant and animal systems1,2,3,4,5,6, and is known to be triggered by the accumulation of double-stranded viral RNA (dsRNA). The viral dsRNAs are then recognized and processed into small interfering RNAs (vsiRNAs) by distinct Dicer-like (DCL) proteins. The 21- and 22-nt vsiRNAs are known to be processed by DCL4 or its surrogate DCL27,8. The 24-nt vsiRNAs are mainly produced by DCL3 during DNA virus (gemini- and pararetroviruses) infection in plant7,9. One strand of the vsiRNA duplex is recruited by specific Argonaute (AGO) proteins within the RNA-induced silencing complexes (RISCs) and directs the complexes for further viral RNA silencing10,11.
During the process of antiviral RNA silencing, the host RNA-dependent RNA polymerases (RDRs) contribute to secondary vsiRNAs generation12. The model plant A. thaliana possesses six RDRs13,14. RDR1 plays an important role in production and amplification of both exogenous vsiRNAs and endogenous viral activated siRNA (vasiRNA) in plants infected with positive-stranded viruses15,16,17. Additionally, RDR1 is involved in plant responses to abiotic stresses18. RDR2 has been found involved in RNA–directed DNA methylation (RdDM) pathway19,20 and required for the development of the female gametophyte21. RDR6 is an important component for the biogenesis of different siRNAs including vsiRNAs22, trans-acting siRNAs (ta-siRNAs)23,24, natural antisense-transcript-derived siRNAs (nat-siRNAs)25, transgene-derived siRNAs26, and several phased or non-phased siRNAs27.
Recent studies on the dicotyledonous model plant A. thaliana and N. benthamiana have demonstrated the significant role of RDR6 in host defense response against some positive-sense single-stranded RNA viruses28,29, as well as viroid30,31. The rdr6 mutant Arabidopsis plants exhibit enhanced susceptibility to the Cucumber mosaic virus (CMV) but not to Turnip mosaic virus (TuMV) or Turnip vein clearing virus (TVCV) infection28. RDR6i N. benthamiana plants are more sensitive to Potato virus X (PVX), Potato virus Y (PVY), CMV with Y satellite29 and Potato spindle tuber viroid (PSTVd)31. Tobacco plants with reduced RDR6 expression exhibit hypersusceptibility to Turnip crinkle virus (TCV) and Tobacco mosaic virus (TMV) in a temperature-dependent manner32. Reduced expression of NbRDR6 also permitted efficient multiplication of TMV32 or PVX29 in the shoot apices. Grafting assays indicates the requirement of NbRDR6 for symptom production induced by Hop stunt viroid (HSVd)30. We also reported previously that down-regulation of rice RDR6 expression in rice plant increased disease symptoms caused by RSV infection comparable to that shown in the wild-type plants infected with the same virus33. Plant reoviruses are major threats to monocotyledonous (monocots) food crops including rice and corn. Therefore, development of new and effective disease management strategies for these viruses is critical for rice and other cereal crop production. In this study, we investigated the function of OsRDR6 for its role in resistance against RDV, a member of genus Phytoreovirus.
RDV is a member of the genus Phytoreovirus, family Reoviridae. It is transmitted by leafhopper in a circulative manner and causes severe dwarf disease symptoms in rice34. The genome of RDV comprises twelve double-stranded RNA segments encoding seven structural proteins and at least seven nonstructural proteins35,36,37,38,39. To date, only a few RDV genome segments have been studied. The nonstructural Pns6 and Pns10 proteins are known as the viral cell-to-cell movement protein39 and viral suppressor of RNA-silencing (VSR)40,41 respectively. The outer capsid protein P2 is a multifunctional protein required for virion entry into insect vector cells42 and is involved in plant hormone (e.g., GA) pathway34,43. Rice encodes five RDRs: OsRDR1, OsRDR2, OsRDR3a, OsRDR3b and OsRDR644. OsRDR1 is required for the production of small RNA in response to DNA-damage in rice45 and for the RNA silencing mediated by Brome mosaic bromovirus (BMV), but not by Wheat dwarf geminivirus (WDV)46. Recent report showed that OsRDR2 did not function in siR441 and siR446 production, which were previously annotated as microRNAs (miRNAs)47. The functions of OsRDR3a and OsRDR3b have not been studied in detail.
Although OsRDR6 is known to play roles in the defense response against dsRNA virus infection in monocot plants, how dsRNA viruses counteract this host defense strategy remains largely unknown. We demonstrated here for the first time that down-regulation of OsRDR6 expression in rice significantly enhanced rice susceptibility to RDV infection but up-expression of OsRDR6 in rice had no effect on its defense against the virus. We also demonstrated that the accumulation of OsRDR6 protein in the OsRDR6 over-expressed lines was suppressed upon RDV infection due to an unidentified mechanism that confers the suppression of translation of the transgene and/or destabilization of the protein. Taken together, our finding presented in this paper provides some new insights into the function of OsRDR6 in defense response against dsRNA virus infection, and the defense and counter-defense reaction between host plant and virus.
Results
Down-regulation of OsRDR6 expression increased rice susceptibility to RDV infection
Our previous study showed that RDV infection in rice reduced OsRDR6 expression41. We also reported in a separate study that OsRDR6AS transgenic rice plants accumulated significantly less amount of OsRDR6 RNA transcripts and were more susceptible to RSV (a single-stranded RNA virus) infection33. In this study, the OsRDR6AS transgenic rice lines were inoculated with RDV via viruliferous leafhopper. By three weeks post virus inoculation (wpi), the RDV-inoculated OsRDR6AS plants exhibited more severe stunting phenotypes (Fig. 1a) than those shown by the RDV-inoculated wild type (WT) rice plants. The infection rates of RDV in the OsRDR6AS transgenic lines were also higher than those in the WT rice plants at various wpi (Fig. 1b). Plants of OsRDR6AS transgenic line B and C showed RDV symptoms as early as 1wpi (Fig. 1b). By 7wpi, the RDV infection rate in the three OsRDR6AS transgenic lines reached above 90% while the infection rate of the WT plants was about 80%, indicating that down-regulation of OsRDR6 expression in rice increased the susceptibility to RDV infection.
Figure 1. RDV-infected OsRDR6AS rice plants exhibited more severe stunting than the RDV-infected wild type (WT) Zhonghua11 plants.
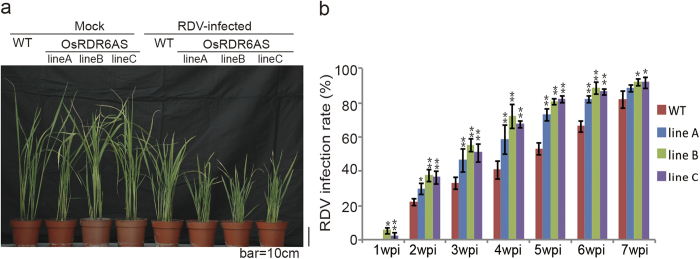
(a) Phenotypes of the RDV-infected OsRDR6AS and WT Zhonghua11 plants at 3 weeks post RDV inoculation (wpi). (b) Time course study of infection rate for RDV-inoculated OsRDR6AS and WT plants. The visual disease symptoms were assessed with 30 individual plants per treatment. * P < 0.05; ** P < 0.01 (Student’s t test). The inoculation assay was repeated three times. The error bars indicate the standard errors.
Accumulation levels of RDV RNAs correlated with RDV symptoms in the inoculated plants
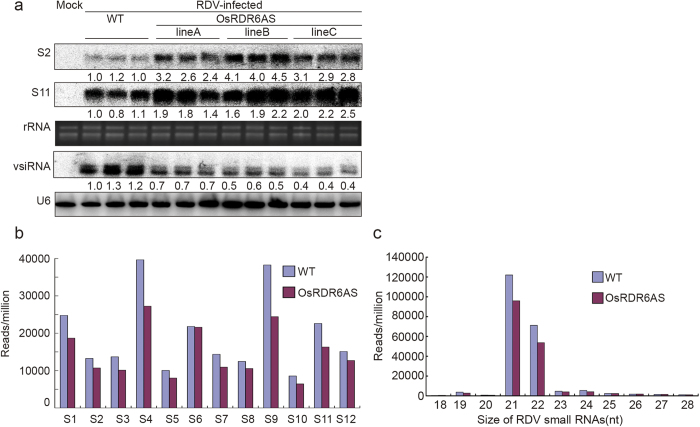
To determine whether RDV symptoms in the inoculated plants were correlated with the levels of RDV RNA accumulation in the infected plants, we analyzed RDV S2 and S11 genomic RNA levels in the assayed plants by Northern blot. Result of the assay showed that the accumulation levels of RDV S2 and S11 RNAs in the inoculated OsRDR6AS transgenic lines were much higher than that in the inoculated control WT plants (Fig. 2a). Further analysis of viral vsiRNAs accumulated in these plants revealed that the levels of RDV vsiRNAs in the inoculated OsRDR6AS transgenic lines were significantly lower than that in the inoculated control plants. This finding indicated that down-regulation of OsRDR6 expression in rice led to a reduction of RDV RNA silencing in the RDV-infected OsRDR6AS transgenic plants.
Figure 2. Accumulation of RDV S2 and S11 genomic RNAs and vsiRNAs in the OsRDR6AS and WT plants.
(a) Northern blot assay of RDV S2 and S11 genomic RNAs and vsiRNAs in the OsRDR6AS and WT plants. Plants showing disease symptoms were harvested at 3 wpi. Three pools of infected plants (>10 plants per pool) were harvested from each treatment and used for total RNA isolation. The rRNA or U6 was used as loading control for the assay. (b) Deep sequencing result showing the relative abundance of vsiRNAs from each RDV RNA. The blue bars represent the results from the WT plants and red bars represent the results from the OsRDR6AS plants. (c) Size distribution of RDV-derived small RNA populations based on the deep sequencing data.
We then analyzed the vsiRNAs from the RDV-infected OsRDR6AS and control WT plants by deep sequencing followed by data analysis using the bioinformatics tool BOAT provided by CBI (the Center of Bioinformatics) as described previously33,48. Result from two independent RDV vsiRNA libraries showed a reduced amount of RDV vsiRNAs in the inoculated OsRDR6AS plants comparable to the control plants except S6 (Table 1, Fig. 2b, Fig. S1). The length and polarity profiles of vsiRNAs from the infected OsRDR6AS plants were similar to that from the wild type plants (Fig 2c). Unlike the previous report for RSV33, the RDV derived vsiRNAs were mostly 21- and 22-nt in length (Fig. 2c). This result suggested that OsRDR6 acted differently during silencing double-stranded RNA viruses and negative-sense single-stranded RNA viruses.
Table 1. Summary of the deep sequencing results for the small RNAs derived from the RDV-infected rice plants.
| Libraries | Replicate 1 | Replicate 2 | ||
| Zhonghua11 | OsRDR6AS | Zhonghua11 | OsRDR6AS | |
| Unique sequencesa | 2,519,939 | 2,171,452 | 2,527,417 | 1,910,802 |
| Total readsa | 8,504,431 | 7,264,974 | 6,756,834 | 6,032,770 |
| Unique RDV vsiRNAa,b | 124,143 | 122,007 | 142,329 | 110,547 |
| Total Reads of RDV vsiRNAd (% of total reads)a,b | 1,886,645 (22.18%) | 1,265,099 (17.41%) | 1,584,898 (23.46%) | 1,071,029 (17.76%) |
| (+)-strands (%)a,c | 59.40% | 61.00% | 60.50% | 62.70% |
| (−)-strands (%)a,d | 40.50% | 39.00% | 39.50% | 37.30% |
aNumber of sequences within the set (18 nt≤ length ≤28 nt).
bSequences with perfect matches to the RDV genome.
cSequences with perfect matches to the (+)-strand RNAs of the RDV genome.
dSequences with perfect matches to (−)-strand RNAs of the RDV genome.
Over-expression of OsRDR6 in rice did not affect RDV infection
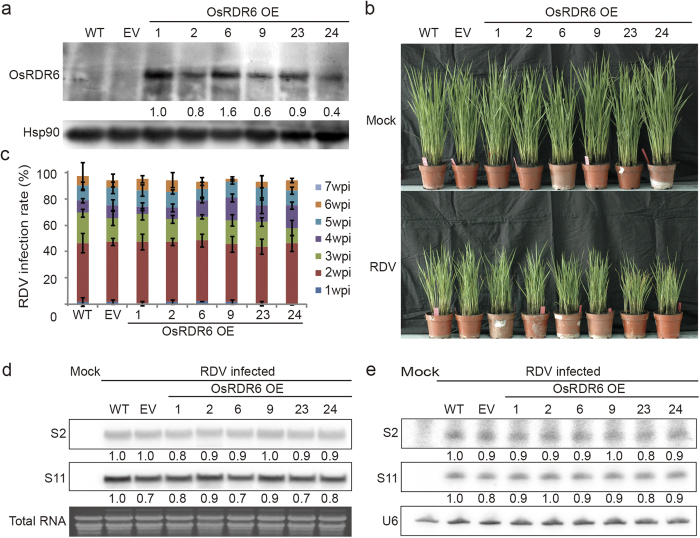
Because silencing OsRDR6 expression in rice resulted in an increased RDV accumulation and stronger disease symptoms in the RDV-infected rice plants, we reasoned that over-expression of OsRDR6 would cause an increased resistance against RDV. To test this hypothesis we generated OsRDR6-over-expressing transgenic rice lines and inoculated them with RDV. Figure S2 showed that transgenic line 1, 2, 6, 23 and 24 contained a single-copy of OsRDR6 transgene while line 9 has two copies (Fig. S2). Western blot showed that all the over-expression (OE) transgenic lines produced high levels of OsRDR6 proteins while this protein in the WT or the empty vector (EV) transformed control rice plants was not detectable with this method (Fig. 3a). Growth of all the OsRDR6 OE lines were comparable to the WT and EV control plants (Fig. 3b top panel). Surprisingly, none of the OsRDR6 OE rice lines showed an increased resistance to RDV infection comparable with the WT and EV control plants (Fig. 3b). In addition, the infection rates of all the RDV-inoculated OsRDR6 OE lines were similar to that of the WT and EV control plants (Fig. 3c).
Figure 3. Over-expression of OsRDR6 did not affect RDV infection in rice.
(a) Western blot analysis of OsRDR6 protein expression in the OsRDR6-over-expression (OsRDR6 OE), empty vector transformed (EV) and the WT rice plants. Rice Hsp90 was served as the loading control. (b) Comparison of RDV infected phenotypes in the OsRDR6 OE, EV and WT plants. The OsRDR6 OE rice plants did not show reduced disease symptoms comparable with the control plants. (c) The infection rates of the OsRDR6 OE, EV and WT plants at various wpi. The infection rate was determined using 30 individual plants per treatment at each wpi. Inoculation assay was repeated three times. The error bars indicate the standard errors. P > 0.05 (Student’s t test). (d) Northern blot assay of RDV S2 and S11 genomic RNA accumulation in the OsRDR6 OE, EV and WT plants. Thirty plants showing disease symptoms were harvested from each treatment at 3 wpi for total RNA isolation. (e) Northern blot assay for RDV siRNAs accumulation in the OsRDR6 OE, EV and WT plants. The plant samples used in (d) were used for this analysis. U6 RNA was used as the loading control.
To further compare the OsRDR6 OE lines with the control plants, we examined RDV S2 and S11 RNA accumulation in these plants through Northern blot assay. Result of the assay showed that the accumulations of S2 and S11 RNA in the RDV-inoculated OsRDR6 OE lines were similar to those in the WT and EV control plants (Fig. 3d). Northern blot analysis of vsiRNAs also showed that the accumulation levels of vsiRNAs in the OsRDR6 OE lines were similar to those in the RDV-inoculated WT and EV control plants (Fig. 3e). This finding indicated that increased OsRDR6 protein accumulation in OE rice plants did not improve its resistance to RDV infection.
RDV infection reduced OsRDR6 expression in the OsRDR6 OE lines
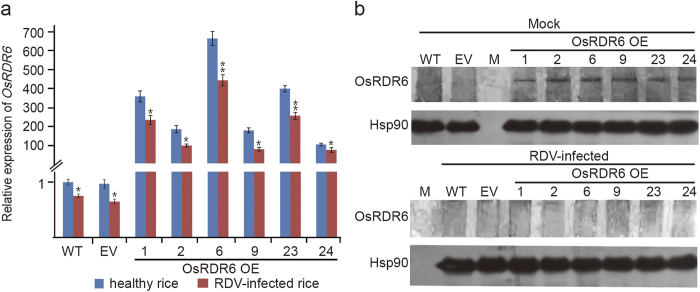
To investigate the effect of RDV infection on OsRDR6 expression in rice plants, we analyzed OsRDR6 mRNA and protein accumulation in the RDV-inoculated and non-inoculated OsRDR6 OE, EV and WT rice plants through quantitative RT-PCR. Results of the assay showed that the levels of OsRDR6 mRNA were much higher in the OE plants than those in the WT or EV control plants (Fig. 4a). Interestingly, RDV infection resulted in decreased levels of OsRDR6 transcripts in all assayed plants. Western blot assay agreed with the quantitative RT-PCR result and showed that the accumulation of OsRDR6 protein in the OsRDR6 OE lines was clearly decreased after RDV infection (Fig. 4b). These data indicated that over-expression of OsRDR6 in rice plant had no significant effect on host response to RDV infection, and the decreased accumulation of OsRDR6 protein in the RDV-infected OE lines might be caused by a translational suppression of OsRDR6 and/or the stability of the protein.
Figure 4. RDV infection reduced the expression of OsRDR6 in the OsRDR6-over-expression rice plants.
(a) Relative expression levels of OsRDR6 RNA transcript in the OsRDR6 OE, EV and WT plants. Relative transcript levels were calculated using the 2−ΔΔC(t) method and OsEF-1a gene transcripts as the internal control. The error bars indicate the standard errors. Asterisks indicate P values compared with healthy rice plants: * P < 0.05; ** P < 0.01 (Student’s t test). (b) Western blot assay of OsRDR6 proteins accumulation in OsRDR6 OE, EV and WT plants. Expression of rice Hsp90 protein was used as the loading control.
Discussion
Many studies have shown that the RDR6-dependent RNA silencing pathway plays an important role in plant defense against virus infection. For example, A. thaliana rdr6 mutant plants showed an increased susceptibility to CMV infection26,28. Further study by Zhu et al. showed that RDR6-dependent silencing of CMV in N. benthamiana was initiated by CMV Satellite RNA-derived small interfering RNA known as satsiR-1249. In other studies, TMV derived vsiRNAs were found to be significantly reduced in the TMV-infected A. thalianardr 6 mutants32,50. RDR6 of N. benthamiana was also shown to regulate defense response against systemic spreading of TCV32 and invasion of apical meristem by Potato virus X29. Our previous study also demonstrated that knockdown of OsRDR6 expression in rice increased its susceptibility to RSV infection33. In addition, RDR6 was reported to play an important role in disease resistance against viroids. For example, silencing RDR6 gene expression in N. benthamiana enhanced disease symptoms caused by HSVd30. Also, the accumulation of PSTVd in the NbRDR6-silenced N. benthamiana plants was increased early after PSTVd infection than that in the non-silenced N. benthamiana plants31. A recent report by Wang et al. showed that over-expression of Gossypium hirsutum L. (cotton) GhRDR6 in N. benthamiana enhanced N. benthamiana resistance to Potato virus Y infection51. Interestingly, results shown in this paper indicated that over-expression OsRDR6 in rice had no clear influence on RDV infection, based on the infection rates of RDV in the OsRDR6 OE, EV and WT control rice plants (Fig. 3c). Our North blot assay supported the above symptom observation and showed that the accumulation levels of RDV S2 and S11 RNAs were similar between the OsRDR6 OE lines and the WT or EV control lines (Fig. 3d). In addition, the accumulation levels of S2 and S11 vsiRNAs in the OsRDR6 OE lines were comparable to those from the control plants (Fig. 3e). Taken together, our results provided new insight into the function of host RDR6 in antiviral RNA silencing during virus infection. We speculate that over-expression of OsRDR6 in rice may alter the expression of other host genes that are also involved in the RNA silencing pathway. It is also possible that dsRNA viruses encode other factors that influence RDR6 gene function and regulate the accumulation of dsRNA viruses in its host plants.
To verify the biological function of OsRDR6 protein in the OE lines, we conducted a complementation assay by over-expressing the full length coding sequence of OsRDR6 and AtRDR6 in the rdr6–11 mutant Arabidopsis23,24 to produce OsRDR6/rdr6–11 and AtRDR6/rdr6–11 transgenic lines, respectively. OsRDR2/rdr6–11 line was produced to serve as a control. Analysis of these lines showed that OsRDR6 and AtRDR6 indeed restored the accumulation of TAS siRNAs52 in the over-expression Arabidopsis transgenic plants whereas the transgenic plants over-expression OsRDR2 did not (Fig. 5). Therefore the inability of OsRDR6 protein in the OE rice line to suppress RDV infection was not due to the authenticity of gene sequence transformed into the rice plants.
Figure 5. Over-expression of OsRDR6 and AtRDR6 in rdr6 mutant A. thaliana restored the accumulation of TAS siRNAs in plants.
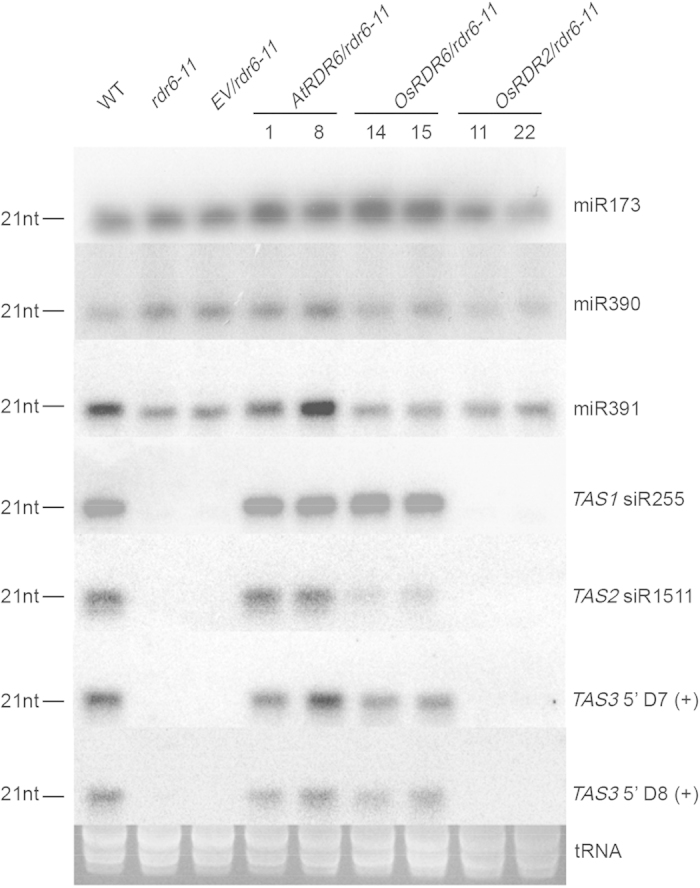
Four TASs [TAS1 siRNA255, TAS2 siRNA1511, TAS3 5'D7 (+) and TAS3 5'D8 (+)] were analyzed using Northern blot assay. Accumulation of miR173, miR390 and miR391 in the OsRDR6/rdr6–11, AtRDR6/rdr6–11 or OsRDR2/rdr6–11 transgenic Arabidopsis plants were also analyzed in the assay. tRNA isolated from each sample was served as the loading control.
The antiviral RNA silencing machinery also contains many other proteins involved in different process including DCLs53,54,55,56 and AGOs56,57,58,59. For instance, recently we found that AGO18 confers broad-spectrum resistance against single-stranded RNA virus RSV and double-stranded RNA virus RDV through a novel mechanism of positive regulation AGO1 homeostasis6. The broad-spectrum resistance of AGO18 provides a potential application of AGO18 in crops. Both transcript and translation levels of AGO18 were increased after RDV infection. While unlike AGO18, the quantitative RT-PCR data shown in Fig. 4a indicated that OsRDR6 mRNA levels were reduced after RDV infection. Although the transcript levels of OsRDR6 in the OE rice lines infected with RDV were reduced to some extent, these expression levels were still over a hundred fold higher than those observed from the control plants. Result of Western blot assay (Fig. 4b top panel) showed that OsRDR6 protein was indeed accumulated in the OE lines but not in the WT or EV transformed control plants before RDV inoculation. However, after RDV inoculation, OsRDR6 protein in the OE lines also became undetectable (Fig. 4b bottom panel). Because RDR6 plays an important role in antiviral RNA silencing pathway, it is also a target of virus-encoded suppressor of RNA silencing (VSRs). For example, Rice yellow stunt rhabdovirus (RYSV) encoded P6 protein was reported to interact with OsRDR6 and AtRDR6, and to repress the synthesis of the secondary siRNAs in both N. benthamiana and rice protoplasts60. Li et al. reported that expression of βC1 by the DNA satellite associated with Tomato yellow leaf curl China virus (TYLCCNV, geminivirus) induced the expression of Nbrgs-CaM leading to a repression of RDR6 expression in N. benthamiana followed by suppressing the TYLCCNV-induced gene silencing61. We also reported that RDV encoded Pns10 suppressed the expression of NbRDR6 in N. benthamiana plant41. Interestingly, in this study, infection of RDV in the OsRDR6-over-expression rice plants significantly reduced the accumulation of OsRDR6 protein. We hypothesized that the RDV encoded Pns10 might play a role in suppression of translation of OsRDR6. To test this hypothesis we performed a binding assay using purified recombinant Pns10 protein and RDR6 mRNA. The results of the assay showed that Pns10 protein did not bind RDR6 mRNA. In addition, Pns10 protein did not interact with OsRDR6 protein in the yeast two-hybrid and co-immunoprecipitation (CO-IP) assays. Whether other uncharacterized RDV encoded proteins contain such a function of suppress translation of OsRDR6 and/or destabilize the protein remains to be determined. To test whether other virus infection of rice also affects OsRDR6 expression, we analyzed the mRNA and protein level of OsRDR6 in RSV infected rice and found that infection of RSV had no effect on OsRDR6 expression. Therefore, we presume that viruses with different genome structures may have different effects on OsRDR6 expression.
Although over-expression of OsRDR6 in rice failed to alter plant resistance to RDV infection, down-regulation of OsRDR6 expression indeed enhanced plant susceptibility to the virus comparable with the WT control plants. Our result agreed with previous reports that OsRDR6 did play an important role in plant defense against dsRNA viruses. Further characterization of dsRNA virus encoded proteins will help us to better understand the mechanism(s) that regulate the expression of OsRDR6 and stability of the protein in the OE transgenic lines upon RDV infection.
Methods
Rice transformation and RDV infection assay
OsRDR6AS transgenic rice lines A to C were from a previously described source33. To generate OsRDR6-over-expression rice, the full-length coding sequence of OsRDR6 was ligated into the PstI site within the binary vector pCam23ACT:OCS62 to produce pCam23-ACT:OCS-OsRDR6. Briefly, the full-length coding sequence of OsRDR6 was digested with HindIII (Promega) from pBS(KS)-OsRDR633. Then the ends made blunt with T4 DNA polymerase (Promega) and digested with XbaI (Promega). pCam23ACT:OCS was digested with PstI (Promega). Then the ends made blunt with T4 DNA polymerase (Promega) and digested with PstI (Promega). The above digested OsRDR6 sequence and pCam23ACT:OCS were ligated by T4 DNA Ligase (NEB). The plasmid was transformed into a widely cultivated rice cultivar (Zhonghua11) in China by Weiming Kaituo Co., Ltd (Beijing, China) through A. tumefaciens-mediated stable transformation procedure. Seeds of T0 generation rice were immersed in water containing G418 antibiotic (10 mg/L) to obtain positive transgenic plants. The OsRDR6 transgenic and empty vector transformed control rice plants were grown in a greenhouse under a 14-h light/10-h dark photoperiod at 24–29 °C. Thirty T1 generation transgenic rice plants from each line were inoculated with RDV using viruliferous insect (Nephotettix cincticeps) as previously described43,48.
OsRDR6 and AtRDR6 functional complementation assay
OsRDR6, OsRDR2 and AtRDR6 were cloned individually into pWM101 to produce pWM101-OsRDR6, pWM101-OsRDR2 and pWM101-AtRDR6. The resulting plasmids were transformed into A. thaliana rdr6–11 mutant plants using the floral-dip method63. Seeds from the transgenic OsRDR6/rdr6–11, OsRDR2/rdr6–11 and AtRDR6/rdr6–11 A. thaliana plants were sterilized and then planted on a 1/2 MS medium containing hygromycin (25 mg/L). The germinated seedlings were transferred to a greenhouse at 10 days post planting. Individual transgenic lines were determined through RT-PCR with specific primers (Table S1). Total RNA was then extracted from more than thirty plants with different transgenes and used for small RNA assay through Northern blot with specific probes (Table S2)
OsRDR6 protein expression, polyclonal antibody production and Western blot assay
A 1221-bp sequence in the 3' end of OsRDR6 open reading sequence was cloned into the expression vector pET-28a(+) prior to transformation into Escherichia coliBL21(DE3) using primers OsRDR6F3-3/OsRDR6R3-3 (Table S1). The transformed cells were cultured at 37 °C and then induced for 4h with 0.5 mM isopropyl-β-d-1-thiogalactoside (IPTG). Cultures of E.Coli were harvested and sonicated at 4 °C. Inclusion bodies were isolated through HiTrap Chelating with the FPLC System as instructed by the manufacture (Amersham). The extracted recombinant proteins were separated in 8% SDS-PAGE gel. Polyclonal antiserium against OsRDR6 was raised in rabbit by ComWin Biotech Co., Ltd (Beijing, China). Plant protein extracts were separated in 8% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes as instructed (Immobilon-P Transfer membrane, MILIIPOR). The membranes were blocked and probed with the anti-OsRDR6 antiserium diluted 1:1,000 in blocking buffer (1×TBS, 0.1% Tween-20, 5%BSA). Anti-Rice Hsp90 was purchased from HuaDa (Beijing, China) diluted 1:2,000 in blocking buffer and used as protein loading controls.
Southern blot analysis
Southern blot assay was similar as previously described64. Briefly, forty microgram total genomic DNA was obtained from each transgenic line, digested overnight with HindIII restriction enzyme and transferred to Hybond-N+ nylon membrane (Amersham) through electrophoresis. Probes specific for the hygromycin-resistance gene (Hyg) were amplified with the Hyg-specific primers Hyg-F/R (Table S1), and labeled using the DIG DNA labeling mix kit as instructed (Roche). Hybridization was carried out overnight at 42 °C in an ULTRAhyb® hybridization buffer (Ambion) and then detected with an anti-DIG antibody conjugated with alkaline phosphatase (Roche).
Non-preference test
Non-preference tests were performed using rice seedlings with different genetic background. The seedlings were inoculated with the virus viaviruliferous leafhopper vector as described previously65.
RT–PCR
Total RNA was extracted from A. thaliana plants using Trizol as instructed (Invitrogen) followed by the DNase treatment using RQ1 RNase-free DNase (Promega). Synthesize of cDNA was conducted using the Super Script III Reverse Transcriptase (Invitrogen) and an oligo d (T) primers instructed by the manufacture. The resulting cDNA was diluted 20 fold with water and subjected to 30 cycles of PCR reaction with gene-specific primers (Table S1). Expression level of AtelF4A was used as an internal control.
Quantitative real-time PCR
The rice plants showing typical disease symptoms were collected at 3 wpi. Total RNA was extracted from more than ten rice plants as a pool. The RNA for the quantitative RT-PCR was treated with RQ1 RNase-free DNase (Promega) for 30 min at 37 °C followed by phenol/chloroform extraction. Reverse transcription was done using two microgram RNA per twenty microliter reaction with the Super Script III Reverse Transcriptase and an oligo d(T) primer. The quantitative PCR was performed using the SYBR Green Real-Time PCR Master Mix as instructed (TOYOBO) and primers specific for OsRDR6 (OsRDR6 P13/P14, Table S1). Expression of OsEF-1a gene was used as an internal control for the assay (using primers OsEF1a F/R, Table S1). Three independent biological replicates were analyzed per treatment. Relative transcription levels were calculated using 2-ΔΔC(t) method.
Determination of viral S2 and S11 genomic RNA accumulation
Accumulation of RDV S2 and S11 genomic RNA was analyzed by Northern blot as described33. Briefly, ten microgram total RNA from each assayed plant was transferred to a Hybond N+ nylon membrane (Amersham) after electrophoresis in a 1.2% (w/v) formaldehyde-denaturing agarose gel. Radioactive probes used for the detection were PCR products amplified with primer sets S2-F1/S2-R1 and S11-F1/S11-R1 (Table S1) and labeled with α−32P-dCTP using the Random Primer DNA Labeling Kit Ver.2 (Takala). The hybridization was performed overnight in the PerfectHybTM Plus Hybridization Buffer (Sigma) at 65 °C. The blotted membrane was washed for 15 min in a 2 ×SSC solution containing 0.1% SDS at 65 °C followed by washing two times (15 min each) in a 0.1×SSC solution containing 0.1% SDS. The detection signal was monitored with a phosphorimager (GE, USA) and quantified using the ImageJ (version 1.44).
Detection of Small RNAs
The rice plants of 30 seedlings showing typical disease symptoms were pooled at 3 wpi. Small RNAs were extracted using RNAzol®RT as instructed (Molecular Research Center, Inc. USA). Ten microgram small RNA from each sample was separated in a 15% denaturing polyacrylamide gel containing 8 M urea and transferred to a Hybond N+ nylon membrane (Amersham). Approximately 500 ng PCR product amplified with the S2-F1/S2-R1 or S11-F1/S11-R1 primer set (Table S1) and labeled with α−32P-dCTP were used as the probes. Hybridization was performed overnight in the PerfectHybTM Plus Hybridization Buffer (Sigma) at 42 °C. The membrane was stripped with 0.1% SDS at 100 °C 10 min three times to remove S11 probe and hybridized with S2 probe.
Small RNA sequencing and bioinformatics analysis
More than fifteen seedlings were harvested from each treatment and pooled for RNA isolation. The procedure of small RNA library construction and Illumina1G sequencing was described previously33,48. Sequences of 18–28 nt small RNAs were mapped to the O. sativa genome sequence (TIGR Rice Annotation Release 5.0, ftp://ftp.plantbiology.msu.edu/pub/data/EukaryoticProjects/osativa/annotationdbs/) and RDV genome sequence (ftp://ftp.ncbi.nih.gov/genomes/Viruses/Ricestripevirusuid14795/)66 using the bioinformatics tool BOAT provided by CBI (the Center of Bioinformatics).
Additional Information
How to cite this article: Hong, W. et al. OsRDR6 plays role in host defense against double-stranded RNA virus, Rice Dwarf Phytoreovirus. Sci. Rep. 5, 11324; doi: 10.1038/srep11324 (2015).
Supplementary Material
Acknowledgments
We thank Dr. Yijun Qi for help with construction of small RNA libraries and data analysis. We thank Dr. R. Scott Poethig for providing seed of rdr6-11 Arabidopsis. This work was supported by grants from 973 program (2014CB138403) and the National Natural Science Foundation of China (31200116, 31420103904).
Footnotes
Author Contributions W.H., D.Q., R.H.S. and L.J. designed and performed the experiments; Y.W. collected and analyzed data; C.H.W., W.H.Z., K.Z. and Y.L. wrote the paper. Y.L. initiated the project.
References
- Ding S. W. & Voinnet O. Antiviral immunity directed by small RNAs. Cell 130, 413–426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 15, 701–707 (2010). [DOI] [PubMed] [Google Scholar]
- Incarbone M. & Dunoyer P. RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18, 382–392 (2013). [DOI] [PubMed] [Google Scholar]
- de Alba A. E. M., Elvira-Matelot E. & Vaucheret H. Gene silencing in plants: a diversity of pathways. Bba-Gene Regul Mech. 1829, 1300–1308 (2013). [DOI] [PubMed] [Google Scholar]
- Li Y., Lu J. F., Han Y. H., Fan X. X. & Ding S. W. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. G. et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4e:05733 10.7554/eLife.05733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T. et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nuleic Acids Res. 34, 6233–6246 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A. et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 (2006). [DOI] [PubMed] [Google Scholar]
- Moissiard G. & Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA 103, 19593–19598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tolia N. H. & Joshua-Tor L. Slicer and the Argonautes. Nat Chem Biol. 3, 36–43 (2007). [DOI] [PubMed] [Google Scholar]
- Ding S. W. & Voinnet O. Antiviral immunity directed by small RNAs. Cell 130, 413–426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. J., Carter S. A., Cole A. B., Cheng N. H. & Nelson R. S. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA. 101, 6297–6302 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M. & Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11, 142–151 (2006). [DOI] [PubMed] [Google Scholar]
- Willmann M. R., Endres M. W., Cook R. T. & Gregory B. D. The functions of RNA-dependent RNA polymerases in Arabidopsis. The Arabidopsis book. 9, e0146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L. et al. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82, 5167–5177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. B. et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107, 484–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M. J. et al. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 111, 14613–14618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T. et al. Expressional and regulatory characterization of Arabidopsis RNA-dependent RNA polymerase 1. Planta 237, 1561–1569 (2013). [DOI] [PubMed] [Google Scholar]
- Xie Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. J., Chen T. & Zhu J. K. Regulation and function of DNA methylation in plants and animals. Cell Res. 21, 442–465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V. et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F. Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. MPMI 23, 1248–1252 (2010). [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H. L. & Poethig R. S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Gene Dev. 18, 2368–2379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M. Y. & Poethig R. S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Gene Dev. 19, 2164–2175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J. H., Verslues P. E., Sunkar R. & Zhu J. K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton A., Rudd S., Angell S. & Baulcombe D. C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553 (2000). [DOI] [PubMed] [Google Scholar]
- Zheng Q. et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. Plos Genet. 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P. et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542 (2000). [DOI] [PubMed] [Google Scholar]
- Schwach F., Vaistij F. E., Jones L. & Baulcombe D. C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G., Martinez G. & Pallas V. Viroid-induced symptoms in Nicotiana benthamiana plants are dependent on RDR6 activity. Plant Physiol. 148, 414–423 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Serio F., de Alba A. E. M., Navarro B., Gisel A. & Flores R. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J. Virol. 84, 2477–2489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F. et al. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79, 15209–15217 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. et al. RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res. 163, 512–519 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu S. et al. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 139, 1935–1945 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Sugawara M. & Kusano T. Rice dwarf phytoreovirus segment S12 transcript is tricistronic in vitro. Virology 191, 992–995 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Sugawara M., Nuss D. L. & Matsuura Y. Polycistronic (tri- or bicistronic) phytoreoviral segments translatable in both plant and insect cells. J. Virol. 70, 8155–8159 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Rice dwarf phytoreovirus segment S11 encodes a nucleic acid binding protein. Virology 240, 267–272 (1998). [DOI] [PubMed] [Google Scholar]
- Zheng H. et al. Assembly of double-shelled, virus-like particles in transgenic rice plants expressing two major structural proteins of rice dwarf virus. J. Virol. 74, 9808–9810 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Rice dwarf phytoreovirus segment S6-encoded nonstructural protein has a cell-to-cell movement function. J. Virol. 78, 5382–5389 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. S. et al. Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol. 79, 13018–13027 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B. et al. Multiple functions of Rice Dwarf Phytoreovirus Pns10 in suppressing systemic RNA silencing. J. Virol. 84, 12914–12923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F. et al. The P2 capsid protein of the nonenveloped rice dwarf phytoreovirus induces membrane fusion in insect host cells. Proc. Natl. Acad. Sci. USA 104, 19547–19552 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F. et al. OsRFPH2-10, a RING-H2 finger E3 ubiquitin ligase, is involved in rice antiviral defense in the early stages of Rice dwarf virus Infection. Mol Plant. 7, 1057–1060 (2014). [DOI] [PubMed] [Google Scholar]
- Zong J., Yao X., Yin J. Y., Zhang D. B. & Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447, 29–39 (2009). [DOI] [PubMed] [Google Scholar]
- Chen H., et al. Both OsRecQ1 and OsRDR1 are required for the production of small RNA in response to DNA-damage in rice. Plos ONE 8, e55252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Analysis of rice RNA-dependent RNA polymerase 1 (OsRDR1) in virus-mediated RNA silencing after particle bombardment. J Gen Plant Pathol. 76, 152–160 (2010). [Google Scholar]
- Yan Y. et al. Small RNAs from MITE-derived stem-loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. Plant J. 65, 820–828 (2011). [DOI] [PubMed] [Google Scholar]
- Du P. et al. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. Plos Pathog. 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. et al. Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3’ untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation. J. Virol. 85, 13384–13397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X. P., Bao F. S. & Xie Z. X. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. Plos ONE 4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. Characterization and functional analysis of GhRDR6, a novel RDR6 gene from cotton (Gossypium hirsutum L.). Bioscience Rep. 32, 139–151 (2012). [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M. Y. & Poethig R. S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. W. RNA-based antiviral immunity. Nat Rev Immunol. 10, 632–644 (2010). [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H. et al. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant cell. 22, 481–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang X., Singh J., Li D. & Qu F. Temperature-dependent survival of Turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires DCL2, AGO2, and HEN1. J. Virol. 86, 6847–6854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. B. et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant cell. 23, 1625–1638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert M., Bhattacharjee S., Mello A. F., Perry K. L. & Moffett P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Ye X. & Morris T. J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA. 105, 14732–14737 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z. et al. Characterization of small interfering RNAs derived from Sugarcane mosaic virus in infected maize plants by deep sequencing. Plos ONE 9, e97013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. Y. et al. Rice yellow stunt rhabdovirus Protein 6 suppresses systemic RNA silencing by blocking RDR6-mediated secondary siRNA synthesis. Mol Plant Microbe In. 26, 927–936 (2013). [DOI] [PubMed] [Google Scholar]
- Li F. F., Huang C. J., Li Z. H. & Zhou X. P. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. Plos Pathog. 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. et al. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139, 296–305 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Sun R., Du P., Jiang L., An D. & Li Y. Heterologous expression of artificial miRNAs from rice dwarf virus in transgenic rice. PCTOC. 116, 353–360 (2013). [Google Scholar]
- Hiroshi Nemoto K. I., Eiji Shimura. The resistance to Rice stripe virus and small brown planthopper in rice variety, IR 50. Breeding Sci. 44, 13–98 (1994). [Google Scholar]
- Zhao S. Q. et al. BOAT: Basic oligonucleotide alignment tool. BMC genomics 10 Suppl 3, S2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.