Abstract
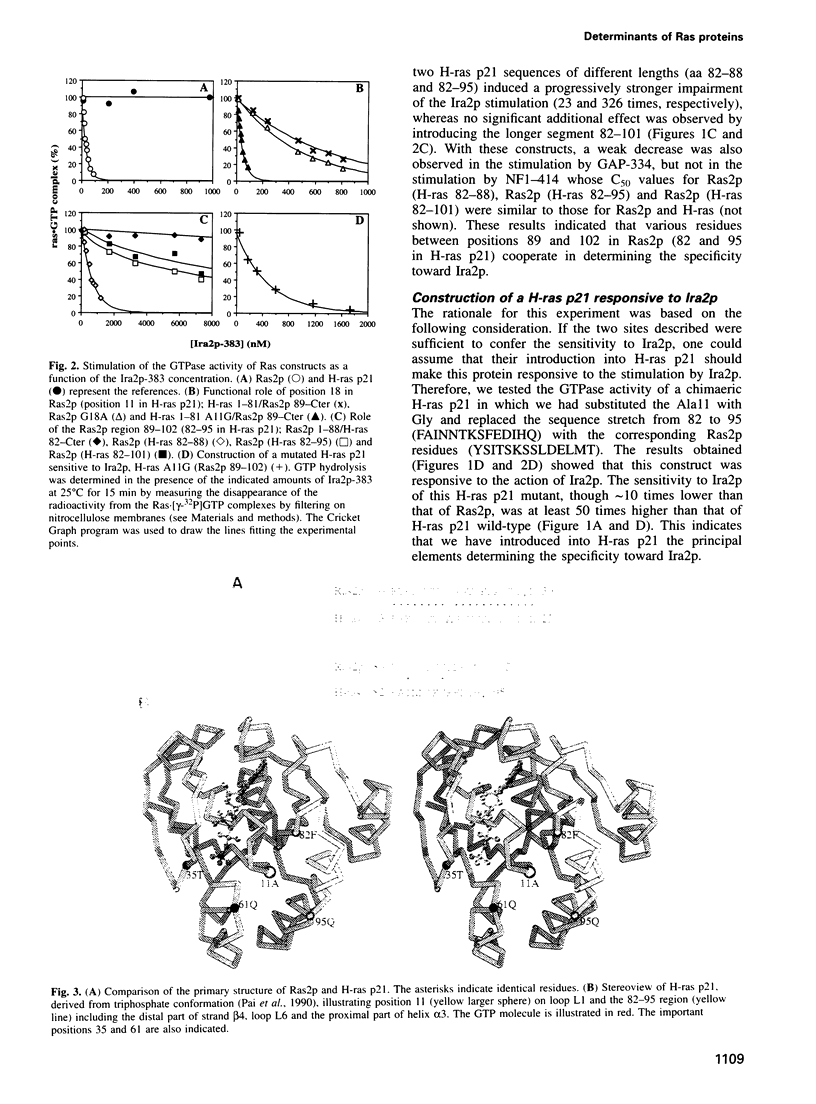
Human and Saccharomyces cerevisiae Ras proteins and their regulators GAP (GTPase activating protein)and GEF (guanine nucleotide exchange factor) display structural similarities and are functionally interchangeable in vivo and in vitro, indicating that the molecular mechanism regulating Ras proteins has been conserved during evolution. As the only exceptions, the two S.cerevisiae GAPs, Ira1p and Ira2p, are strictly specific for yeast Ras proteins and cannot stimulate the GTPase of mammalian Ras. This study searches for the reasons for the different sensitivity to Ira2p of human H-ras p21 and yeast Ras2p. Construction of H-ras/Ras2p chimaeras showed that Gly18 of Ras2p (Ala11 of H-ras p21) is an important determinant for the specificity of Ira2p, revealing for the first time a function for this position. A second even more crucial determinant was found to be the 89-102 region of Ras2p (82-95 of H-ras p21) including the distal part of strand beta4, loop L6 and the proximal part of helix alpha3. It was possible to construct Ras2p's resistant to Ira2p but still sensitive to human p120-GAP and, conversely, a H-ras p21 sensitive to Ira2p. This work helps clarify specific aspects of the conserved molecular mechanism of interaction between Ras proteins and their negative GAP regulators.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boguski M. S., McCormick F. Proteins regulating Ras and its relatives. Nature. 1993 Dec 16;366(6456):643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Chardin P., Camonis J. H., Gale N. W., van Aelst L., Schlessinger J., Wigler M. H., Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993 May 28;260(5112):1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- Gideon P., John J., Frech M., Lautwein A., Clark R., Scheffler J. E., Wittinghofer A. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol Cell Biol. 1992 May;12(5):2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet E., Parrini M. C., Bernardi A., Martegani E., Parmeggiani A. Properties of the catalytic domain of CDC25, a Saccharomyces cerevisiae GDP/GTP exchange factor: comparison of its activity on full-length and C-terminal truncated RAS2 proteins. Biochem Biophys Res Commun. 1994 Mar 15;199(2):497–503. doi: 10.1006/bbrc.1994.1256. [DOI] [PubMed] [Google Scholar]
- Jacquet E., Vanoni M., Ferrari C., Alberghina L., Martegani E., Parmeggiani A. A mouse CDC25-like product enhances the formation of the active GTP complex of human ras p21 and Saccharomyces cerevisiae RAS2 proteins. J Biol Chem. 1992 Dec 5;267(34):24181–24183. [PubMed] [Google Scholar]
- Maekawa M., Li S., Iwamatsu A., Morishita T., Yokota K., Imai Y., Kohsaka S., Nakamura S., Hattori S. A novel mammalian Ras GTPase-activating protein which has phospholipid-binding and Btk homology regions. Mol Cell Biol. 1994 Oct;14(10):6879–6885. doi: 10.1128/mcb.14.10.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S. The effector interactions of p21ras. Trends Biochem Sci. 1993 Jul;18(7):250–254. doi: 10.1016/0968-0004(93)90175-m. [DOI] [PubMed] [Google Scholar]
- Martegani E., Vanoni M., Zippel R., Coccetti P., Brambilla R., Ferrari C., Sturani E., Alberghina L. Cloning by functional complementation of a mouse cDNA encoding a homologue of CDC25, a Saccharomyces cerevisiae RAS activator. EMBO J. 1992 Jun;11(6):2151–2157. doi: 10.1002/j.1460-2075.1992.tb05274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta H., Holden J., Sizeland A., D'Abaco G. The residues of Ras and Rap proteins that determine their GAP specificities. J Biol Chem. 1991 Jun 25;266(18):11661–11668. [PubMed] [Google Scholar]
- Nur-E-Kamal M. S., Sizeland A., D'Abaco G., Maruta H. Asparagine 26, glutamic acid 31, valine 45, and tyrosine 64 of Ras proteins are required for their oncogenicity. J Biol Chem. 1992 Jan 25;267(3):1415–1418. [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini M. C., Jacquet E., Bernardi A., Jacquet M., Parmeggiani A. Properties and regulation of the catalytic domain of Ira2p, a Saccharomyces cerevisiae GTPase-activating protein of Ras2p. Biochemistry. 1995 Oct 24;34(42):13776–13783. doi: 10.1021/bi00042a008. [DOI] [PubMed] [Google Scholar]
- Polakis P., McCormick F. Structural requirements for the interaction of p21ras with GAP, exchange factors, and its biological effector target. J Biol Chem. 1993 May 5;268(13):9157–9160. [PubMed] [Google Scholar]
- Schweighoffer F., Faure M., Fath I., Chevallier-Multon M. C., Apiou F., Dutrillaux B., Sturani E., Jacquet M., Tocque B. Identification of a human guanine nucleotide-releasing factor (H-GRF55) specific for Ras proteins. Oncogene. 1993 Jun;8(6):1477–1485. [PubMed] [Google Scholar]
- Tanaka K., Lin B. K., Wood D. R., Tamanoi F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):468–472. doi: 10.1073/pnas.88.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990 Aug;10(8):4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D. R., Poullet P., Wilson B. A., Khalil M., Tanaka K., Cannon J. F., Tamanoi F. Biochemical characterization of yeast RAS2 mutants reveals a new region of ras protein involved in the interaction with GTPase-activating proteins. J Biol Chem. 1994 Feb 18;269(7):5322–5327. [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Zhang K., Papageorge A. G., Martin P., Vass W. C., Olah Z., Polakis P. G., McCormick F., Lowy D. R. Heterogeneous amino acids in Ras and Rap1A specifying sensitivity to GAP proteins. Science. 1991 Dec 13;254(5038):1630–1634. doi: 10.1126/science.1749934. [DOI] [PubMed] [Google Scholar]



