Abstract
The inbred mouse C57BL/6J is the reference strain for genome sequence and for most behavioral and physiological phenotypes. However the International Knockout Mouse Consortium uses an embryonic stem cell line derived from a related C57BL/6N substrain. We found that C57BL/6N has lower acute and sensitized response to cocaine and methamphetamine. We mapped a single causative locus and identified a non-synonymous mutation of serine to phenylalanine (S968F) in Cytoplasmic FMR interacting protein 2 (Cyfip2) as the causative variant. The S968F mutation destabilizes CYFIP2 and deletion of the C57BL/6N mutant allele leads to acute and sensitized cocaine response phenotypes. We propose CYFIP2 is a key regulator of cocaine response in mammals and present a framework to utilize mouse substrains to discover novel genes and alleles regulating behavior.
The reference mouse strain, C57BL/6J, was established in 1921 and has been maintained at the Jackson Laboratory since 1948 (1). In 1951 a colony of C57BL/6J was shipped to NIH and C57BL/6N became a second major source. Large-scale projects use different C57BL/6 substrains, including the International Knockout Mouse Consortium (IKMC) which uses C57BL/6N ES cells (2, 3), and the Mouse Genome Sequencing Consortium and Allen Brain Atlas which use C57BL/6J (4, 5). Although most behavioral measurements to date have been carried out in the C57BL/6J substrain, the IKMC will shift the emphasis to the C57BL/6N substrain. While behavioral differences have been noted in C57BL/6 substrains, their genetic basis has not been elucidated (6, 7). Therefore it becomes important to understand the genetic causes of phenotypic differences between C57BL/6N and C57BL/6J.
Drug addiction is characterized by loss of control over drug consumption and behaviors associated with drug seeking. Even though drugs of abuse fall into different pharmacological categories, they all act on the mesolimbic reward pathway in the brain (8). Long-term structural and functional changes in neuronal circuitry are thought to be a key feature of addiction (9). We investigated in detail a difference in cocaine response between the C57BL/6N and C57BL/6J mouse substrains.
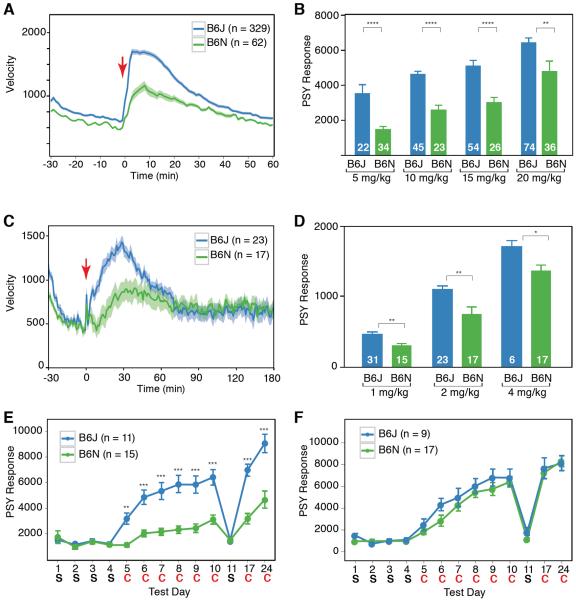
We characterized C57BL/6N and C57BL/6J animals for locomotor response to cocaine. Due to the nonlinear nature of drug response we carried out testing at multiple doses. C57BL/6N had a 45% [1-standard deviation (SD)] lower acute response to cocaine and methamphetamine at multiple doses (Fig. 1A–D, fig.S1). In psychomotor sensitization, repeated administration of the stimulus elicits a progressively larger behavioral response. It is regulated by experience-dependent neuronal plasticity and thought to be a key event leading to addiction (10, 11). At 10mg/kg C57BL/6J sensitized to cocaine much more efficiently than C57BL/6N (compare day 5 to 10, Fig1E) although both strains sensitized to similar levels at a higher 15mg/kg dose (Fig. 1F).
Fig. 1.
Acute and sensitized cocaine response, measured by locomotor hyperactivity, is lower in C57BL/6N (B6N) substrain than in C57BL/6J (B6J). (A) Baseline locomotor velocity data of B6J (blue) and B6N (green) mice were measured for 30 minutes and then injected with cocaine (20 mg/kg, red arrow) and recorded for further 60 minutes. (B) Cocaine dose response for B6J and B6N. (C) Decreased methamphetamine response in B6N. Animals were injected with methamphetamine (2mg/kg, red arrow) and recorded for 3 hours. (D) Methamphetamine dose response for B6J and B6N. Mice were treated with 1mg/kg, 2mg/kg and 4mg/kg of the drug. Average response 60 minutes post injection is shown. (E, F) B6N have a lower sensitized response to cocaine at 10mg/kg (E) but respond similarly at 15mg/kg dose (F). Mice were measured for baseline and post injection response for one hour and injected with saline(S) or cocaine(C). 30-minute post injection response is shown. All data are shown as mean ± SEM, the number of animals in each group is indicated; *p<0.05,**p<0.01,***p<0.001,****p<0.0001 from Tukey post hoc test following two way ANOVA (B,D) or pair wise t-test (E,F).
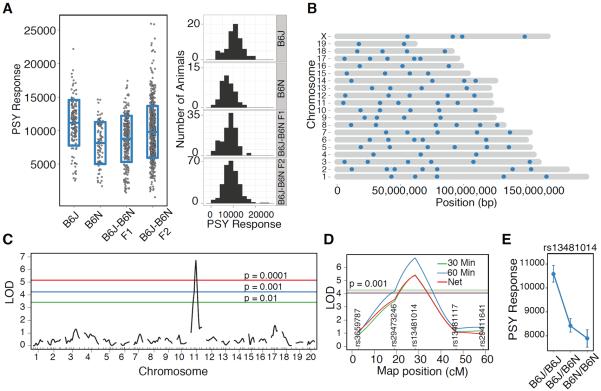
To determine which genetic loci contribute to reduced cocaine response seen in C57BL/6N we carried out quantitative trait locus (QTL) analysis (fig.S2). Parental strains, F1, and F2 animals were tested concurrently. C57BL/6N had approximately a 1SD lower response to cocaine. The F1 animals exhibited cocaine response similar to C57BL/6N indicating dominance of the C57BL/6N allele and the F2 progeny were intermediate in response when compared with the parental strains (Fig.2A). These differences were seen in all our measures and both sexes (fig.S3,S4).
Fig. 2.
QTL on chromosome 11 regulates cocaine response. (A) Hyperactivity following intraperitoneal injection of cocaine (20 mg/kg) was quantitated. B6J (n = 73), B6N (n = 44), F1 (n = 124), F2 (n = 244) were tested. The blue box represents mean ± 1SD range and histogram shows normal distribution of the measures. (B) Locations of polymorphic markers between B6J and B6N. (C) Genome wide QTL scan revealed a single highly significant QTL on chromosome 11. The significance thresholds are established through 100,000 permutation tests. (D) The chromosome 11 QTL is significant for multiple cocaine response measures. (E) Genotype effect plot at maker rs13481014 shows the B6N allele has lower response than B6J allele, and the F1 is intermediate. Data are mean ± SEM.
To perform QTL analysis, we identified polymorphic markers between C57BL/6J and C57BL/6N (supplementary materials, fig.S5, Table S1, S2). QTL analysis for cocaine response yielded a single QTL on chromosome 11 (log of odds (LOD) 6.8) (Fig.2C). This peak is highly significant based on permutation tests (Fig.2C) and is specific to cocaine response and was not seen for baseline activity (fig.S6). The peak linkage occurs at marker rs13481014 for 30 minute, 60 minute and net response (Fig.2D) and the genotype effect plot indicates that the B6N/B6N allele elicits a lower response than the B6J/B6J allele. The F1 response is similar to C57BL/6N implying dominance of B6N allele (Fig.2E). This QTL accounts for 11% of the total phenotypic variance (genetic, environmental and error) and 61% of the genetic variance (fig.S7). The QTL support interval translates to a 22Mb interval between 35Mb–57Mb of chromosome 11 (12) (supplementary materials, fig.S8).
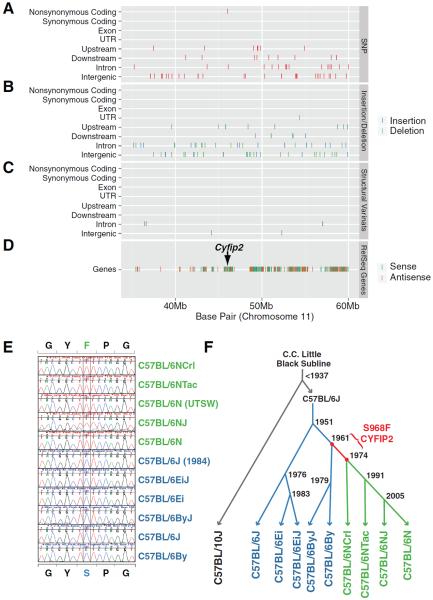
Phenotypic differences between related strains may occur because of three possibilities: residual heterozygosity at the time of separation, genetic contamination following separation, or genetic drift. Genotyping indicated that inbreeding was essentially complete and there was no genetic contamination or residual heterozygosity in C57BL/6N and C57BL/6J substrains (13). Because C57BL/6N and C57BL/6J diverged 62 years ago, there may be a limited number of polymorphisms accumulated through genetic drift that may account for the phenotypic difference. To identify the causative mutation we performed whole-genome sequencing runs and combined the data with published and unpublished C57BL/6NJ sequence for 99.8 fold coverage of chromosome 11 (TableS3) (14). We reanalyzed the datasets with identical parameters (fig.S9,Table S4,S5) and found a single non-synonymous SNP in the QTL interval (Fig.3A, fig.S10). No indels or structural variants cause protein-coding changes (Fig.3B,C). This non-synonymous SNP changes G to A at 46,036,117bp of chromosome 11 and produces a serine 968 to phenylalanine (S968F) missense mutation in Cyfip2 (Fig.3D). This variant had a high Polyphen2 score of 0.957/1 and was predicted to be damaging (15) (fig.S11). CYFIP1 and CYFIP2 are highly conserved proteins implicated in fragile X-mental retardation protein (FMRP) function and regulation of actin polymerization through the WAVE regulation complex (WRC) (16, 17). Cyfip2 is widely expressed throughout the brain whereas Cyfip1 is more restricted (fig.S12). FMRP is the most common cause of mental retardation in humans, and WAVE complex members have been shown to regulate neuronal connectivity and behavior in mice and Drosophila (18–23). Because many C57BL/6 substrains are readily available with information about the time of separation from the founder colony we constructed a phylogenetic timeline of the S969F variation (Fig.3E, F). The Cyfip2 polymorphism was fixed in the C57BL/6N colony sometime between 1961 and 1974 and is present in most commercially available sources of C57BL/6N including Charles River (NCrl) and Taconic (NTac) (Fig.3F).
Fig. 3.
Next generation sequencing identifies a single nonsynonymous polymorphism in Cyfip2. Classification of SNPs (A), indels (B), and structural variants (C) sequencing reveals only a single SNP (top row of A) in Cyfip2 (D) that changes Serine 968 to phenylalanine in B6N. Only the QTL support interval is shown. (E, F) Comparison of C57BL/6 substrains shows that S968F variant was fixed in B6N lineage between 1961 and 1974.
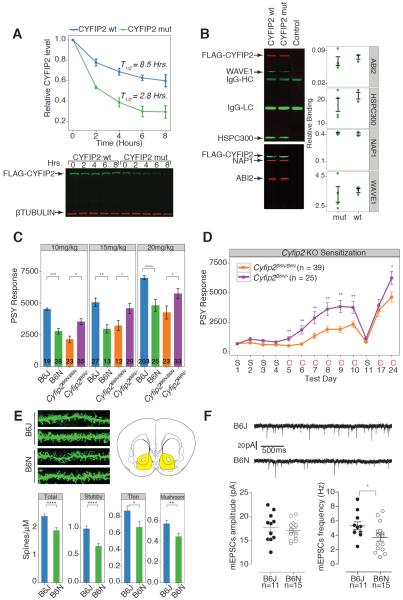
The Cyfip gene family is highly conserved in metazoans and multiple sequence alignment indicated CYFIP2 S968 is 100% conserved in orthologs from C. elegans to humans (fig.S13A). The crystal structure of CYFIP1 has been solved as part of a pentameric 400-kilodalton WRC (16). Molecular modeling of the S969F mutation using the WRC homology model revealed that the mutation leads to substantial steric hindrance with adjacent residues (fig.S13B,S14). We measured protein stability following cycloheximide treatment to estimate the relative half-life of wild type (WT) and mutant CYFIP2. The WT and mutant proteins had half-lives of 8.5 and 2.8 hours respectively, indicating that the S968F mutation greatly destabilizes CYFIP2 (Fig.4A) (sup. results). Stably transfected HEK293 cell lines expressing equivalent levels of the WT and mutant CYFIP2 proteins were used for biochemical and proteomic analyses. Quantitative co-immunoprecipitation (IP) assays showed no difference in interaction between the WT and mutant CYFIP2 with NAP1, WAVE1, ABI2 and HSPC300, the core components of the WRC (Fig 4B). We did not observe differences in our proteomic analysis (fig.S15) and saw no interaction of WT or mutant CYFIP2 with FMRP, FXR1 or FXR2 under our conditions.
Fig. 4.
CYFIP2 S968F causes destabilization and knockout analyses of Cyfip2 shows heterozygous mice have cocaine response phenotypes. (A) Protein stability assay shows CYFIP2 S968F is less stable. 293T cells transfected with FLAG-CYFIP2 (green bands) were treated cycloheximide for the indicated times and western blot was performed (bottom). ß-tubulin was used as control (red bands). Replicate experiments were quantitated and half-life was calculated using nonlinear one phase decay. K, the half-file parameter is highly significant p < 0.0001 (top panel). (B) Quantitative IP experiments show CYFIP2 S968F interacts with WAVE complex members. IP (left) was conducted in replicate and binding was quantitated (right). (C) Mice generated from ES cells harboring the knockout first allele of Cyfip2 from the IKMC are on B6N background (designated Cyfip2B6N/B6N for WT and Cyfip2B6N/− for heterozygous knockout). The homozygous deletion is lethal at birth and therefore not shown. Cyfip2B6N/B6N animals have lower cocaine response than the Cyfip2B6N/− mice. (D) The Cyfip2 heterozygous mice show higher sensitized response to cocaine. Cyfip2B6N/B6N (orange), Cyfip2B6N/− (purple) were injected with saline (S) or cocaine (C, 10 mg/kg). Pairwise comparisons for genotype for each day were significant as indicated. (E) Diolistic labeling of nucleus accumbens neurons shows decrease in dendritic spine density in B6N. Sample images are shown with representative location of neurons (yellow on coronal section of brain). Quantitation and classification of spines are shown. At least 6 animals were used in each group with over 10 images collected from each animal. (F) B6N has a decrease in frequency of AMPAR mEPSCs. Sample traces of mEPSCs in NAc shell MSNs from B6J and B6N mice. Dot plots for mEPSC amplitude and frequency. Data are mean ± SEM and Student's t-test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Two-way ANOVA with Tukey post hoc test (A).
To generate a second mutant allele of Cyfip2, we produced mice using ES cells carrying the `knockout first' (null) allele from the IKMC (3) and maintained them in C57BL/6N background (fig.S16). The homozygous knockouts die within a few hours of birth, however the heterozygous mice are viable with no overt phenotypes. Because the ES cells used in IKMC are of C57BL/6N origin, the WT mice carry the mutant C57BL/6N alleles of Cyfip2 (designated Cyfip2B6N/B6N) and the heterozygotes carry one copy of the C57BL/6N allele (designated Cyfip2B6N/−). We compared mice heterozygous for the knockout allele with the homozygous WT mice for cocaine response (Fig.4C). As expected the C57BL/6J mice have lower cocaine response than C57BL/6N mice at all doses (Fig.4C). The Cyfip2B6N/B6N mice also have low response to cocaine similar to C57BL/6N, however heterozygous Cyfip2B6N/− mice have higher cocaine response than the homozygous Cyfip2B6N/B6N mice at all three doses (Fig.4C). The deletion of one mutant allele alleviates the severity of the phenotype seen with two mutant alleles and explains the dominant phenotype of the F1 progeny and genotype effect plot of linked QTL maker seen previously (Fig.2A, E). The sensitized response is also higher in heterozygous mice (Cyfip2B6N/−), confirming that CYFIP2 regulates acute and sensitized response to cocaine (Fig.4D). We did not see any changes in cocaine metabolism between the two strains (fig.S17).
Next, we explored the functional effect of the CYFIP2 S968F mutation. Drug induced structural plasticity is thought to be key in addiction. We measured and classified dendritic spine frequency of medium spiny neurons in the nucleus accumbens. C57BL/6N have lower number of total spines with significantly lower spines of each class (Fig.4E). Because dendritic spines are the major sites of excitatory postsynaptic current we predicted a deficit in excitatory glutamatergic signaling in C57BL/6N. We measured the frequency and amplitude of mini-excitatory postsynaptic signaling currents (mEPSC) in the nucleus accumbens shell and found a clear decrease in frequency, but not amplitude, of mEPSC (Fig.4F). Lowered frequency of mEPSCs is a plausible functional consequence of reduced dendritic spines although we cannot rule out other adaptations.
Our work has three major implications. First, we identify a mutation at the nucleotide level utilizing QTL analysis and provide evidence that CYFIP2 is a regulator of acute and sensitized cocaine response. Second, with over 20 commercially available C57BL/6 substrains, we describe a framework to utilize mouse substrains as a rich genetic source for discovering new genes and alleles that regulate behavior. If other substrains such as C3H/He and DBA/2 are included, there may be hundreds of mouse substrains available for analysis. Third, care must be taken when comparing new behavioral data from C57BL/6N substrains with existing data from C57BL/6J.
Supplementary Material
Acknowledgments
We thank A.Khan, W.Khan for genotyping help, A.M.Wissman, J.Gibson, and K.Huber help with diolistics, C.Olker for behavioral testing, C.Cowan for advice on sensitization, M.Strobel for C57BL/6 substrain DNA, S.Padrick, B.Chen and M.Rosen for WRC reagents and advice, T.Keane (Sanger Centre) for access to sequencing data, N.Kumar and I.Kornblum for technical help, and O.Hermanson and P.Lowrey for advice on manuscript. V.K. was funded through NRSA F32DA024556 from NIDA. NIH grants U01MH61915 (J.S.T.). J.S.T. is an Investigator, S.H.Y. and V.K. were associates in the HHMI.
References and Notes
- 1.Morse HC. Origins of Inbred Mice. Academic Press Inc; 1979. [Google Scholar]
- 2.Pettitt SJ, et al. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skarnes WC, et al. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterston RH, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Lein ES, et al. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 6.Bryant CD, et al. J Neurogenet. 2008;22:315–331. doi: 10.1080/01677060802357388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuo N, et al. Front Behav Neurosci. 2010;4:29. doi: 10.3389/fnbeh.2010.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koob GF, Volkow ND. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luscher C, Malenka RC. Neuron. 2011;69:650–663. doi: 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson TE, Berridge KC. Brain Res Brain Res Rev. 1993;18:247–291. doi: 10.1016/0165-0173(93)90013-p. [DOI] [PubMed] [Google Scholar]
- 11.Nestler EJ. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 12.Cox A, et al. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, et al. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keane TM, et al. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzhubei IA, et al. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, et al. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napoli I, et al. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Qurashi A, et al. Neural Dev. 2007;2:18. doi: 10.1186/1749-8104-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schenck A, et al. Dev Biol. 2004;274:260–270. doi: 10.1016/j.ydbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Soderling SH, et al. J Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soderling SH, et al. Proc Natl Acad Sci U S A. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, et al. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 23.Bogdan S, Grewe O, Strunk M, Mertens A, Klambt C. Development. 2004;131:3981–3989. doi: 10.1242/dev.01274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.