Abstract
Basidioascus undulatus is a soil basidiomycete belonging to the order Geminibasidiales. The taxonomic status of the order was unclear as originally it was only tentatively classified in the class Wallemiomycetes. The fungi in Geminibasidiales have an ambiguously defined sexual cycle. In this study, we sequenced the genome of B. undulatus to gain insights into its sexuality and evolutionary origins. The assembled genome draft was approximately 32 Mb in size, had a median nucleotide coverage of 24X, and contained 6123 predicted genes. Previous morphological descriptions of B. undulatus relied on interpretation of putative sexual structures. In this study, nuclear staining and confocal microscopy showed meiosis occurring in basidia and genome analysis confirmed the existence of genes involved in meiosis and mating. Using 35 protein-coding genes extracted from genomic information, phylogenomic and molecular dating analyses confirmed that B. undulatus indeed belongs to a lineage distantly related to Wallemia while retaining a basal position in Agaricomycotina. These results, combined with differences in septal pore morphology, led us to move the order Geminibasidiales out of the Wallemiomycetes and into the new class Geminibasidiomycetes cl. nov. Finally, the concept of Agaricomycotina is emended to include both Wallemiomycetes and Geminibasidiomycetes.
Keywords: Agaricomycotina, Basidiomycota, Geminibasidiomycetes, septal pore ultastructure, Wallemiomycetes
INTRODUCTION
Using morphological characters, Matsushima (2003) described Basidioascus undulatus as the only species in the new genus Basidioascus based on a single strain isolated from the tropical rainforest soil in Cape Tribulation National Park, Queensland, Australia. Matsushima (2003) photographed fertile structures of B. undulatus, which he interpreted as asci giving rise to ascospores, bearing hooks that could be either croziers or clamps. The genus was named Basidioascus after what he interpreted as “basidia-like asci” and the species epithet undulatus was given to recall “ascospores with a wavy wall” (Matsushima 2003). Also, B. undulatus produced a geotrichum-like asexual morph in culture, characterized by chains of aseptate arthroconidia. MycoBank (Robert et al. 2013) and Ainsworth & Bisby’s Dictionary of the Fungi (Kirk et al. 2008) classified B. undulatus in Saccharomycetes because the asexual morph was assumed to be a Geotrichum (a genus typified by an asexual morph and usually associated with sexual morphs in Galactomyces or Dipodascus)
During a survey of heat resistant fungi in Canadian soils, Nguyen et al. (2013) isolated nine additional strains of B. undulatus, and a second species of Basidioascus, named B. magus. A third yeast-like species, B. persicus, was recently described from soil in Iran (Nasr et al. 2014). Soil appears to be the main habitat for Basidioascus species and their distribution is probably broad. However, their ecological role is currently unknown, but they are presumably saprobic as are many soil inhabiting fungi (Domsch et al. 1980).
Surprisingly, phylogenetic analyses with rDNA sequences showed that Basidioascus was related to Wallemiomycetes (Basidiomycota) rather than Saccharomycetes (Ascomycota) (Nguyen et al. 2013). This finding initiated a revision of its taxonomy and a re-interpretation of its morphology as a basidiomycete. The structures identified as asci and ascospores by Matsushima (2003) are reinterpretted as thick-walled basidiospores, and the subtending cell as a basidium that usually produces a single basidiospore. Most unusual was that the basidia appeared to be forcibly discharged, leaving them collapsed with the basidiospore still attached by a long, cylindrical sterigma (Nguyen et al. 2013). The species of Basidioascus, and of its sister genus Geminibasidium (G. donsium and G. hirsutum), were classified in the new order Geminibasidiales (Nguyen et al. 2013). The Geminibasidiales are a phylogenetic sister group to Wallemiales and were placed tentatively under the class Wallemiomycetes (Nguyen et al. 2013). Wallemiales currently includes a single genus Wallemia with three species: W. sebi, W. muriae, and W. ichthyophaga (Zalar et al. 2005). The phylogenetic placement of Wallemiomycetes in the fungal kingdom was at first ambiguous (Matheny et al. 2006) because only a few protein coding genes were used in phylogenetic analyses and because ribosomal genes did not provide robustly supported conclusions. However, a few recent studies, through phylogenomic analyses with a large number of protein coding genes, demonstrate that this lineage is an early diverging one within Agaricomycotina (Padamsee et al. 2012, Zajc et al. 2013).
In this study, our first objective was to gain further insight into the sexuality of B. undulatus because the structures referred to as basidia and basidiospores were only putatively identified as such (Nguyen et al. 2013). For this purpose, we performed nuclear staining on these presumed sexual structures and observed them with laser confocal microscopy. Further, we sequenced the genome of B. undulatus and looked for genes involved in meiosis and mating to support our findings. Our second objective was to resolve the tentative placement of Geminibasidiales in Wallemiomycetes. We conducted phylogenomic analysis using 35 single copy protein-coding genes from the B. undulatus genome and we performed a molecular clock analysis to date the divergence of B. undulatus from Wallemia species and other fungi. The third objective was to investigate the septal pore morphology, which has proved significant in basidiomycete systematics, especially at class rank and particularly in lineages of Agaricomycotina (van Driel et al. 2009). We imaged the septal pore of B. undulatus, G. donsium, and W. sebi using transmission electron microscopy to support our interpretation of the higher classification of the Geminibasidiales.
MATERIALS AND METHODS
Growth, DNA extraction and sequencing
The ex-type strain of Basidioascus undulatus (DAOM 241956) was inoculated in 2 % malt extract broth in an Erlenmeyer flask on an orbital shaker at 25 °C for 2 wk. The broth culture was transferred to two 50 mL Falcon tubes and centrifuged at 10000 × g for 5 min. The liquid was decanted, leaving only the fungal tissue. The fungal tissue was frozen in liquid nitrogen and crushed with a sterile pestle. DNA was extracted with the OmniPrep kit (G-Biosciences, St Louis, MO) following the manufacturer’s instructions. DNA quality and quantity were verified with Qbit (Life Technologies, Burlington, Canada). Whole-genome sequencing (101 base pairs (bp) paired-end) was performed on an Illumina HiSeq 2500 with TrueSeq V3 chemistry at the National Research Council Canada facilty in Saskatoon (Saskatchewan).
Genome assembly and annotation
The quality of the reads was checked with the program FastQC v. 0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Using fastx_trimmer (part of the FASTX-Toolkit v. 0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/), eight bases from the 5’ end were trimmed to yield reads of 93 bp in length of higher quality. De novo assembly was performed using SPAdes v. 3.0 (Bankevich et al. 2012) with the BayesHammer error correction (Nikolenko et al. 2013) and mismatch correction enabled (parameters: --careful and k=21, 31, 41, 51, 61, 71, 81, 91). Final contigs were assembled into scaffolds with SSPACE v2.0 (Boetzer et al. 2011) (parameters: -x 1 -m 45 -o 10 -t 0 -r 0.7 -k 5 -a 0.7 -n 10 -z 2000 -T 16 -p 1) and contigs shorter than 2000 bp were discarded. Assembly statistics were generated with QUAST v. 2.3 (Gurevich et al. 2013). The assembly was checked by aligning the corrected reads onto the scaffolds using Bowtie2 v2.1.0 (Langmead & Salzberg 2012). Alignments produced by Bowtie2 in SAM format were converted to sorted BAM format by SAMtools v. 0.1.19 (Li et al. 2009) and statistics for coverage were generated with Qualimap v. 0.8.1 (Garcia-Alcalde et al. 2012). To benchmark the completeness of our genome assembly, CEGMA v. 2.5 (Parra et al. 2007) was run on the scaffolds to detect the percentage of conserved eukaryotic genes (CEGs).
Genome annotation was performed following established guidelines (Haas et al. 2011). Repeats in scaffold sequences were masked with RepeatMasker v. 4.0.5 (http://www.repeatmasker.org) (parameters: -no_is -species fungi) using the Repbase libraries (http://www.girinst.org/). The masked scaffolds were used as input for the MAKER2 v. 2.10 (Holt & Yandell 2011) genome annotation pipeline. In the MAKER2 pipeline, the GeneMark-ES v. 2.3e (Borodovsky & Lomsadze 2011) ab initio gene prediction tool was enabled and the NCBI RefSeq protein sequences were aligned to the genome using exonerate v. 2.2.0 (Slater & Birney 2005). Predicted gene models exhibiting strong evidence by exon alignment were exported as protein sequences and coding nucleotide sequences (CDS). Predicted gene models lacking evidence from exon alignment were discarded in downstream analyses. To determine function, the protein sequences were used as input for InterProScan 5RC6 (Jones et al. 2014) (parameters: -dp -f -t p -iprlookup -pa -goterms) and were also compared to the manually curated protein data set from UniProt/Swiss-Prot by blastp v. 2.2.28+. The results in XML format from blastp v. 2.2.28+ and InterProScan were loaded into Blast2GO v. 2.7.1 (Conesa et al. 2005) and merged to create an annotation table (available from the first author on request). The gene models with BLAST hits having e-value of less than 1.0E-100 and mean similarity hit of ≥ 70% were assumed to be orthologs and they were given names following recommended conventions (http://www.uniprot.org/docs/proknameprot). Ribosomal RNA’s were predicted by RNAmmer v. 1.2 (Lagesen et al. 2007). Data files are publicly available at NCBI (Genome Accession No. JTLS00000000 version JTLS01000000; BioProject Accession No. PRJNA247992) and JGI MycoCosm portal (Grigoriev et al. 2014).
Identification of meiosis and mating genes
BLAST was used for finding the Basidioascus undulatus mating and meiosis genes. The protein sequences predicted from evidence-supported gene models, determined above, were formatted into a local BLAST database with makeblastdb v. 2.2.28+ (Camacho et al. 2009).
Genes previously determined to be involved in meiosis in Saccharomyces cerevisiae and Cryptococcus neoformans (Halary et al. 2011) were chosen as input queries (e-value cut off < 1.0E-05) for the B. undulatus protein BLAST database using blastp v. 2.2.28+. Saccharomyces cerevisiae and C. neoformans were chosen because they are well studied genetically and all meiosis-specific proteins are present (Halary et al. 2011).
Mating genes were located with the protein domains identified by InterProScan, and by blastp v. 2.2.28+ using known mating genes in Saccharomyces cerevisiae as blastp input queries (e-value cut off < 1.0E-05).
Phylogenomics and molecular dating
Protein sequences of selected fungi (Agaricus bisporus var. bisporus (H97) v. 2.0 (Morin et al. 2012), Alternaria brassicicola (Ohm et al. 2012), Arthrobotrys oligospora ATCC 24927 (Yang et al. 2011), Aureobasidium pullulans var. pullulans EXF-150 (Gostincar et al. 2014), Auricularia subglabra v. 2.0 (Floudas et al. 2012), Botrytis cinerea v. 1.0 (Staats and van Kan 2012), Coccidioides immitis RS (Sharpton et al. 2009), Coprinopsis cinerea (Stajich et al. 2010), Cryptococcus neoformans var. neoformans JEC21 (Loftus et al. 2005), Dacryopinax sp. DJM 731 SSP1 v. 1.0 (Floudas et al. 2012), Fomitiporia mediterranea v. 1.0 (Floudas et al. 2012), Fomitopsis pinicola FP-58527 SS1 v. 3.0 (Floudas et al. 2012), Malassezia globosa (Xu et al. 2007), Mixia osmundae IAM 14324 v. 1.0 (Toome et al. 2014b), Monacrosporium haptotylum CBS 200.50 (Meerupati et al. 2013), Neurospora crassa OR74A v. 2.0 (Galagan et al. 2003), Penicillium chrysogenum Wisconsin 54-1255 (van den Berg et al. 2008), Puccinia striiformis f. sp. tritici PST-130 (Cantu et al. 2011), Pyronema confluens CBS 100304 (Traeger et al. 2013), Rhizophagus irregularis DAOM 181602 v. 1.0 (Tisserant et al. 2013), Saccharomyces cerevisiae S288C (Goffeau et al. 1996), Sclerotinia sclerotiorum v. 1.0 (Amselem et al. 2011), Taphrina deformans (Cisee et al. 2013), Tilletiaria anomala (Toome et al. 2014a), Tremella mesenterica Fries v. 1.0 (Floudas et al. 2012), Trichoderma atroviride v. 2.0 (Kubicek et al. 2011), Tuber melanosporum (Martin et al. 2010), Ustilago maydis (Kamper et al. 2006), Wallemia ichthyophaga EXF-994 (Zajc et al. 2013), and W. sebi v. 1.0 (Padamsee et al. 2012)) were downloaded from the JGI MycoCosm portal (Grigoriev et al. 2014) and formatted into separate BLAST databases with makeblastdb v. 2.2.28+. The 246 reliable single copy ortholog protein data set from FUNYbase (Marthey et al. 2008) was downloaded. Only amino acid sequences coming from nuclear genes yielding a topological score of > 90 % (Marthey et al. 2008) were considered for our phylogenomic analysis (information on exact genes chosen are available from the first author on request). These amino acid sequences were used as blastp v. 2.2.28+ search queries (e-value threshold < 1.0E-05) against the protein databases built from data downloaded from JGI MycoCosm described above. Protein sequences were aligned with T-Coffee v10.00.r1613 (Notredame et al. 2000) (parameters: t_coffee sequence.fasta -output score_ascii, aln) and poorly aligned regions and columns containing gaps were automatically discarded (parameters: t_coffee -other_pg seq_reformat -in sequence.aln -struc_in sequence.score_ascii -struc_in_f number_aln -action +use_cons +keep ‘[8-9]’ +rm_gap 1 > sequence.best.aln). The alignments were concatenated and converted to PHYLIP format with SeaView v. 4.5.3 (Gouy et al. 2010). Three independent phylogenomic analyses were performed with the parallelized version of PhyloBayes 3 (pb_mpi v1.4) (Lartillot et al. 2013, Lartillot et al. 2009) using the CAT-GTR model (Lartillot & Philippe 2004). Analyses were stopped when convergence was attained (effective size > 100 and maxdiff < 0.1 determined with the programs bpcomp (parameters: -x 1000 50 run1 run2 run3) and tracecomp (parameters: -x 1000 run1 run2 run3), which are part of pb_mpi software package). A lognormal ‘relaxed clock’ molecular dating analysis was performed with the non-parallelized version of PhyloBayes 3 (pb v3.2e) with a birth-death prior using the tree topology generated from the converged Bayesian analysis above (parameters: -d combined.phy -T bpcomp.con.tre -r outgroup.txt -cal calib.txt -ln -bd md1). Rhizophagus irregularis (Glomeromycota) was specified as the outgroup. Date constraints previously determined were used as calibrations (Hibbett et al. 1997, Smith et al. 2004, Taylor & Berbee 2007, Berbee & Taylor 2010, Prieto & Wedin 2013); the exact calibrations are available from the first author on request. Chronograms and statistics were obtained with readdiv (parameters: -x 1000 50 md1), which is part of PhyloBayes 3.
Confocal laser scanning microscopy
To study nuclear behaviour and to look for indicators of meiosis, Basidioascus undulatus DAOM 241956 was grown on corn meal agar (CMA, Acumedia Manufacturers, Lansing, MI) for 1 wk and mounted in DNA stains: DAPI-Fluoromount-GTM mounting medium (EMS, Hatfield, PA) or aqueous SYTO 9 (25 μM) (Life Technologies, Burlington, ON). Samples were visualized under confocal laser scanning microscopy using an LSM 510 DUO (Carl Zeiss MicroImaging, Göttingen, Germany) with a Plan-Apochromat 40×/1.4 Oil DIC objective and electronic zoom 4. An excitation diode laser (405 nm) and emission light (420–700 nm) were used for DAPI. An excitation Argon laser (488 nm) and emission light (505–550 nm) were used for SYTO 9. Images were captured using ZEN 2009 Imaging Software (Carl Zeiss MicroImaging).
Transmission electron microscopy
Actively growing hyphae of Basidioascus undulatus DAOM 241956, Geminibasidium donsium DAOM 241966, and Wallemia sebi CBS 633.66 were prepared for transmission electron microscopy using cryo-preparation methods. Hyphae were grown on thin, sterile, deionized dialysis membrane segments overlaying appropriate media at 23 ºC. The leading edge of growing mycelia and supporting membranes were trimmed with a sharp razor blade to approximately 5 × 5 mm and after 30‒40 min (time to recover from trimming) were removed from the agar surface and immediately cryo-fixed by rapid plunging into liquid pro‐pane cooled to –186 ºC with liquid nitrogen (Hoch 1986, Roberson & Fuller 1988, McDaniel & Roberson 2000). After rapid freezing, the samples were freeze-substituted in 1 % glutaraldehyde (w/v) and 1 % tannic acid (w/v) in anhydrous acetone at −85°C for 72 h. After washing in cold acetone (−85 °C), the samples were warmed slowly to room temperature in 1 % OsO4 (w/v) in acetone, washed in acetone, and infiltrated and flat em‐bedded on glass slides in Spurr’s resin (Spurr 1969). Using phase contrast optics (100×), we examined the slides for well-preserved hyphae and mounted the selected cells on resin blocks (Howard & O’Donnell 1987) then hand-trimmed them. Selected hyphae were sec‐tioned using a Leica Ultracut R ultramicrotome (Leica Microsystems, Bannockburn, IL), collected on copper grids, and post-stained for 10 min in 2 % uranyl acetate in 50 % ethanol and for 5 min in Sato’s lead citrate (Hanaichi et al. 1986). Sections were then examined using a JEOL 1200EX (JEOL, Tokyo,) transmission electron microscope equipped with a SIA L3C CCD camera (SIA, Duluth, GA). Measurements from captured images were made with ImageJ (Schneider et al. 2012).
RESULTS
Genome sequencing, assembly and annotation
Short-read Illumina sequencing generated approximately 12 million paired end reads (six million reads in the forward (R1) direction, and six million reads in the reverse (R2) direction) of 101 bp in length each. After trimming to 93 bp, about 1.1 Gb of data was assembled de novo to yield a genome assembly size of about 32 Mb. The GC content in Basidioascus undulatus was 58 %. The final assembly contained 2992 scaffolds and the longest scaffold was 97 Kb. The nucleotide coverage varied for each scaffold but it was 28X on average and the median nucleotide coverage was 24X. The N50 statistic was 15 Kb. According to CEGMA, 81 % and 88 % of complete and partial CEGs, were detected respectively.
A total of 13935 gene models were detected ab initio using GeneMark-ES, but only 6123 gene models were supported by evidence from protein alignment to the NCBI RefSeq fungal protein data. We kept the set of 6123 gene models with evidence for further manual annotation and used it in downstream analyses. Only 3681 of these (60 %) were considered complete because they contained a start and stop codon while the remaining 2442 (40 %) lacked either a start codon, a stop codon, or both.
Meiosis and meiosis specific genes
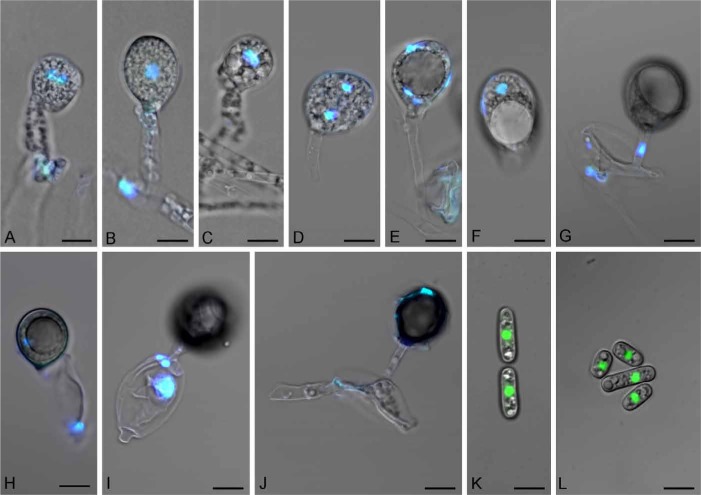
To determine whether the putative sexual structures identified by Nguyen et al. (2013) truly represented basidia, we followed the fungus’s ontogeny and performed nuclear staining by laser confocal microscopy (Fig. 1). Three nuclear divisions normally occur during basidiospore maturation: meiosis I, meiosis II, followed by four different patterns of post-meiotic mitosis (Duncan & Galbraith 1972). We observed dikaryotic nuclei in the basidia (Fig. 1A), karyogamy (Fig. 1B), anaphase I (Fig. 1C), and the telophase I stage where the basal lateral projection was collapsed (Fig. 1D). Four nuclei from telophase II (Fig. 1E) were seen in both non-discharged and discharged basidia (Fig. 1F). The migration of a nucleus through the sterigma (Fig. 1G) into the basidiospore (Fig. 1H) was observed as the basidiospore matured. There were three remaining nuclei in the collapsing basidium (Fig. 1I) and they eventually degenerated at a later stage when the basidium completely collapsed (Fig. 1J). Whole arthroconidia became stained instead of just their nuclei when DAPI was used. A single nucleus could be spotted in each arthroconidium when stained with SYTO 9 (Fig. 1K–L).
Fig. 1.
Basidioascus undulatus (DAOM 241956) sexual and asexual structures stained with DAPI (A–J) and SYTO 9 (K–L) and imaged with confocal microscopy. A. Dikaryotic basidium (2 nuclei). B. Karyogamy. C. Anaphase I. D. Telophase I and collapsed basal lateral projection. E. Telophase II (4 nuclei). F. Ejected basidium (4 nuclei). G–H. Maturation of a basidiospore on a basidium and the migration of a nucleus through the sterigma and into the basidiospore. I. Collapsing basidium with the three remaining nuclei. J. A totally collapsed basidium and probably one nucleus inside the mature basidiospore. K–L. Single nuclei inside arthroconidia. Bar = 5 μm.
Our microscopic observations correlated with our analysis of the genome to detect meiotic genes (Table 1). All meiosis-specific genes, as defined in Malik et al. (2008) and Halary et al. (2011), were found in the genome.
Table 1.
List of known and putative meiosis genes in the genome of Saccharomyces cerevisiae, Cryptococcus neoformans and Basidioascus undulatus. Meiosis specific proteins are highlighted in red.
| Process | Gene name | Saccharomyces cerevisiaea | Cryptococcus neoformansb | Basidioascus undulatus vs. Saccharomyces cerevisiae | Basidioascus undulatus vs. Cryptococcus neoformans | Basidioascus undulatus | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus ID | Locus ID | Score | E-value | % identity | % positive | Locus ID | Score | E-value | % identity | % positive | Probable Locus ID | |||
| DSB generation | SPO11 | YHL022C | XM_767420.1 | 102794F55C | 72.8 | 0.0 | 25.46 | 43.54 | 102794F55C | 191 | 0.0 | 32.92 | 51.49 | 102794F55C |
| REC107/MEI2 | YJR021C | Absent | Not detected | NA | Absent or not detected | |||||||||
| MEI4 | YER044C | Absent | Not detected | NA | Absent or not detected | |||||||||
| REC102 | YLR329W | Absent | Not detected | NA | Absent or not detected | |||||||||
| REC104 | YHR157W | Absent | Not detected | NA | Absent or not detected | |||||||||
| REC114 | YMR133W | Absent | Not detected | NA | Absent or not detected | |||||||||
| SKI8 | YGL213C | XP_567964.1 | Not detected | A5C17B7D33 | 79.3 | 0.0 | 40 | 54.29 | A5C17B7D33 | |||||
| MER1 | YNL210W | Absent | Not detected | NA | Absent or not detected | |||||||||
| HFM1/MER3 | YGL251C | XP_774045.1 | 268E1B92EF | 286 | 0.0 | 30.69 | 50.14 | 268E1B92EF | 285 | 0.0 | 29.55 | 48.66 | 268E1B92EF | |
| NAM8/MRE2 | YHR086W | XP_568215.1 | 012CC612A1 | 88.6 | 0.0 | 27.24 | 46.24 | C3284824DB | 211 | 0.0 | 75.57 | 83.21 | Conflict | |
| Removal of | MRE11 | YMR224C | XP_571170.1 | 550A163039 | 365 | 0.0 | 40.59 | 58.02 | 550A163039 | 629 | 0 | 56.55 | 74.34 | 550A163039 |
| Spo11 | RAD50 | YNL250W | XP_771929.1 | 1CAAD52FF0 | 543 | 0.0 | 30.88 | 52.16 | 1CAAD52FF0 | 869 | 0 | 38.91 | 61.16 | 1CAAD52FF0 |
| XRS2/NBS1 | YDR369C | Absent | Not detected | NA | Absent or not detected | |||||||||
| SAE2/COM1 | YGL175C | Absent | Not detected | NA | Absent or not detected | |||||||||
| Strand invasion | RAD51 | YER095W | XP_567016.1 | D36F2DAE01 | 481 | 0.0 | 69.28 | 85.58 | D36F2DAE01 | 533 | 0 | 77.71 | 84.46 | D36F2DAE01 |
| DMC1 | YER179W | XP_772121.1 | 1B6A6FD704 | 376 | 0.0 | 58.13 | 75.94 | 1B6A6FD704 | 388 | 0.0 | 67.13 | 80.28 | 1B6A6FD704 | |
| RAD52 | YML032C | XP_569087.1 | EA2B481C23 | 186 | 0.0 | 51.9 | 74.05 | EA2B481C23 | 237 | 0.0 | 60.57 | 78.29 | EA2B481C23 | |
| RAD54 | YGL163C | XP_570462.1 | 4E6C655AE5 | 525 | 0.0 | 61.26 | 78.21 | 4E6C655AE5 | 599 | 0 | 71.22 | 82.93 | 4E6C655AE5 | |
| RDH54 | YBR073W | Absent | ED83E37023 | 476 | 0.0 | 42.43 | 60.41 | NA | ED83E37023 | |||||
| RFA1 | YAR007C | XP_775959.1 | 1512E55FF2 | 350 | 0.0 | 41.02 | 62.08 | 1512E55FF2 | 489 | 0.0 | 51.93 | 70.39 | 1512E55FF2 | |
| RFA2 | YNL312W | XP_776149.1 | C06D199EC7 | 95.9 | 0.0 | 37.78 | 55.56 | C06D199EC7 | 174 | 0.0 | 40.51 | 61.18 | C06D199EC7 | |
| RFA3 | YJL173C | Absent | Not detected | NA | Absent or not detected | |||||||||
| SAE3 | YHR079C-A | Absent | Not detected | NA | Absent or not detected | |||||||||
| RAD55 | YDR076W | Absent | Not detected | NA | Absent or not detected | |||||||||
| DNA damage | PCH2 | YBR186W | XP_567632.1 | 1586D154C9 | 177 | 0.0 | 36.88 | 53.49 | 1586D154C9 | 418 | 0.0 | 47.79 | 66.81 | 1586D154C9 |
| checkpoint | MEC1 | YBR136W | XP_568889.1 | E568B50733 | 483 | 0.0 | 24.05 | 43.61 | 87D911C03D | 713 | 0 | 61.72 | 76.24 | Conflict |
| RAD17 | YOR368W | XP_569244.1 | Not detected | D5120AB0E3 | 171 | 0.0 | 34.76 | 48.66 | D5120AB0E3 | |||||
| RAD24 | YER173W | Absent | D5120AB0E3 | 54.3 | 0.000 | 27.48 | 43.51 | NA | D5120AB0E3 | |||||
| DDC1 | YPL194W | Absent | Not detected | NA | Absent or not detected | |||||||||
| Regulation of | MLH1 | YMR167W | XP_571158.1 | F0BB260AD1 | 361 | 0.0 | 53.03 | 70.03 | F0BB260AD1 | 642 | 0 | 51.99 | 66.51 | F0BB260AD1 |
| crossover | MLH3 | YPL164C | XP_570272.1 | BC2D40152B | 115 | 0.0 | 23.47 | 44.13 | BC2D40152B | 178 | 0.0 | 37.67 | 57.88 | BC2D40152B |
| frequency | MSH4 | YFL003C | XP_773414.1 | CAE76A5567 | 347 | 0.0 | 31.07 | 50.64 | CAE76A5567 | 546 | 0 | 39.76 | 59.04 | CAE76A5567 |
| MSH5 | YDL154W | XP_566842.1 | A5F9B81D55 | 304 | 0.0 | 30.46 | 52.12 | A5F9B81D55 | 436 | 0.0 | 39.91 | 58.36 | A5F9B81D55 | |
| SGS1 | YMR190C | XP_776787.1 | FAB5DF8B5F | 526 | 0.0 | 45.14 | 64.24 | FAB5DF8B5F | 597 | 0 | 43.47 | 60.52 | FAB5DF8B5F | |
| MEI5 | YPL121C | Absent | Not detected | NA | Absent or not detected | |||||||||
| MUM2 | YBR057C | Absent | Not detected | NA | Absent or not detected | |||||||||
| NDJ1 | YOL104C | Absent | Not detected | NA | Absent or not detected | |||||||||
| RAD1 | YPL022W | XP_772313.1 | 29F5BE17B9 | 258 | 0.0 | 33.14 | 50.95 | 29F5BE17B9 | 374 | 0.0 | 40.1 | 56.48 | 29F5BE17B9 | |
| Rad2 | YGR258C | XP_566738.1 | 41955FBDBD | 248 | 0.0 | 44.04 | 67.51 | 41955FBDBD | 355 | 0.0 | 54.75 | 70.82 | 41955FBDBD | |
| Synaptonemal | HOP1 | YIL072W | 50255507 | D659917BB2 | 141 | 0.0 | 31.54 | 56.54 | D659917BB2 | 170 | 0.0 | 34.16 | 50 | D659917BB2 |
| complex | HOP2 | YGL033W | 50255433 | Not detected | Not detected | Absent or not detected | ||||||||
| MND1 | YGL183C | XP_772550.1 | 5756414F12 | 62 | 0.0 | 20.94 | 43.59 | 5756414F12 | 115 | 0.0 | 37.13 | 51.05 | 5756414F12 | |
| ZIP1 | YDR285W | Absent | 2C075CC8D6 | 204 | 0.0 | 79.7 | 86.47 | NA | 2C075CC8D6 | |||||
| ZIP2 | YGL249W | Absent | Not detected | NA | Absent or not detected | |||||||||
| ZIP3 | YLR394W | Absent | Not detected | NA | Absent or not detected | |||||||||
| Zip4/Spo22 | YIL073C | 134115038 | Not detected | Not detected | Absent or not detected | |||||||||
| DNA Repair | HTA1 | YDR225W | XP_567962.1 | 2C075CC8D6 | 204 | 0.0 | 79.7 | 86.47 | 2C075CC8D6 | 130 | 0.0 | 57.98 | 73.11 | 2C075CC8D6 |
| HTA2 | YBL003C | XP_569065.1 | 2C075CC8D6 | 206 | 0.0 | 80.45 | 87.22 | F84A4F188E | 205 | 0.0 | 79.39 | 87.79 | Conflict | |
| RED1 | YLR263W | Absent | Not detected | NA | Absent or not detected | |||||||||
| SMC5 | YOL034W | XP_570071.1 | 8388BCCBA3 | 362 | 0.0 | 27.24 | 48.17 | 8388BCCBA3 | 529 | 0.0 | 32.52 | 51.9 | 8388BCCBA3 | |
| SMC6 | YLR383W | XP_775824.1 | 13E0CFCD07 | 340 | 0.0 | 27.58 | 47.32 | 13E0CFCD07 | 476 | 0.0 | 29.98 | 50.65 | 13E0CFCD07 | |
| EXO1 | YOR033C | XP_777034.1 | 36DFC1EC56 | 276 | 0.0 | 43.67 | 65.33 | 36DFC1EC56 | 389 | 0.0 | 54.49 | 71.07 | 36DFC1EC56 | |
| HRR25 | YPL204W | XP_570121.1 | 22D0CDA5D9 | 464 | 0.0 | 65.33 | 83.28 | 22D0CDA5D9 | 594 | 0 | 95.22 | 98.63 | 22D0CDA5D9 | |
| RAD23 | YEL037C | XP_777611.1 | 0FBB054DC3 | 54.7 | 0.000 | 40.54 | 60.81 | 0FBB054DC3 | 91.3 | 0.0 | 60.81 | 78.38 | 0FBB054DC3 | |
| Mismatch repair | MSH2 | YOL090W | XP_567098.1 | E449052477 | 746 | 0 | 43.93 | 63.77 | E449052477 | 1087 | 0 | 58.26 | 73.21 | E449052477 |
| MSH3 | YCR092C | XP_569494.1 | 7F6ED5B406 | 354 | 0.0 | 29.13 | 49.57 | 7F6ED5B406 | 790 | 0 | 45.31 | 65.61 | 7F6ED5B406 | |
| MSH6 | YDR097C | XP_772722.1 | 122F99619E | 672 | 0 | 36.44 | 56.87 | 122F99619E | 1026 | 0 | 51.24 | 66.83 | 122F99619E | |
| MLH2 | YLR035C | Absent | 8C03A290CE | 86.7 | 0.0 | 31.93 | 53.61 | NA | 8C03A290CE | |||||
| PMS1 | YNL082W | 57225775 | 8C03A290CE | 176 | 0.0 | 48.65 | 67.03 | 8C03A290CE | 236 | 0.0 | 60.51 | 75.38 | 8C03A290CE | |
| Resolution of | MMS4 | YBR098W | 58260752 | Not detected | Not detected | Not detected or absent | ||||||||
| recombination | MUS81 | YDR386W | XP_777360.1 | E55E7AB0BD | 84 | 0.0 | 30.49 | 49.78 | E55E7AB0BD | 101 | 0.0 | 32.39 | 50.7 | E55E7AB0BD |
| intermediates | SLX1 | YBR228W | XP_567159.1 | A74C037F01 | 83.2 | 0.0 | 52.44 | 68.29 | A74C037F01 | 108 | 0.0 | 61.54 | 74.36 | A74C037F01 |
| TOP1 | YOL006C | XP_572925.1 | DB366D3081 | 439 | 0.0 | 49.78 | 66.89 | DB366D3081 | 518 | 0.0 | 56.71 | 70.56 | DB366D3081 | |
| TOP2 | YNL088W | XP_566700.1 | 4AE1080675 | 1197 | 0 | 52.97 | 71.47 | 4AE1080675 | 1491 | 0 | 66.48 | 79.24 | 4AE1080675 | |
| TOP3 | YLR234W | XP_773035.1 | BCF17A4E2A | 266 | 0.0 | 43.67 | 60.54 | BCF17A4E2A | 518 | 0.0 | 57.95 | 70.91 | BCF17A4E2A | |
| SLX4 | YLR135W | Absent | Not detected | Not detected or absent | ||||||||||
| SLX5 | YDL013W | Absent | Not detected | Not detected or absent | ||||||||||
| SLX8 | YER116C | Absent | 52369A1C42 | 52.4 | 0.000 | 35.9 | 48.72 | 52369A1C42 | ||||||
| Nonhomologous | YKU70 | YMR284W | XP_573016.1 | AD4E738F04 | 115 | 0.0 | 24.21 | 42.14 | AD4E738F04 | 228 | 0.0 | 32.28 | 47.76 | AD4E738F04 |
| end joining | YKU80 | YMR106C | XP_568810.1 | 1FC448F440 | 60.8 | 0.000 | 30.29 | 48.57 | 1FC448F440 | 241 | 0.0 | 28.83 | 46.23 | 1FC448F440 |
| DNL4 | YOR005C | XP_572602.1 | 9206958189 | 223 | 0.0 | 27.5 | 43.38 | E8D29BB570 | 902 | 0 | 71.5 | 83.49 | Conflict | |
| LIF1 | YGL090W | Absent | Not detected | NA | Absent or not detected | |||||||||
| Other | MSC1 | YML128C | XP_570348.1 | 6453C9DC91 | 53.1 | 0.000 | 22.02 | 45.83 | 6453C9DC91 | 168 | 0.0 | 34.33 | 51.12 | 6453C9DC91 |
| MSC7 | YHR039C | XP_773481.1 | AE68F812D8 | 179 | 0.0 | 26.57 | 46.65 | AE68F812D8 | 185 | 0.0 | 28.6 | 45.59 | AE68F812D8 | |
| MSC3 | YLR219W | Absent | Not detected | NA | Absent or not detected | |||||||||
| MSC6 | YOR354C | Absent | Not detected | NA | Absent or not detected | |||||||||
| SRS2 | YJL092W | Absent | C8820A864B | 273 | 0.0 | 28.95 | 44.65 | NA | C8820A864B | |||||
| MPS3 | YJL019W | Absent | Not detected | NA | Absent or not detected | |||||||||
| REC8 | YPR007C | 134090540 | Not detected | 130D4D9937 | 65.1 | 0.0 | 38.04 | 59.78 | 130D4D9937 | |||||
| Mcd1/Rad21 | 6320201b | 58266400 | 130D4D9937 | 68.6 | 0.0 | 35.05 | 57.73 | 130D4D9937 | 142 | 0.0 | 32.61 | 46.65 | 130D4D9937 | |
| SMC1 | BAA09230.1b | XP_568851.1 | 82DC381A41 | 534 | 0.0 | 31.89 | 54.54 | 82DC381A41 | 894 | 0 | 44.85 | 64.74 | 82DC381A41 | |
| SMC2 | P38989.1b | XP_572171.1 | 68E0241B20 | 516 | 0.0 | 42.44 | 63.29 | 68E0241B20 | 720 | 0 | 58.48 | 70.98 | 68E0241B20 | |
| SMC3 | CAA74655.1b | XP_570201.1 | 10D8B5FBE4 | 248 | 0.0 | 41.53 | 60.17 | 10D8B5FBE4 | 404 | 0.0 | 57.95 | 72.56 | 10D8B5FBE4 | |
| SMC4* | EDV09389.1b | XP_571168.1 | C71F05C621 | 895 | 0 | 41.41 | 60.31 | C71F05C621 | 1367 | 0 | 56.6 | 73.61 | C71F05C621 | |
| SCC3 | P40541.1b | XP_567136.1 | Not detected | Not detected | Absent or not detected | |||||||||
| PDS5 | Q04264.1b | XP_567466.1 | 062A151CC3 | 326 | 0.0 | 24.85 | 45.12 | 062A151CC3 | 681 | 0 | 35.27 | 55.63 | 062A151CC3 | |
a These LocusID’s are from Saccharomyces Genome Database (http://www.yeastgenome.org/).
b These LocusID’s are from NCBI Protein database (http://www.yeastgenome.org/).
Mating genes
We looked for mating genes such as those encoding for homeodomain proteins, G-protein coupled pheromone receptor, high mobility group (HMG) DNA binding proteins, mitogen-activated protein kinases (MAPK, MAPKK, MAPKKK) and the subunits of the trimeric GTPase protein (Gα, Gβ, Gγ) (James et al. 2013). We followed the Saccharomyces cerevisiae standard for gene names. Most of the mating response genes and the pheromone processing genes (Table 2) were detected in the B. undulatus genome. All of the mating genes detected were located on different scaffolds, except for KSS1 and FUS3, which co-occurred on scaffold 2837. The presence of most mating genes suggests that a mating type locus exists, but its structure and gene order could not be determined because our assembled genome was too fragmented.
Table 2.
List of mating genes.
| Process | Gene name | Saccharomyces cerevisiaea | Basidioascus undulatus vs. Saccharomyces cerevisiae | ||||
|---|---|---|---|---|---|---|---|
| Locus ID | Locus ID | Score | E-value | % identity | % positive | ||
| Mating response | STE3 | YKL178C | 601070A992 | 84.7 | 0.0 | 25.17 | 44.37 |
| KSS1 | YGR040W | 03DD35A234 | 341 | 0.0 | 58.5 | 72.45 | |
| FUS3 | YBL016W | 03DD35A234 | 355 | 0.0 | 57.48 | 74.83 | |
| STE7 | YDL159W | C56EA11F93 | 209 | 0.0 | 49.32 | 66.06 | |
| STE11 | YLR362W | A726A87E58 | 319 | 0.0 | 52.6 | 67.86 | |
| GPA1 | YHR005C | 00D128FF38 | 243 | 0.0 | 55 | 75.5 | |
| STE4 | YOR212W | 51579617AB | 195 | 0.0 | 43.67 | 61.22 | |
| STE18 | YJR086W | Not detected | |||||
| STE20 | YHL007C | ABF41DDAEF | 375 | 0.0 | 59.26 | 76.09 | |
| STE12 | YHR084W | Not detected | |||||
| FAR1 | YJL157C | Not detected | |||||
| STE5 | YDR103W | Not detected | |||||
|
| |||||||
| Pheromone processing | KEX1 | YGL203C | C1EB8324D8 | 249 | 0.0 | 34.79 | 51.2 |
| KEX2 | YNL238W | 1DAB65F8AC | 480 | 0.0 | 47.45 | 63.53 | |
| STE13 | YOR219C | C51BE5DE56 | 131 | 0.0 | 42.11 | 63.16 | |
| RAM1 | YDL090C | B544DBFB4B | 169 | 0.0 | 37.09 | 53.31 | |
| RAM2 | YKL019W | B2B303A077 | 82.4 | 0.0 | 30.73 | 45.31 | |
| RCE1 | YMR274C | Not detected | |||||
| STE24 | YJR117W | Not detected | |||||
| STE14 | YDR410C | CC90F4A946 | 99.8 | 0.0 | 45.45 | 58.68 | |
| AXL1 | YPR122W | 98D2221B58 | 129 | 0.0 | 30.38 | 51.9 | |
| STE6 | YKL209C | ED8249ED54 | 359 | 0.0 | 25.6 | 43.36 | |
a: These LocusID’s are from Saccharomyces Genome Database (http://www.yeastgenome.org/).
Septal pore morphology
We imaged the septal pores of Wallemia sebi, Basidioascus undulatus, and Gemmibasidium donsium, all currently classified in Wallemiomycetes, using identical fixation methods to strengthen comparisons (Fig. 2). Basidioascus undulatus, G. donsium and W. sebi all have a dolipore septum characteristic of Agaricomycotina. The pore swelling of B. undulatus and G. donsium is electron dense, but is not in W. sebi. The adseptal tubular extensions that arise from sheets of endoplasmic reticulum in W. sebi were obvious (Fig. 2E–F) but they were either absent or unclear in B. undulatus (Fig. 2A–B) and G. donsium (Fig. 2C–D). Electron-dense regions were evident near the septal pore in all three fungi, but this area was denser in W. sebi than in B. undulatus and G. donsium. An electron-dense septal pore occlusion extending across the septal pore was observed in all three fungi. This occlusion was non-membranous in W. sebi, which agrees with previously published findings (Terracina 1974, Padamsee et al. 2012) and the W. sebi data on the AFTOL Structural and Biochemical Database (Celio et al. 2007). The septal pore occlusion in B. undulatus and G. donsium had a membrane, making it more clearly defined compared to that of W. sebi. Furthermore, this occlusion was almost twice as wide in B. undulatus and G. donsium (~0.12 μm) compared with W. sebi (~0.06 μm). Also, the septal pore of W. sebi had striations that appeared to be fine fibrils vertically arranged at the pore opening, but these were absent in B. undulatus and G. donsium. We did not see a septal pore cap in our sections of W. sebi, B. undulatus or G. donsium.
Fig. 2.
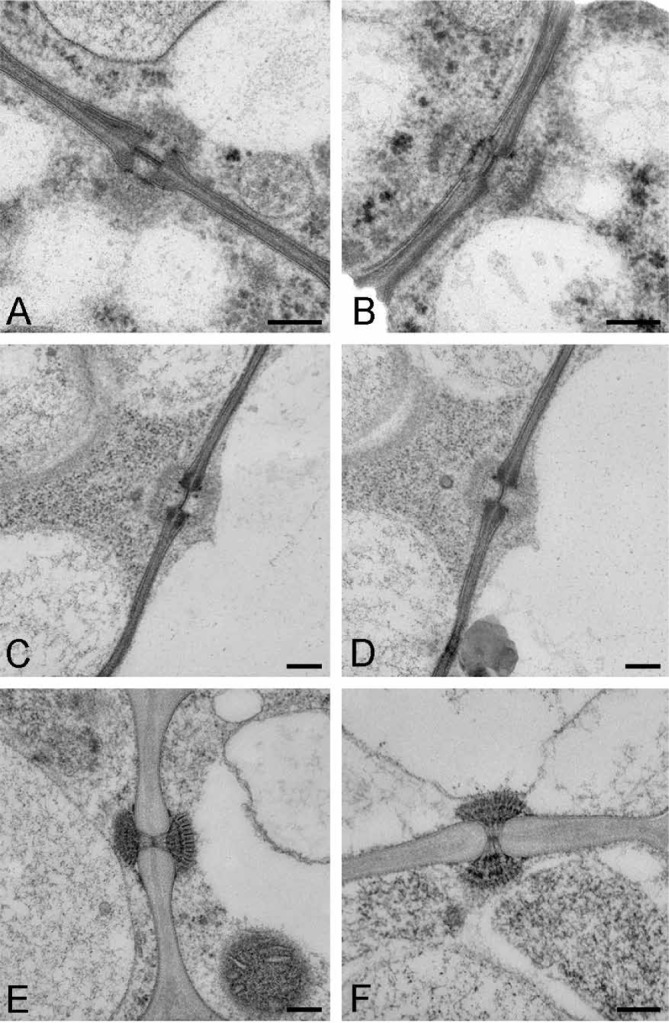
Transmission electron micrographs showing septal pore morphology. A–B. Basidioascus undulatus (DAOM 241956 ex-type). C–D. Geminibasidium donsium (DAOM 241966 ex-type). E–F. Wallemia sebi (CBS 633.66). Bar = 20 μm.
Phylogenomic analysis and molecular dating
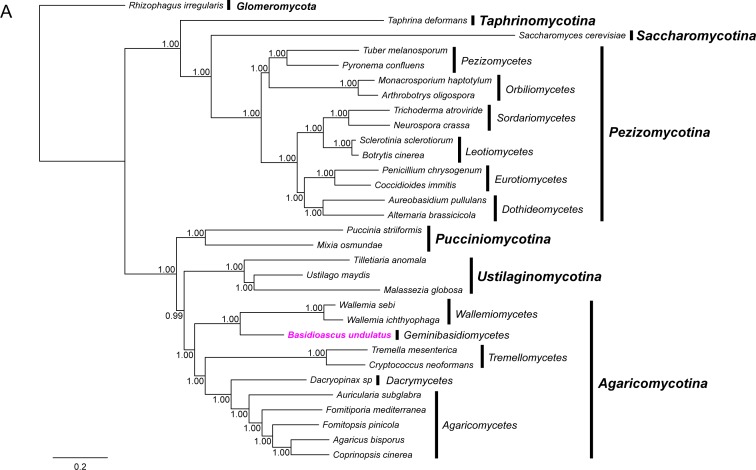
Because Basidioascus is currently classified in Wallemio-mycetes, and the phylogenetic position of the fungi classified in this class has been unstable in different analyses as a result of sparse taxon sampling, low gene sampling and conflicts in certain genealogies (Zalar et al. 2005, Matheny et al. 2006), we performed a phylogenomic analysis using protein sequences from 35 single copy protein-coding genes to study its phylogenetic position in a more robust fashion (Fig. 3A). Our results were similar to those obtained from the rDNA phylogenetic analysis in Nguyen et al. (2013), showing Basidioascus as a distant lineage to Wallemia, with a posterior probability of 1.00. The results were also similar to those of Padamsee et al. (2012) and Zajc et al. (2013), where the Wallemiomycetes were shown to be an early diverging lineage of Agaricomycotina.
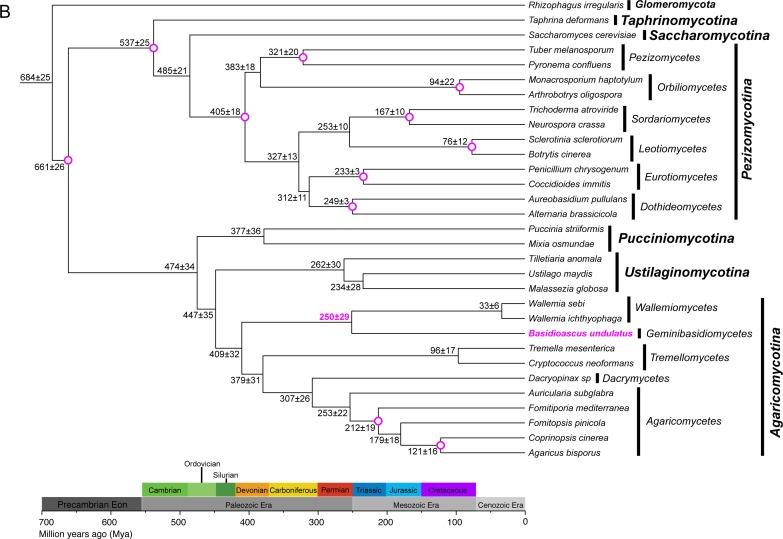
Fig. 3.
Phylogenetic trees resulting from phylogenomic analysis and molecular dating. A. Consensus topology and branch lengths from analyses of concatenated amino acid sequences from 35 single copy genes with a total of 10129 data columns. Gapped and poorly aligned sites were removed. Analyses were performed with the CAT-GTR model. Posterior probabilities are shown at the nodes of the tree. Scale bar indicates expected changes per site. B. Chronogram resulting from a lognormal relaxed molecular clock analysis with the birth-death prior. The mean divergence time with standard error are shown at each node. Circled nodes were pre-calibrated before the analysis. The paleontological periods, in million years ago (Mya), are shown as a scale at the bottom. Rhizophagus irregularis (Glomeromycota) was used as the outgroup.
We dated the divergence of Basidioascus from other lineages using some reliable fossil data of basidiomycetes (Hibbett et al. 1997, Smith et al. 2004) and the latest calculated calibration points (Berbee & Taylor 2010, Prieto & Wedin 2013) (Fig. 3B). Under these assumptions, the split between B. undulatus and Wallemia was estimated at 250±29 Mya, and that between W. sebi and W. ichthyophaga was about 33±6 Mya.
DISCUSSION
Genome sequencing and annotation
Basidioascus undulatus is the first fungus in Geminibasidiales with a sequenced genome. The 1.1 Gb of sequence data gave a median nucleotide coverage of 24X and a final assembly that contained 2992 scaffolds, which is similar to what has been produced in many recent whole genome sequencing studies (e.g. Sims et al. 2014). Wallemia sebi and W. ichthyophaga were the closest known relatives to B. undulatus. Despite being considered close relatives, the genome assembly size of B. undulatus (32 Mb) was larger than W. sebi (9.8 Mb) (Padamsee et al. 2012) and W. ichthyophaga (9.6 Mb) (Zajc et al. 2013). Also, the GC content of B. undulatus was higher (58 %) compared to W. sebi (40 %) and W. ichthyophaga (45 %) (Table 3). Based on these results, the fungi of these two genera are more distantly related than initially suspected.
Table 3.
Genome assembly and annotation statistics of Basidioascus undulatus compared to Wallemia sebi and W. Ichthyophaga.
| Descriptive statistic | B. undulatus | W. sebi | W. icthyophaga | |
|---|---|---|---|---|
| Genome assembly | ||||
| Sequencing platform | Illumina Hi-Seq | Illumina Hi-Seq & 454 | Illumina Hi-Seq | |
| Total number of reads | 12 Milliona | — | — | |
| Read length after trimming | 93 bp | — | — | |
| Data size | 1.1 Gb | — | 3.7 Gb | |
| Assembly size | 32 Mbc | 9.8 Mb | 9.6 Mb | |
| Estimated Percent GC | 58%c | 40% | 45% | |
| Nucleotide coverage | 24Xb | 71X | >270X | |
| Number of contigs | 3058 | 114 | 95 | |
| Number of scaffolds | 2992 | 56 | 82 | |
| Longest scaffold size | 97 Kbc | 900 Kb | 790 Kb | |
| N50 | 15 Kbc | 340 Kb | 440 Kb | |
|
| ||||
| Genome annotation | ||||
| Percent core eukaryotic genes detected | 88%d | — | — | |
| All predicted gene models | 13935e | 5284 | 4863 | |
| Evidence supported gene models | 6123e | — | — | |
| Evidence supported complete gene models | 3681 | — | — | |
Statistics were found by the following programs: (a) FastQC, (b) qualimap, (c) QUAST, (d) CEGMA, and (e) MAKER.
We were able to detect 88 % of partial core eukaryotic genes (CEGs). We assumed that downstream annotation with this genome assembly should, in theory, reveal close to the same proportion of the total number of genes contained in B. undulatus.
Many gene prediction tools are available, but GeneMark-ES was selected because it was shown previously to be accurate at detecting genes in fungal genomes (Ter-Hovhannisyan et al. 2008). GeneMark-ES considers branch point sequences in the intron model, which guides lariat formation during splicing, providing greater accuracy at locating intron boundaries. Because RNA data were not acquired in our study, we chose to use GeneMark-ES, which does not require RNA data for algorithm training. Nevertheless, genome annotation based on genome sequences alone has limitations. First, without RNA data, CDS predictions cannot be validated and genes from splice sites that use donor and acceptor sequences other than the canonical GT–AG introns cannot be predicted. The frequencies of the non-canonical GC–AG introns are 1.0–1.2 % in some ascomycetes (Rep et al. 2006) and could be as high as 3 % in basidiomycetes (Misiek & Hoffmeister 2008). To compensate for the absence of RNA data, the fungal NCBI RefSeq protein sequence data set (13 March 2014 release), which contains curated protein sequences from completed fungal genome projects, was aligned to our B. undulatus genome using the program “exonerate”. In the MAKER2 pipeline, aligning protein sequences to predicted genes on the genome generates ‘evidence’ expressed as the Annotation Evidence Distance (AED) score for exons (eAED) (Eilbeck et al. 2009). The eAED score is a metric that measures how a predicted gene determined from GeneMark-ES agrees with the protein alignment evidence from exonerate while accounting for protein reading frames shifts. This score is helpful for assessing annotation quality of a predicted gene (Eilbeck et al. 2009). Overall, 13 935 genes were predicted by GeneMark-ES but only 6123 of those genes were supported by protein alignment evidence. For each of the 6123 predicted genes, we made further functional annotation with a BLAST search against the UniProt/Swiss-Prot manually curated data set and an InterProScan analysis. Using this information, 452 genes were manually annotated with confidence. Future releases of the B. undulatus genome with RNA sequence data would increase the number of annotated genes and allow discovery and validation of completely novel genes or splice variants.
Meiosis
We used genomic information in combination with confocal microscopy to gain further understanding of sexuality in Basidioascus undulatus. Fungal taxonomists infer function of meiosporangia in newly discovered fungi based on morphological similarities to proven meiosporangia in similar or related fungi. Proof of meiosis by nuclear staining is not usually required. In our case, B. undulatus is different morphologically, ontogenetically, and phylogenetically from known basidiomycetes and it was difficult to assess whether previous morphological interpretations by Nguyen et al. (2013) were correct without nuclear staining experiments. We suggested previously that the clavate structures found on somatic hyphae were basidia (Nguyen et al. 2013). These basidia were deciduous, forcibly discharged, and had a basal lateral projection that eventually collapsed. Sterigmata grew on the basidium, and a basidiospore developed at the tip of each sterigma. To verify these interpretations, we visualized these putative sexual structures with nuclear staining and confocal microscopy. Our observations suggested that meiosis occurred in the clavate structures (Fig. 1) providing the evidence for previous interpretation of them as basidia. Post-meiotic mitosis in the basidium was not seen but it was difficult to visualize nuclei in the mature basidiospores. Although we saw evidence of meiosis with microscopy, we also confirmed the presence of meiotic genes in the genome (Table 1). We did not detect HOP2 among the 10 meiosis specific genes. The HOP2 gene product is involved in preventing synapsis between non-homologous chromosomes during meiotic double-strand break repair (Malik et al. 2008). The function of HOP2 could be served by another unknown gene or gene product in B. undulatus, or perhaps our genome assembly was not complete enough to detect it. With 88 % of CEGs detected, about 1 out of every 10 genes would not be detected during the annotation procedure.
Basidioascus undulatus produces cells attached in chains, which are presumed to be arthroconidia (Nguyen et al. 2013). These arthroconidia were indeed single-nucleated (Fig. 1K–L) and therefore represent an asexual morph of B. undulatus. Given these results, B. undulatus indeed exhibits both sexual and asexual morphs in culture.
Mating
The genes located at the MAT locus orchestrate the fungal mating process and determine the sex of individuals (Lee et al. 2010). In the fungal kingdom, mating is most comprehensively studied in Saccharomyces cerevisiae. Therefore, we used what is known about mating in S. cerevisiae as a model to guide our interpretations of mating genes in the B. undulatus genome.
Homeodomain (HD) transcription factors control the expression of pheromone and pheromone receptor genes. Normally, dimerization occurs between two paralogous HD transcription factors, HD1 and HD2, to form HD1-HD2 complex. In Ustilago maydis, this is termed the bE/bW heterodimer, and in S. cerevisiae the a1/α2 complex (Lee et al. 2010). We identified the HD1 (8946F71C4C) and HD2 (AF24C22F02) proteins in B. undulatus.
Pheromone production and processing are also important in mating. All orthologs for pheromone processing (KEX1, KEX2, STE13, RAM1, RAM2, STE14, AXL11, STE6) were detected in B. undulatus except for the RCE1/STE24 genes responsible for cleavage of the pheromone peptide (Table 2).
Upon release into the environment, pheromones must bind to pheromone receptors to initiate a signaling cascade involving the mitogen-activated protein kinase (MAPK) pathway to turn on mating genes (Jones & Bennett 2011). MAPK pathways regulate the activity of high mobility group (HMG) DNA binding proteins, which are transcription factors regulating pheromone responsive genes in the MAT loci (Hartmann et al. 1996). Most of the mating response genes were detected (STE3, KSS1, FUS3, STE7, STE11, GPA1, STE4, STE20) (Table 2) except the G-protein γ subunit (STE18), the transcription factor STE12, the coordinator of MAPK pathway STE5, and the cell cycle control FAR1 protein. We identified four HMG DNA binding proteins (B013112742, C8B77D01D8, C1AD40EA22, and 9CC40B0A26).
We were unable to isolate single arthroconidia or successfully germinate basidiospores so mating tests were not possible. Thus, it is unknown whether B. undulatus is homothallic or heterothallic. Wallemia sebi apparently has a bipolar mating system (Padamsee et al. 2012) but no discernable mating type locus could be identified in the genome of W. ichthyophaga (Zajc et al. 2013). Our genome assembly was too fragmented to determine the structure of the mating type locus conclusively and therefore the mating system in B. undulatus remains uncharacterized until more sequencing is done. The detection of a near complete cellular machinery for the mating response, pheromone processing, and meiosis suggests that B. undulatus is capable of outcrossing. However, future experiments with gene knockout mutants will be needed to validate these identified genes.
Higher classification and divergence of Geminibasidiales
Basidioascus undulatus and Geminibasidium donsium have been classified in Geminibasidiales, a sister order to Wallemiales. Geminibasidiales was placed tentatively in the class Wallemiomycetes (Nguyen et al. 2013). The main reason for this initial inconclusive placement was the absence of septal pore ultrastructure data and that only ribosomal genes could be used to make a phylogenetic analysis with enough taxon sampling to reach a conclusion. More genomes are available now and phylogenomic analysis with protein coding genes is more feasible and informative.
Our phylogenomic analysis shows the same relationships previously determined with rDNA, where Wallemia and Basidioascus are distantly related sister groups that occupy a basal phylogenetic position in Agaricomycotina (Fig. 3A). The distance of this relationship is also reflected in differences in GC content and genome size between B. undulatus and Wallemia discussed above. According to the sampling and assumptions made in our analyses, molecular dating suggests that the split between B. undulatus and W. sebi occurred 250 Mya (Fig. 3B), which is around the same time that Sordariomycetes split from Leotiomycetes (253 Mya) and Exobasidiomycetes from the Ustilaginomycetes and Malasseziomycetes (262 Mya), but we also note that not all nodes corresponding to class ranks in our analysis represent divergences at around 250 Mya. If our dates are accurate, it means Wallemia and B. undulatus shared a common ancestor until a divergence event occurred between the Permian and Triassic period. Interestingly, it was during this period that Earth experienced its most severe extinction event (Sahney & Benton 2008). The split between W. sebi and W. ichthyophaga was dated at 33±6 Mya, an estimate that differs from previous analyses; a 11.9 Mya split was proposed by Zajc et al. (2013), but with different methods, taxon sampling, and calibrations used for the molecular dating analysis. These estimates should all be considered approximate because the fungal fossil record is rather limited and many studies generate radically different age estimates for the same divergence events (Lücking et al. 2009).
Before sequence data began to dominate fungal systematics, morphological characters of the septa and septal pore caps were considered important for classifying fungi at the class or higher taxonomic levels (Khan & Kimbrough 1982, Oberwinkler & Bandoni 1982, Müller et al. 2000, van Driel et al. 2009) and septal pore morphology is still considered essential for delimiting classes in Basidiomycota. However, the overall septal pore morphologies of B. undulatus and G. donsium are similar and there are significant differences when compared to W. sebi (see p. 219 above; Fig. 2). The septal pore cap is probably not a reliable or practical feature for the classification of Wallemia, Basidioascus and Geminibasidium because it is sometimes present or absent in Wallemia (Padamsee et al. 2012, D.J. McLaughlin pers. comm.) and was not seen in B. undulatus or G. donsium.
Given the results obtained from phylogenomics analysis, molecular dating, and septal pore ultrastructure, we propose to classify Geminibasidiales in a new class Geminibasidiomycetes. Although Geminibasidium could not be included in the phylogenomic analysis and molecular dating because its genome is not yet sequenced, we are confident from previous rDNA phylogenetic analysis (Nguyen et al. 2013), morphological characters, and similarities in the septal pore morphology (Fig. 2) that Geminibasidium will remain a close sister group to Basidioascus. Previous phylogenies position Wallemiomycetes at the base of Basidiomycota (Matheny et al. 2006, Zalar et al. 2005), as a sister group to or as the earliest diverging lineage of Agaricomycotina (Padamsee et al. 2012, Nguyen et al. 2013, Zajc et al. 2013). The classification for the Wallemiomycetes is shown as incertae sedis in the Basidiomycota in the NCBI taxonomy (Federhen 2012), MycoBank (Robert et al. 2013), and the Encyclopedia of Life (http://www.eol.org). In the taxonomic section below, we emend the description of the subphylum Agaricomycotina to include the classes Geminibasidiomycetes and Wallemiomycetes along with the Agaricomycetes, Tremellomycetes, and Dacrymycetes, which are well delimited classes of Agaricomycotina (Hibbett 2007). By formally connecting the basal Geminibasidiomycetes and Wallemiomycetes to the Agaricomycotina, we eliminate the uncertain status of these taxa. The subphylum name Wallemiomycotina (MycoBank no. MB 550364) was published recently to accommodate Wallemia (Doweld 2014). There was no discussion of the rationale behind this proposal and we consider it unnecessary to include Wallemiomycetes (with or without the Geminibasidiomycetes) in its own subphylum.
TAXONOMY
Agaricomycotina Doweld, Prosyllabus: lxxxvii (2001).
MycoBank MB560553
Homonym: Agaricomycotina R. Bauer et al., Mycol. Prog. 5: 45 (2006).
Description: Members of the Basidiomycota that have a cell wall carbohydrate composition with a dominance of glucose and presence of xylose and having a type B secondary structure of the 5S RNA (Bauer et al. 2006). Fungi that belong to this subphylum are classified in the classes Agaricomycetes, Dacrymycetes, Tremellomycetes, Wallemiomycetes or Geminibasidiomycetes.
Notes: Although Agaricomycotina Dowell (2001) is the oldest name, it is the concept of proposed by Bauer et al. (2006) (i.e. Hymenomycetes sensu Swann & Taylor 1995) that is widely used today. We modified it above only to explicitly state all of the accepted classes within Agaricomycotina. The cell wall of B. persicus was composed mostly of glucose and xylose (Nasr et al. 2014), and assuming that this is the same for other species of Basidioascus and Geminibasidium, this placement for the class Geminibasidiomycetes is appropriate. The cell wall carbohydrate composition of Wallemia species is unknown.
Wallemiomycetes Zalar et al., Antonie van Leeuwenhoek 87: 322 (2005).
MycoBank MB501496
Description: Class of xerophilic basidiomycetes belonging to the subphylum Agaricomycotina. These fungi produce basauxic anamorphs and do not produce basidiomata in culture. Species have dolipore septa with adseptal tubular extensions that arise from sheets of endoplasmic reticulum that form the septal pore cap. The septal pore cap is sometimes absent. The septal pore has an electron-dense non-membranous septal pore occlusion and striations that are oriented vertically.
Type order: Wallemiales.
Notes: This description is altered to exclude characters of Geminibasidiales previously added to the concept of Wallemiomycetes by Nguyen et al. (2013). Information about the septal pore morphology was added, as detailed in Padamsee et al. (2012).
Geminibasidiomycetes H.D.T. Nguyen & Seifert, cl. nov.
MycoBank MB811680
Description: Class of xerotolerant basidiomycetes belonging to the subphylum Agaricomycotina. Basidiomata not produced in culture. Basidia arising from somatic hyphae or from swollen basidium-bearing cells (primary cells) with a basal lateral projection occurring either on the basidium or the swollen primary cell. Basidiospores symmetrical on sterigma, not forcibly discharged, and brown at maturity. Arthroconidial and/or yeast-like asexual morphs sometimes produced. Species have a dolipore septum that is electron-dense at the pore swelling with an electron-dense membranous septal pore occlusion. Some species are heat resistant.
Type order: Geminibasidiales.
ACKNOWLEDGEMENTS
We thank Adrian Pelin and Zaky Adam for discussions on genome assembly, Michael Li for advice on genome annotation with MAKER2, Christine Lowe for guidance on doing phylogenic analyses with PhyloBayes, Keith Bradnam for running CEGMA, Charlotte Grace and Karen Fisher for help with TEM, Jeff Cullis for reviewing an early draft of the manuscript, Robert Riley and Igor Grigoriev for processing and uploading genomic data to the JGI MycoCosm portal, and the staff at the DNA Technologies Laboratory at the National Research Council Canada (Saskatoon) for library construction and Illumina sequencing. The US National Science Foundation (NSF) provided support through a collaborative research grant to R.W.R. (DEB-0732503). The Ontario government provided funding support through the Ontario Graduate Scholarship (OGS) to H.D.T. Nguyen.
REFERENCES
- Amselem J, Cuomo CA, van Kan JA, Viaud M, Benito EP, et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genetics 7 (8): e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R, Begerow B, Sampaio JP, Weiss M, Oberwinkler F. (2006) The simple-septate basidiomycetes: a synopsis. Mycological Progress 5: 41–66. [Google Scholar]
- Berbee ML, Taylor JW. (2010) Dating the molecular clock in fungi – how close are we? Fungal Biology Reviews 24: 1–16. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579. [DOI] [PubMed] [Google Scholar]
- Borodovsky M, Lomsadze A. (2011) Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Current Protocols in Bioinformatics: unit–4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, Govindarajulu M, Kozik A, Wang M, Chen X, et al. (2011) Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS ONE 6 (8): e24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celio GJ, Padamsee M, Dentinger BT, Bauer R, McLaughlin DJ. (2007) [“2006”] Assembling the Fungal Tree of Life: constructing the structural and biochemical database. Mycologia 98: 850–859. [DOI] [PubMed] [Google Scholar]
- Cisse OH, Almeida JM, Fonseca A, Kumar AA, Salojarvi J, et al. (2013) Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. mBio 4 (3): e00055-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Domsch KH, Gams W, Anderson TH. (1980) Compendium of Soil Fungi. London: Academic Press. [Google Scholar]
- Doweld AB. (2014) Wallemiomycotina. Index Fungorum 73: 1. [Google Scholar]
- Duncan EG, Galbraith MH. (1972) Post-meiotic events in the Homobasidiomycetidae. Transactions of the British Mycological Society 58: 387–392. [Google Scholar]
- Eilbeck K, Moore B, Holt C, Yandell M. (2009) Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S. (2012) The NCBI Taxonomy database. Nucleic Acids Research 40 (Database issue): D136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, et al. (2012) The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336: 1715–1719. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Garcia-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Gotz S, et al. (2012) Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28: 2678–2679. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, et al. (1996) Life with 6000 genes. Science 274: 546, 563–567. [DOI] [PubMed] [Google Scholar]
- Gostincar C, Ohm RA, Kogej T, Sonjak S, Turk M, et al. (2014) Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics 15: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, et al. (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research 42 (Database issue):D699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29 (8): 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Zeng Q, Pearson MD, Cuomo CA, Wortman JR. (2011) Approaches to fungal genome annotation. Mycology 2: 118-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halary S, Malik SB, Lildhar L, Slamovits CH, Hijri M, et al. (2011) Conserved meiotic machinery in Glomus spp., a putatively ancient asexual fungal lineage. Genome Biology and Evolution 3: 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaichi T, Sato T, Iwamoto T, Malavasi-Yamashiro J, Hoshino M, et al. (1986) A stable lead by modification of Sato’s method. Journal of Electron Microscopy 35: 304–306. [PubMed] [Google Scholar]
- Hartmann HA, Kahmann R, Bolker M. (1996) The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO Journal 15: 1632–1641. [PMC free article] [PubMed] [Google Scholar]
- Hibbett D, Grimaldi D, Donoghue M. (1997) Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of Homobasidiomycetes. American Journal of Botany 84: 981–991. [PubMed] [Google Scholar]
- Hibbett DS. (2007) [“2006”] A phylogenetic overview of the Agaricomycotina. Mycologia 98: 917–925. [DOI] [PubMed] [Google Scholar]
- Hoch HC. (1986) Freeze-substitution of fungi. In: Ultrastructure Techniques for Microorganisms (Aldrich HC, Todd WJ, eds): 183–212. New York: Plenum Press. [Google Scholar]
- Holt C, Yandell M. (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard RJ, O’Donnell KL. (1987) Freeze substitution of fungi for cytological analysis. Experimental Mycology 11: 250–269. [Google Scholar]
- James TY, Sun S, Li W, Heitman J, Kuo HC, et al. (2013) Polyporales genomes reveal the genetic architecture underlying tetrapolar and bipolar mating systems. Mycologia 105: 1374–1390. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SK, jr, Bennett RJ. (2011) Fungal mating pheromones: choreographing the dating game. Fungal Genetics and Biology 48: 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444 : 97–101. [DOI] [PubMed] [Google Scholar]
- Khan SR, Kimbrough JW. (1982) A reevaluation of the basidiomycetes based upon septal and basidial structures. Mycotaxon 15: 103–120. [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. (2008) Ainsworth & Bisby’s Dictionary of the Fungi. 10th edn Wallingford: CABI Publishing. [Google Scholar]
- Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, et al. (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biology 12 (4):R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, et al. (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. (2004) A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Molecular Biology and Evolution 21: 1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. (2013) PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Systematic Biology 62: 611–615. [DOI] [PubMed] [Google Scholar]
- Lee SC, Ni M, Li W, Shertz C, Heitman J. (2010) The evolution of sex: a perspective from the fungal kingdom. Microbiology and Molecular Biology Reviews 74: 298–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, et al. (2005) The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking R, Huhndorf S, Pfister DH, Plata ER, Lumbsch HT. (2009) Fungi evolved right on track. Mycologia 101: 810–822. [DOI] [PubMed] [Google Scholar]
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., jr (2008) An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS ONE 3 (8): e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthey S, Aguileta G, Rodolphe F, Gendrault A, Giraud T, et al. (2008) FUNYBASE: a FUNgal phYlogenomic dataBASE. BMC Bioinformatics 9: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, et al. (2010) Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464: 1033–1038. [DOI] [PubMed] [Google Scholar]
- Matheny PB, Gossmann JA, Zalar P, Kumar TK, Hibbett DS. (2006) Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Canadian Journal of Botany 84: 1794–1805. [Google Scholar]
- Matsushima T. (2003) Basidioascus gen. nov. Matsushima Mycological Memoirs 10: 98–104. [Google Scholar]
- McDaniel DP, Roberson RW. (2000) Intracellular motility and mechanisms of control during hyphal tip growth in Allomyces. Fungal Genetics and Biology 31: 223–234. [DOI] [PubMed] [Google Scholar]
- Meerupati T, Andersson KM, Friman E, Kumar D, Tunlid A, et al. (2013) Genomic mechanisms accounting for the adaptation to parasitism in nematode-trapping fungi. PLoS Genetics 9 (11): e1003909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiek M, Hoffmeister D. (2008) Processing sites involved in intron splicing of Armillaria natural product genes. Mycological Research 112: 216–224. [DOI] [PubMed] [Google Scholar]
- Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, et al. (2012) Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proceedings of the National Academy of Sciences, USA 109: 17501–17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller WH, Stalpers JA, van Aelst AC, de Jong MDM, van der Krift TP, et al. (2000) The taxonomic position of Asterodon, Asterostroma and Coltricia inferred from the septal pore cap ultrastructure. Mycological Research 104:1485–1492. [Google Scholar]
- Nasr S, Soudi MR, Nasrabadi SM, Nikou MM, Salmanian AH, et al. (2014) Basidioascus persicus sp. nov., a yeast-like species of the order Geminibasidiales isolated from soil. International Journal of Systematic and Evolutionary Microbiology 64: 3046–3052. [DOI] [PubMed] [Google Scholar]
- Nguyen HDT, Nickerson NL, Seifert KA. (2013) Basidioascus and Geminibasidium: a new lineage of heat-resistant and xerotolerant basidiomycetes. Mycologia 105: 1231–1250. [DOI] [PubMed] [Google Scholar]
- Nikolenko SI, Korobeynikov AI, Alekseyev MA. (2013) BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14 (Suppl 1): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Oberwinkler F, Bandoni RJ. (1982) A taxonomic survey of the gasteroid, auricularioid Heterobasidiomycetes. Canadian Journal of Botany 60: 1726–1750. [Google Scholar]
- Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, et al. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathogens 8 (12): e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padamsee M, Kumar TK, Riley R, Binder M, Boyd A, et al. (2012) The genome of the xerotolerant mold Wallemia sebi reveals adaptations to osmotic stress and suggests cryptic sexual reproduction. Fungal Genetics and Biology 49: 217–226. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Prieto M, Wedin M. (2013) Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS ONE 8 (6): e65576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Duyvesteijn RG, Gale L, Usgaard T, Cornelissen BJ, et al. (2006) The presence of GC-AG introns in Neurospora crassa and other euascomycetes determined from analyses of complete genomes: implications for automated gene prediction. Genomics 87: 338–347. [DOI] [PubMed] [Google Scholar]
- Roberson RW, Fuller MS. (1988) Ultrastructural aspects of the hyphal tip of Sclerotium rolfsii preserved by freeze substitution. Protoplasma 146: 143–149. [Google Scholar]
- Robert V, Vu D, Amor AB, van de Wiele N, Brouwer C, et al. (2013) MycoBank gearing up for new horizons. IMA Fungus 4: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahney S, Benton MJ. (2008) Recovery from the most profound mass extinction of all time. Proceedings of the Royal Society of London, Biological Sciences 275 : 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, et al. (2009) Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Research 19: 1722–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. (2014) Sequencing depth and coverage: key considerations in genomic analyses. Nature Reviews Genetics 15: 121–132. [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E. (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SY, Currah RS, Stockey RA. (2004) Cretaceous and Eocene poroid hymenophores from Vancouver Island, British Columbia. Mycologia 96:180–186. [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. Journal of Ultrastructure Research 26 (1):31–43. [DOI] [PubMed] [Google Scholar]
- Staats M, van Kan JA. (2012) Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryotic Cell 11: 1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Wilke SK, Ahren D, Au CH, Birren BW, et al. (2010) Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proceedings of the National Academy of Sciences, USA 107: 11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann EC, Taylor JW. (1995) Phylogenetic perspectives on basidiomycete systematics: evidence from the 18s rRNA gene. Canadian Journal of Botany 73 (S1): 862–868. [Google Scholar]
- Taylor JW, Berbee ML. (2007) [“2006”] Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98: 838–849. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. (2008) Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Research 18: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracina FC. (1974) Fine structure of the septum in Wallemia sebi. Canadian Journal of Botany 52: 2587–2590. [Google Scholar]
- Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, et al. (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proceedings of the National Academy of Sciences, USA 110: 20117–20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toome M, Kuo A, Henrissat B, Lipzen A, Tritt A, et al. (2014a) Draft genome sequence of a rare smut relative, Tilletiaria anomala UBC 951. Genome Announcements 2 (3): e00539-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toome M, Ohm RA, Riley RW, James TY, Lazarus KL, et al. (2014b) Genome sequencing provides insight into the reproductive biology, nutritional mode and ploidy of the fern pathogen Mixia osmundae. New Phytologist 202: 554–564. [DOI] [PubMed] [Google Scholar]
- Traeger S, Altegoer F, Freitag M, Gabaldon T, Kempken F, et al. (2013) The genome and development-dependent transcriptomes of Pyronema confluens: a window into fungal evolution. PLoS Genetics 9 (9): e1003820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, et al. (2008) Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nature Biotechnology 26: 1161–1168. [DOI] [PubMed] [Google Scholar]
- van Driel KG, Humbel BM, Verkleij AJ, Stalpers J, Muller WH, et al. (2009) Septal pore complex morphology in the Agaricomycotina (Basidiomycota) with emphasis on the Cantharellales and Hymenochaetales. Mycological Research 113: 559–576. [DOI] [PubMed] [Google Scholar]
- Xu J, Saunders CW, Hu P, Grant RA, Boekhout T.>, et al. (2007) Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proceedings of the National Academy of Sciences, USA 104: 18730–18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang L, Ji X, Feng Y, Li X, et al. (2011) Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathogens 7 (9): e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajc J, Liu Y, Dai W, Yang Z, Hu J, et al. (2013) Genome and transcriptome sequencing of the halophilic fungus Wallemia ichthyophaga: haloadaptations present and absent. BMC Genomics 14: 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalar P, de Hoog GS, Schroers HJ, Frank JM, Gunde-Cimerman N. (2005) Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie van Leeuwenhoek 87: 311–328. [DOI] [PubMed] [Google Scholar]