SUMMARY
Research on the human microbiome has established that commensal and pathogenic bacteria can influence obesity, cancer, and autoimmunity through mechanisms mostly unknown. We found that a component of bacterial biofilms, the amyloid protein curli, irreversibly formed fibers with bacterial DNA during biofilm formation. This interaction accelerated amyloid polymerization and created potent immunogenic complexes that activated immune cells, including dendritic cells, to produce cytokines such as Type I interferons, which are pathogenic in systemic lupus erythematosus (SLE). When given systemically, curli-DNA composites triggered immune activation and production of autoantibodies in lupus-prone and wild-type mice. We also found that the infection of lupus-prone mice with curli-producing bacteria triggered higher autoantibody titers compared to curli-deficient bacteria. These data provide a mechanism by which the microbiome and biofilm-producing enteric infections may contribute to the progression of SLE and point to a potential molecular target for treatment of autoimmunity.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a complex autoimmune disease in which both genetic and environmental triggers contribute to disease onset and to the production of autoantibodies to double stranded DNA (dsDNA) and nuclear proteins (Elkon and Stone, 2011; Morel, 2010; Moser et al., 2009). Infection is an important environmental trigger for lupus flares and a major cause of morbidity and mortality in SLE patients (Petri, 1998). In fact, 20–55% of deaths in SLE patients are attributable to infections and up to 23% of hospitalizations are due to infectious complications (Barrera-Vargas et al., 2014; Fessler, 2002). Bloodstream infections, urinary tract infections, soft tissue infections and pneumonia are common in SLE patients. Frequent Salmonella infections have been reported, especially those caused by Salmonella enterica serovar Typhimurium and serovar Enteritidis (Gerona and Navarra, 2009). Moreover, Salmonella behave more aggressively in SLE patients: instead of causing localized gastroenteritis, infection may result in bacteremia or complications in soft tissues with high mortality rates (Costa-Reis et al., 2013; Lim et al., 2001; Pablos et al., 1994; Tsao et al., 2002). Although bacterial infections are thought to contribute to SLE pathogenesis by inducing cell death and inflammation, the exact mechanisms by which bacteria contribute to SLE pathogenesis remain unknown.
Biofilms are bacterial communities embedded in an extracellular matrix (ECM), which protects bacteria from environmental stresses including antibiotics (Lopez et al., 2010; O’Toole et al., 1999). Biofilms are formed on many biotic and abiotic surfaces including the mucosal surfaces of the human body and indwelling medical devices. Several infections are associated with biofilms including UTI, otitis media, and periodontal diseases (Bjarnsholt, 2013) suggesting that the human immune system is exposed to biofilm components throughout life.
Amyloids are proteins with a conserved beta sheet structure. Mammalian amyloids accumulate in tissues during various debilitating human diseases including Alzheimer’s Disease (Schnabel, 2010). Several bacterial species actively produce amyloid proteins in biofilms including those of S. Typhimurium and E. coli. Amyloid fibers of the latter enteric bacteria are termed curli, which are encoded by the csg gene cluster composed of csgBAC and csgDEFG operons. Production and polymerization of bacterial CsgA protein leads to the generation of the bacterial amyloid curli (Barnhart and Chapman, 2006; Chapman et al., 2002; Romling et al., 1998; Wang et al., 2007). Curli fibers are expressed during various enteric infections including sepsis, gastroenteritis and UTI (Bian et al., 2000; Humphries et al., 2003; Kai-Larsen et al., 2010). We have previously shown that curli fibers induce immune activation by triggering the Toll-Like Receptor (TLR) 2-TLR1 heterocomplex (Rapsinski et al., 2013; Tukel et al., 2010; Tukel et al., 2005; Tukel et al., 2009). In addition, many bacterial biofilms contain extracellular DNA (eDNA) which acts to stabilize the biofilm matrix (Whitchurch et al., 2002).
A number of reports have suggested that DNA complexed with a protein antigen can induce lupus-like disease (Desai et al., 1993; Lande et al., 2007). Human prototypic amyloidogenic peptides, prion fragment and amyloid-β 1–42 can directly bind to DNA (Di Domizio et al., 2012b; Jimenez, 2010). Immunization with amyloid fibers complexed with nucleic acids, in the presence of the classic adjuvant CFA, can induce autoantibodies in non-autoimmune mice within 12 weeks (Di Domizio et al., 2012a). Although groundbreaking, the amyloid-DNA composites used in the latter study were made from human serum albumin and salmon sperm DNA – an artificial composite that the immune system would not normally be exposed to. Moreover, the administration of CFA together with amyloid-DNA makes it unclear whether amyloid-DNA complexes are simply autoantigens or they may act as adjuvants to stimulate the autoimmune response. Therefore, we asked whether the natural amyloid fibers from bacterial biofilms might be bound tightly enough to biofilm-derived DNA to create immuno-stimulatory amyloid-DNA composites and investigated the role of bacterial amyloid-DNA composites in lupus pathogenesis and induction of autoimmunity.
Using a multidisciplinary approach we found that curli fibers bound tightly to eDNA in biofilms. Curli-DNA composites were powerful immune stimulators, acting synergistically to activate innate and adaptive immunity and triggered autoantibody production. Finally, we showed that infection with curli-producing bacteria accelerated the generation of autoantibodies in lupus-prone mice, suggesting that infections with amyloid producing bacteria may be important environmental triggers for the progression of lupus.
RESULTS
Curli-DNA composites in S. Typhimurium biofilms
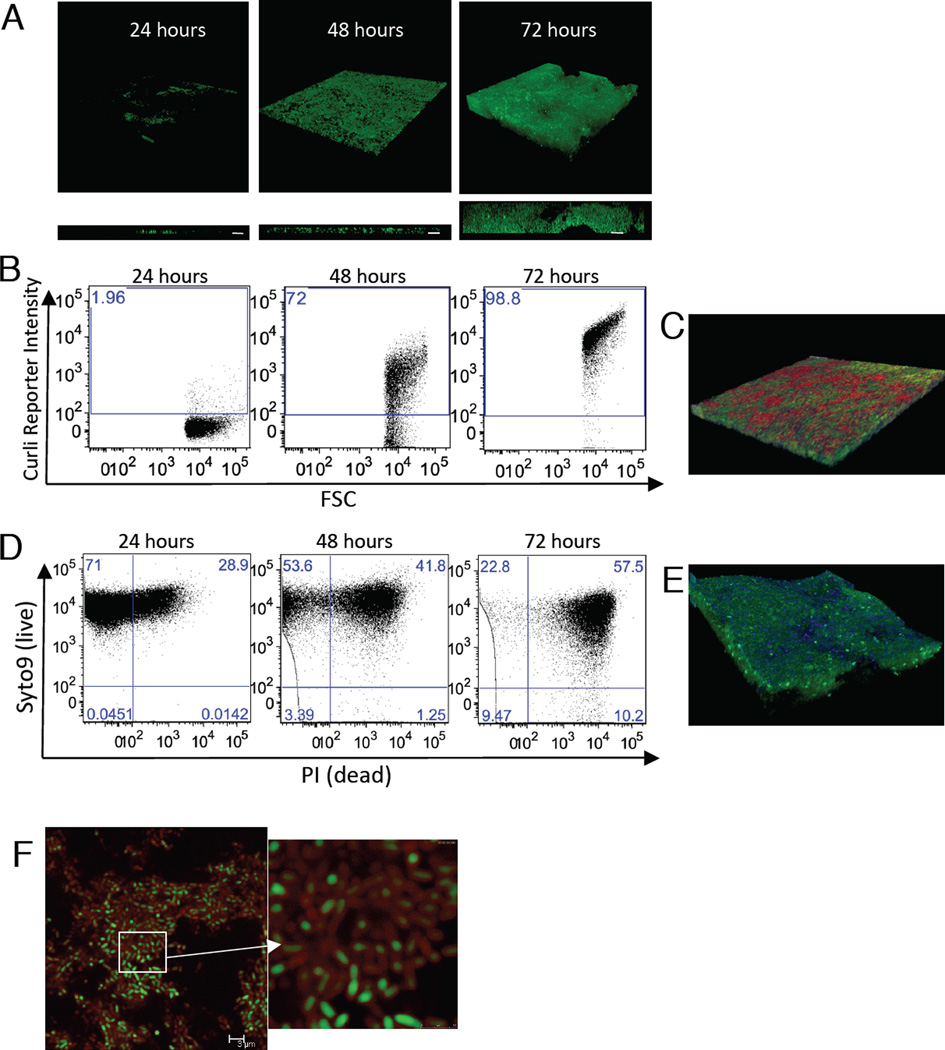
Curli fibers are conserved among the members of Enterobactericeae family (Barnhart and Chapman, 2006). Here, we studied the biofilms of S. Typhimurium as a model organism, since Salmonella is an important cause of infection and morbidity in SLE patients (Gerona and Navarra, 2009). S. Typhimurium forms pellicle biofilms at the air-liquid interface. When grown on microscope slides in 50 ml conical tubes for 72 hours, the biofilm thickness reached up to 33 microns (Figures. 1A, S1A). Under the same experimental conditions, a csgBA mutant that could not express curli monomers failed to form a pellicle biofilm, indicating that curli is required for S. Typhimurium biofilm (Figures S1A, S1B). We monitored the kinetics of curli expression in the biofilms by flow cytometry using a S. Typhimurium strain carrying a plasmid in which PcsgBA promoter drives the expression of green fluorescence protein (GFP) (PcsgBA::gfp). Curli expression started within 48 hours after inoculation, as more than 70% of the bacteria expressed GFP. By 72 hours, GFP was detected in virtually the entire population of the biofilm, suggesting that most bacteria express curli (Figure 1B). We also visualized curli fibers in biofilms by staining the pellicle with an amyloid specific stain, Congo Red (CR) (Figure 1C).
Figure 1. Salmonella biofilms contain curli amyloids and extracellular DNA (eDNA).
(A) 3D projection (upper) and side view (lower) of Confocal Laser Scanning Microscopy images of GFP expressing S. Typhimurium (green) pellicle biofilms during 72-hour time course of growth. White bars represent 10 µm. (B) Curli expression was monitored by Flow cytometry (FC) for 72 hours in S. Typhimurium containing a PcsgBA::gfp reporter plasmid. (C) 3D projection of CLSM images of S. Typhimurium biofilm with PcsgBA::gfp reporter plasmid at 72 hours. Biofilm was stained with Congo Red (red). (D) FC analysis of cell death in Salmonella biofilms using live/dead staining with Syto 9 and PI. (E) 3D projection of CLSM images of a 72-Salmonella biofilm with PcsgBA::gfp reporter plasmid stained with PI (pseudo colored blue). (F) CLSM image of a 72-hour S. Typhimurium biofilm with PcsgBA::gfp reporter plasmid stained with PI. Red “corona” of extracellular DNA around the bacteria is visible. All data shown are representative samples of three independent experiments.
We hypothesized that the nucleic acids found within the bacterial biofilms (Johnson et al., 2013; Whitchurch et al., 2002) could be derived from dying bacteria. To monitor cell death in the biofilm, we stained the bacteria in the pellicle with a live (Syto9)/dead (PI) stain. Approximately 10% of the bacteria in the biofilm were determined dead (Syto9 negative/PI positive) by 72 hours. Unexpectedly, a large fraction of the population – approximately 57%-showed positivity for both stains, suggesting that these cells were dying. However, this result was puzzling because the bacterial enumeration did not reveal a decrease in the bacterial numbers in the pellicle biofilm (Figures 1D, S1C). To further analyze this phenomenon, we stained the biofilm of a S. Typhimurium strain carrying the reporter PcsgBA::gfp with PI to visualize curli expression and DNA release. Green cells were surrounded by a red corona, demonstrating that eDNA was localized around live, curli-expressing cells and indicating that the high percentage of PI positivity in Figure1D is not due to cell death. We also observed high DNA staining of channels within the biofilm (Figure 1E and F). Therefore, eDNA, either released by dying bacteria or actively released into the ECM, is an important component of the S. Typhimurium biofilm pellicle (Figure S1D).
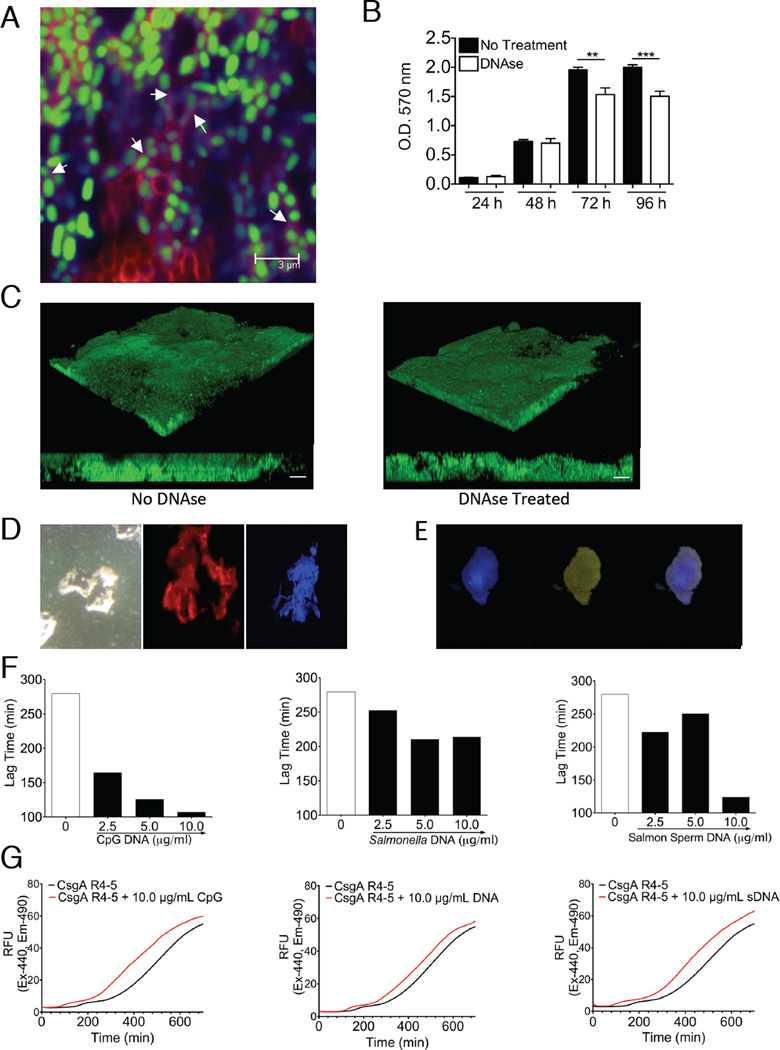
We found the amyloid specific dye, Congo Red, to be incompatible with nucleic acid dyes PI and DAPI but not with Toto-1. Therefore, we stained the S. Typhimurium biofilms using Congo Red (red) and Toto-1 (blue) and showed that the amyloids and DNA co-localized (magenta) within S. Typhimurium biofilms (Figure 2A). Treatment of S. Typhimurium biofilms with DNase did not reduce the biofilm mass as dramatically as reported for P. aeruginosa biofilms, which do not contain amyloids (Whitchurch et al., 2002) (Figure 2B and 2C). Due to the resistance of amyloids to enzymatic breakdown, we hypothesized that the incorporation of eDNA into the curli fibers rendered eDNA resistant to DNase. To further investigate the interactions of curli and eDNA, we purified curli from S. Typhimurium biofilms (Collinson et al., 1991). It is important to note that the purification process of curli includes multiple cycles of DNase and RNase treatments, boiling in SDS, and overnight electrophoresis through a preparative SDS polyacrylamide gel to eliminate any contaminants. When we stained such purified curli individually with Congo Red, or nucleic acid specific dyes, curli stained positive with PI and Hoescht 33258, indicating the presence of nucleic acids in the purified curli fibers (Figure 2D, center and right panel). Birefringence, a characteristic of amyloids stained with Congo Red, was visible under polarizing microscopy (Figure 2D, left panel). Purified curli stained positive with ethidium bromide, a DNA specific dye, confirming the presence of DNA in the fibers. Further treatment of purified curli with DNase or RNase did not remove the nucleic acids from the fibers unless fibers were broken down into its monomeric units by hexaflouroisopropanol (HFIP) and DMSO treatment (Figure S2). Using phenol-chloroform extraction we were able to extract an average of 639 ng (SD = 200) of DNA per 100 µg of purified curli that had extensively been treated with DNase. Finally, purified curli was co-stained with Thioflavin T (ThT), another amyloid specific stain, and Hoescht33258. Purified curli fibers stained positive with both dyes (Figure 2E). Thus, bacterial eDNA is protected from degradation by its close association with amyloid fibers.
Figure 2. eDNA forms complexes with curli in biofilms accelerating the amyloid polymerization process and limiting its degradation by DNAse.
(A) CLSM image of GFP expressing S. Typhimurium (green) biofilm grown for 72 hours and stained with Congo Red (red) and TOTO-1 Iodide (blue). (B) Crystal violet assay of S. Typhimurium biofilms grown in 96-well plates with or without 3 hour DNAse treatment. Data shown is average and SE from 5 wells. Experiments were completed in triplicate. (C) 3D reconstructions (upper) and side view (lower) CLSM images of GFP expressing Salmonella biofilms with and without DNAse treatment for 3 hours after biofilm was formed showing minimal change in biofilm thickness after treatment with DNAse. White bars on side views represent 10 µm. Images are representative of three independent experiments. (D) Microscopy of purified curli amyloids stained with Congo Red and visualized under polarized microscope (left panel), propodium iodide (center panel) and Hoescht 33258 (right panel) and visualized under fluorescent microscope. (E) Microscopy of purified curli fibers co-stained with Hoescht 33258 (left panel), ThT (center panel) and merged (right panel). (F) Calculated polymerization lag times of synthetic peptides consisting of the amino acid sequence of the fourth and fifth repeats of CsgA (CsgA R4-5) in the presence of increasing concentrations of CpG (left), genomic DNA from Salmonella (middle), and salmon sperm DNA (right). (G) Representative ThT amyloid polymerization assay curves for the polymerization of the CsgA R4-5 in the presence and absence of CpG (left), Salmonella genomic (middle), and salmon sperm (right) DNA. Three independent experiments were completed with duplicate wells for each concentration of DNA in each experiment. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DNA accelerates the polymerization of curli fibers
Nucleic acids promote the polymerization of a human amyloid precursor protein (Di Domizio et al., 2012b). Amyloid polymerization can be monitored using ThT and it shows a sigmoidal polymerization curve with a lag phase followed by exponential growth of amyloid fibers, which eventually reaches a plateau as all monomers are consumed (Naiki et al., 1991). To determine the effects of DNA on curli polymerization, we utilized a synthetic peptide corresponding to the fourth and fifth repeats of the CsgA monomer of curli (CsgAR4-5), which is devoid of any DNA contamination, and we performed in vitro polymerization assays using ThT in the presence or absence of increasing concentrations of the synthetic oligonucleotide, CpG, and of Salmonella genomic DNA. Addition of DNA accelerated the polymerization of CsgAR4-5 by decreasing the lag phase of polymerization by an average 172 minutes with CpG DNA and 65 minutes with Salmonella genomic DNA. We think that the difference in the polymerization rates is due to the DNA fragment size in each DNA source. Since enteric bacterial biofilms form in the gastrointestinal tract of humans (Bollinger et al., 2007; Macfarlane et al., 2011; Macfarlane and Dillon, 2007; von Rosenvinge et al., 2013), we also asked whether this acceleration also occurred with eukaryotic DNA. We used salmon sperm DNA and found that it also accelerated curli polymerization by shortening the time in lag phase of polymerization by an average of 155 minutes (Figure 2F–G). These findings suggest a mechanism by which DNA provides structural stability to biofilms, by complexing with amyloids and promoting their polymerization. Furthermore, the fact that eukaryotic DNA also enhanced amyloid polymerization suggests that bacteria could incorporate eukaryotic DNA (e.g. released during tissue damage) into its biofilm structure. This process may generate highly immunogenic molecules since both amyloids and DNA can activate innate immune receptors (Hemmi et al., 2000; Tukel et al., 2009).
Curli-DNA composites are major immune stimulators in S. Typhimurium biofilms
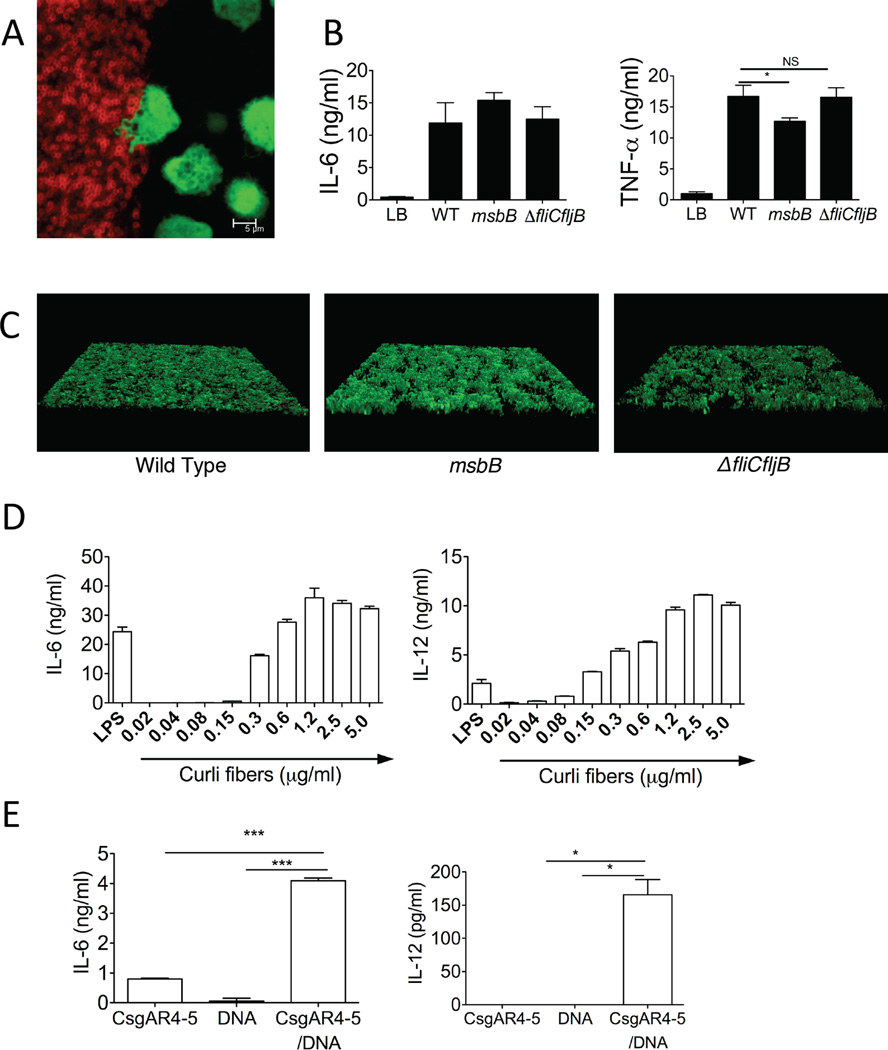
Interactions of bacterial biofilms with the immune system are poorly characterized. Here, we sought to elucidate these interactions using an in vitro model of conventional dendritic cells (cDCs), the pivotal initiators of the immune response that are stimulated via pattern recognition receptors (PRRs) (Gallo and Gallucci, 2013; Iwasaki and Medzhitov, 2004). We incubated bone marrow-derived dendritic cells, expressing GFP, on S. Typhimurium biofilms. Biofilms were allowed to mature for 72 hours prior to addition of cDCs. The cDCs appeared to send dendrites into the biofilm (Figure 3A, S3A–B) and phagocytose bacteria (Movies S1 and S2). Moreover, cDCs produced high amounts of proinflammatory cytokines including IL-6 and TNF-α (Figure 3B). These results indicate that cDCs can uptake antigens and be activated by the impervious bacterial biofilm.
Figure 3. Curli-DNA composites are danger signals that activate dendritic cells.
(A) GFP expressing cDCs (green) interact with S. Typhimurium biofilms stained with Congo Red (red). Image shown is representative of three independent experiments. (B) Cytokine production by cDCs incubated for 6 hours on 72 hour pre-formed biofilms from wild type, msbB, and ΔfliCfljB Salmonella. (C) 3D reconstructions of CLSM images of GFP expressing WT (Left Panel), msbB (Center Panel), and ΔfliCfljB (Right Panel) Salmonella biofilms showing equivalent growth of these biofilms in these conditions. (D) Proinflammatory cytokine production from cDCs stimulated with a dose titration of curli-DNA composites isolated from Salmonella msbB mutant biofilms. Cytokine production was compared to LPS stimulation (100ng/ml). Data in (B)(D) is representative of three independent experiments, performed with three independent bone marrow-derived cultures from 3 different mice and each contained quadruplicate wells for each bacterial strain. (E) Synthetic amyloid peptide CsgA R4-5 was polymerized in the presence or absence of Salmonella genomic DNA and used to stimulate cDCs. Cytokine production in response to 20µg/ml CsgA R4-5, 20 ng/µl genomic DNA, or 20µg/ml CsgA R4-5 fibrillized in the presence of 20ng/µl genomic DNA is shown as Average and SEM, n=5. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Lipid A and monomers of S. Typhimurium flagella are well-known immune system activators that trigger TLR4-MD2-CD14 and TLR5, respectively (Gewirtz et al., 2001; Takeuchi et al., 1999). To determine the importance of LPS and flagellin in the stimulatory effect of biofilms, we established biofilms with a S. Typhimurium msbB mutant that expresses a tetra-acylated form of lipid A, which does not signal through the TLR4-MD2-CD14, and with a S. Typhimurium fliCfljB mutant that lacks flagella. After 48 hours, the biofilms formed by the mutant bacteria were comparable to the biofilms formed by wild type S. Typhimurium. (Figures 3C, S3C). When cDCs were exposed to the biofilms, cDCs produced equally high amounts of IL-6 in response to the biofilms formed by wild type S. Typhimurium, msbB mutant and fliCfljB mutants (Figure 3B). Although there was a statistically significant difference in the amount of TNFα produced by cDCs in response to the msbB mutant biofilm when compared to the wild type S. Typhimurium and the fliCfljB mutant biofilms, cDCs still produced high amounts of TNFα in response to the three biofilms (Figure 3B). Thus, although LPS and flagellin are important for the interaction of immune cells with planktonic S. Typhimurium, components other than LPS and flagellin exist in the S. Typhimurium biofilm that activate DCs.
Next, we sought to determine whether curli-DNA composites are involved in DC response to curli-containing biofilms. However, since a curli-deficient S. Typhimurium does not form biofilms (Figure S1B), we treated cDCs with a dose titration of purified curli fibers. To avoid contamination with LPS, we purified curli fibers from the msbB mutant and found that even nanogram quantities of curli induced significant amounts of IL-6 and IL-12 in cDCs. Interestingly, curli was very potent in inducing the Th1 cell-promoting cytokine IL-12 even when compared to the dose of LPS that showed the maximal cytokine response in cDCs (Figure 3D).
To determine the role of DNA in the stimulatory activity of curli-DNA composites from biofilms, we attempted to remove the DNA from the purified curli and found that we could not unless we dissociated curli into its monomers. However, this treatment damaged the DNA (Figure S2). To overcome this pitfall, we stimulated cDCs with the synthetic CsgAR4-5 peptide polymerized in the presence or absence of bacterial genomic DNA. DNA alone and CsgAR4-5 polymerized in the absence of genomic bacterial DNA induced minimal IL-6 and IL-12 production by cDCs. However, CsgAR4-5 polymerized in the presence of bacterial DNA induced significant amounts of IL-6 and IL-12, suggesting that curli and DNA synergize to activate innate immune cells (Figure 3E). The fact that the CsgAR4-5-DNA composites elicited much lower amounts of cytokines than the purified curli suggests that the natural aggregation of full-length curli may be required for its full activity.
Curli-DNA composites stimulate the Type I interferon response
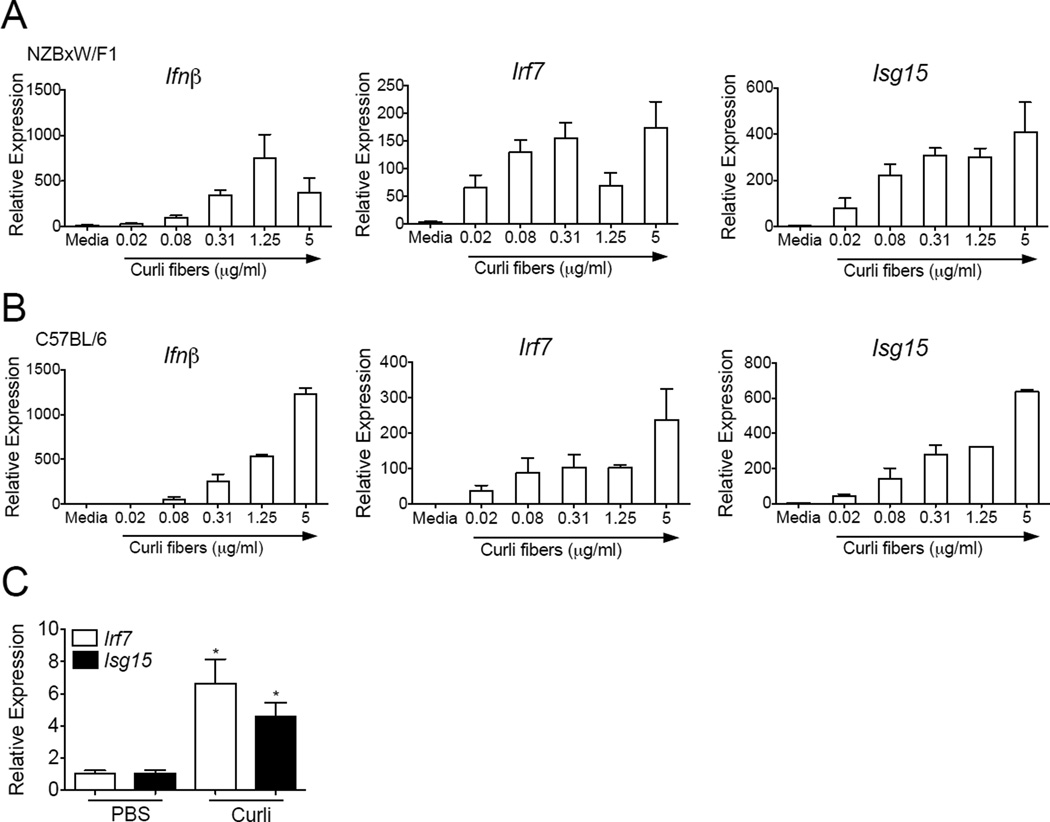
Type I Interferons (IFNs) are important stimulators of the innate and adaptive immune response and are pathogenic in SLE (Elkon and Stone, 2011). Type I IFNs can activate dendritic cells as an exogenous stimulus (Gallucci et al., 1999) and they mediate PRR-dependent activation of DCs through an autocrine feedback (Hoebe et al., 2003). Here we tested whether the curli-DNA composites purified from S. Typhimurium msbB biofilms stimulate DC activation and found that curli-DNA composites were potent inducers of Type I IFNs and IFN Stimulated Genes (ISGs) in vitro in cDCs from lupus-prone NZBxW/F1 and non-lupus prone C57BL/6 mice (Figures 4A and 4B). Furthermore, in vivo injection of curli-DNA composites also resulted in the up-regulation of the ISGs, Isg15 and Irf7, in splenic DCs from young pre-diseased NZBxW/F1 lupus prone mice (Figure 4C). These results indicate that curli-DNA composites are potent danger signals that stimulate autoimmune-prone innate immune cells and may trigger autoimmunity by providing an adjuvant effect through the stimulation of the pathogenic IFN response.
Figure 4. Curli-DNA composites elicit strong type I interferon responses from cDCs in vitro.
mRNA expression of interferon beta, Ifnβ, and two IFN-stimulated genes (ISGs), Irf7 and Isg15, from cDCs generated in vitro for 6–7 days from bone marrow of NZBxW/F1 mice (A) and C57BL/6 mice (B) and stimulated for 6 hours with curli/DNA composites isolated from Salmonella biofilms. All of the conditions were normalized against the control (untreated DCs in medium only) for each strain in each experiment. Results are average of three independent experiments, (C) ISG mRNA expression in NZBxW/F1 splenic CD11c+ DCs isolated by magnetic beads 20 hours post i.p. injection of 50µg of curli. *, P < 0.05.
Curli-DNA composites promote autoimmunity
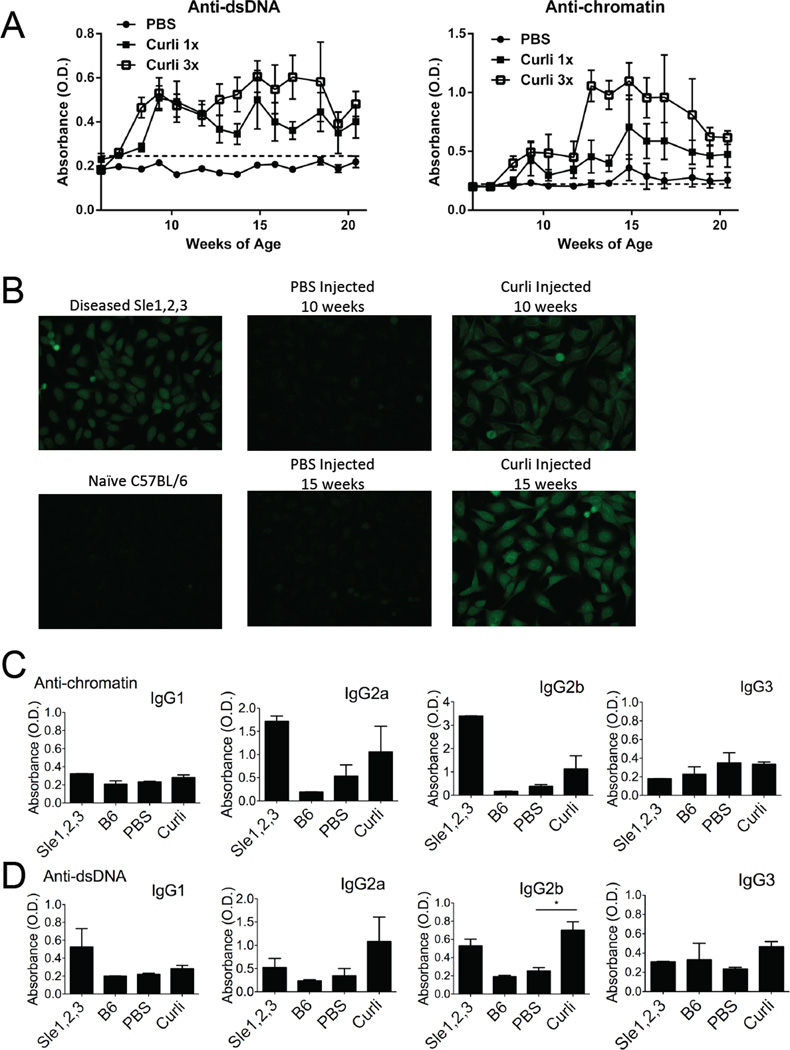
Next, we asked whether the naturally occurring curli-DNA composites found in bacterial biofilms could induce autoimmunity in the absence of any added adjuvant. Since we found that curli-DNA composites stimulate the production of Type I IFNs (Figure 4), which accelerate disease onset in lupus-prone mice (Mathian et al., 2005), we hypothesized that exposure to curli-DNA composites may accelerate autoimmunity. We therefore injected 6-week-old, pre-diseased, NZBxW/F1 mice i.p. with PBS or 50 µg of curli-DNA composites once a week, or three times a week; and monitored autoantibody production. Mice injected with PBS remained negative for autoantibodies up to 20 weeks of age. Mice injected with curli-DNA composites rapidly produced anti-dsDNA and anti-chromatin autoantibodies, a hallmark of lupus autoimmunity (Figure 5A). As a protein control, we injected 50 µg of Bovine Serum Albumin (BSA) i.p. into NZBxW/F1 age and gender matched mice and found that injections with BSA did not induce development of autoantibodies (Figure S4), suggesting specificity for the autoimmunogenicity of curli.
Figure 5. Curli/DNA composites accelerate systemic autoimmunity in lupus-prone NZBxW/F1 mice.
(A) Anti-dsDNA and anti-chromatin autoantibodies were measured by ELISA in sera from NZBxW/F1 mice injected with PBS thrice a week, curli once a week (1×), or curli thrice a week (3×). Optical density (O.D.) indicates ELISA color change and the presence of anti-dsDNA or anti-chromatin autoantibodies. Error bars indicate SEM, n=5 per group. The dotted horizontal line indicates cutoff for positivity, calculated as two standard deviations above the average of sera from naïve C57BL/6 mice. (B) Representative antinuclear antibody assay (ANA) of sera from mice as in Figure 5A at the indicated ages. Hep-2 cells were incubated with sera and FITC-conjugated anti-IgG Abs. Antibody isotype determinations of anti-chromatin (C) and anti-dsDNA (D) antibodies. Serum from 10 month old diseased Sle1,2,3 mouse was utilized as positive control and from naïve C57BL/6 mouse was used as negative control. *, P < 0.05.
Other adjuvants, including TLR ligands, can accelerate murine lupus (Hang et al., 1983; Jorgensen et al., 2006). However, curli-DNA composites induce autoantibody production very quickly and consistently, with all mice becoming positive for anti-dsDNA within two weeks of the first injection. We confirmed lupus autoimmunity in curli-DNA treated mice by antinuclear antibody test (ANA). Curli-DNA treated mice became ANA positive (Figure 5B), with an initial perinuclear-endoplasmic reticulum staining pattern; this pattern then evolved into a nuclear staining pattern by 15 weeks of age, resembling the pattern expressed by aged diseased Sle1,2,3 mice, another strain of mice that spontaneously develop lupus (Morel et al., 1997). Furthermore, when we injected curli-DNA composites into SvJ-129 mice, which do not develop lupus spontaneously but are genetically predisposed to autoimmunity (Bygrave et al., 2004), we found that these mice rapidly produced both anti-dsDNA and anti-chromatin autoantibodies (Figure S5), confirming that bacterial amyloid-DNA composites are sufficient to trigger autoimmunity.
We next analyzed the subclasses of autoantibodies that are induced by curli-DNA composites. We found that anti-dsDNA and anti-chromatin autoantibodies were mostly of immunoglobulin G (IgG) subclass 2a and 2b, as in diseased Sle1,2,3 lupus-prone mice (Figures 5C–D). Since IgG2a and IgG2b subclasses are generally considered pathogenic in systemic autoimmunity (Clynes et al., 1998; Ehlers et al., 2006), our results indicate that curli-DNA composites promote the production of pathogenic autoantibodies.
Curli-DNA composites induce autoimmunity in wild-type mice
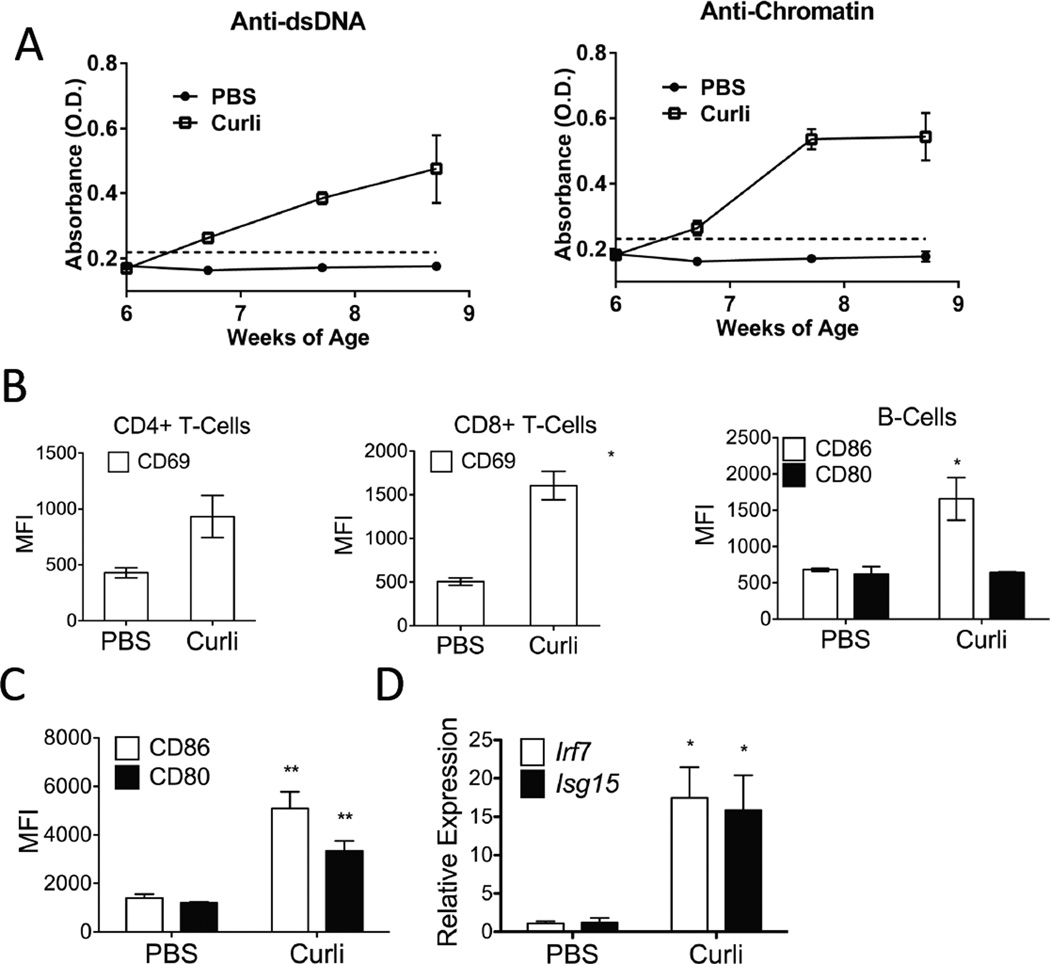
To test whether naturally occurring curli-DNA composites induce autoimmunity in mice not prone to lupus, we injected B6 mice i.p. with curli (50µg per injection) three times a week and found anti-dsDNA and anti-chromatin autoantibodies as soon as two weeks after injection (Figure 6A), following the same kinetics seen in NZBxW/F1 and SvJ-129 mice. These results indicate that the direct exposure of the immune system to the danger signal curli-DNA composites is sufficient to break self-tolerance in non-autoimmune mice.
Figure 6. Curli/DNA composites induce autoantibody production and immune cell activation in WT Mice.
(A) Anti-dsDNA and anti-chromatin autoantibody ELISAs from C57BL/6 mice injected with PBS or curli thrice a week (3×). As in Fig. 5, optical density (O.D.) indicates ELISA color change and the presence of anti-dsDNA or anti-chromatin. Error bars indicate SEM, n=5 per group. (B) Flow cytometry analysis of the expression of activation markers in T-cells and B-Cells and (C) in DCs from spleens of C57BL/6 mice 20 hours post i.p. curli injection; (D) mRNA expression of two interferon-stimulated genes (ISGs), Irf7 and Isg15, from C57BL/6 splenic CD11c+ DCs isolated with magnetic beads from the same mice. Results are shown as averages and standard deviation, n=3 per group, *, P < 0.05.
To investigate the effects of curli-DNA composites on adaptive immune cells in vivo, we injected mice i.p. with either 50µg of curli-DNA composites or PBS and analyzed markers of activation in splenic populations 24 hours later. Injection of curli-DNA composites up-regulated the expression of the co-stimulatory molecule CD86 on CD19+ B cells and the activation marker CD69 on both CD4+ and CD8+ T cells (Figure 6B), suggesting that curli-DNA composites stimulate polyclonal activation of T and B cells. It also caused an increase of CD11b+Ly6C+ inflammatory myeloid cells in the spleen (Figure S6). Moreover, curli-DNA composite injection induced DC activation in vivo, as splenic CD11c+ DCs up-regulated both co-stimulatory molecules CD86 and CD80 (Figure 6C) and highly expressed the ISGs, Irf7 and Isg15, (Figure 6D). Thus, bacterial amyloids, and in particular the curli-DNA composites, represent a class of danger signals that stimulate innate immune cells and induce polyclonal activation of adaptive immunity.
Infection with curli-competent bacteria accelerates autoimmunity
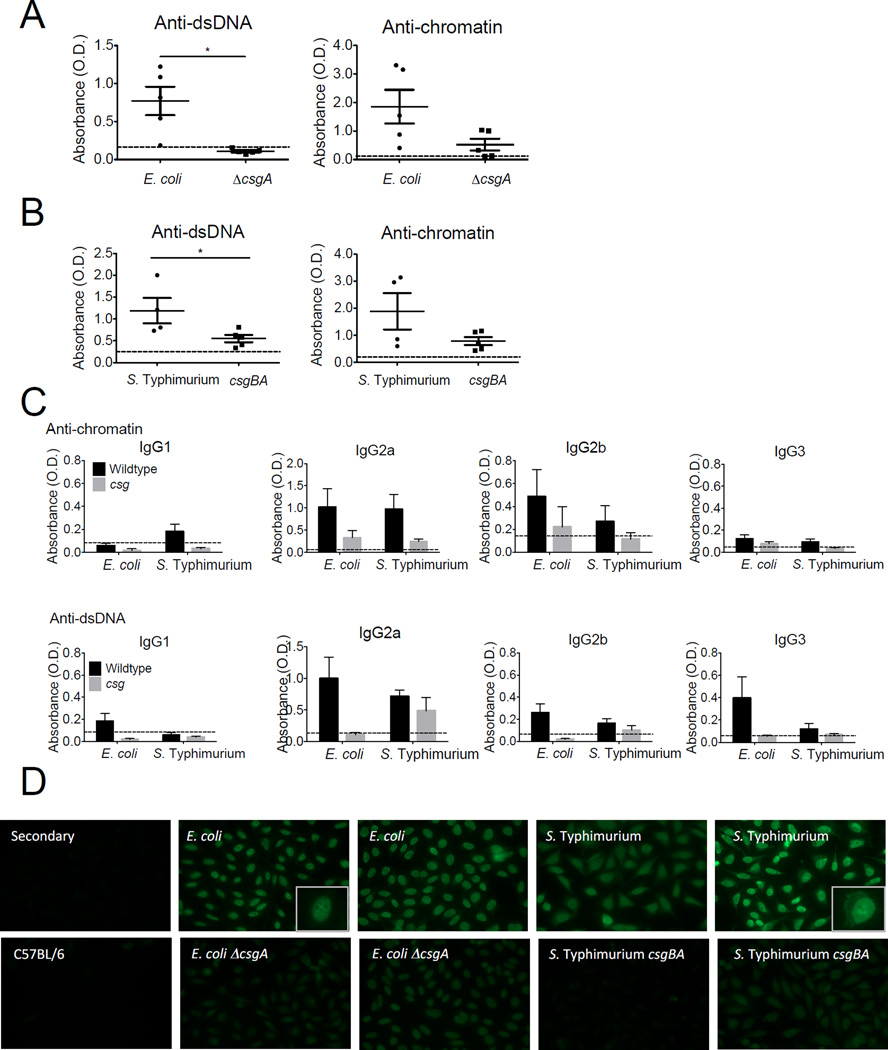
We next asked whether curli-expressing bacteria, either commensal or pathogenic, promote autoimmunity in lupus-prone NZBxW/F1 mice. We injected young pre-diseased NZBxW/F1 mice i.p. with either wild type (curli-expressing) or curli mutant commensal E. coli or virulent S. Typhimurium. Systemic route of infection (i.p.) was chosen to mimic the propensity of SLE patients to develop systemic infections including bacteremia. We injected E. coli once a week for 4 weeks or S. Typhimurium once every two weeks for 8 weeks. We spaced Salmonella injections every two weeks to prevent mouse death. We then stopped injections and let mice recover for at least 4 weeks. We found that mice infected with curli-competent bacteria, both E. coli and Salmonella, developed high titers of autoantibodies (Figure 7A–B), similar to those induced by isolated curli-DNA composites (Figure 5). Mice exposed to curli-deficient bacteria also developed autoantibody titers albeit at lower amounts compared to curli-competent strains. This could be due to other danger signals expressed by the bacteria such as LPS, which was previously shown to trigger autoantibody production in mice (Fournie et al., 1974). The fact that higher amounts of autoantibodies were produced upon infections with curli-competent bacteria indicates that curli is a major autoimmunogen in these bacteria (Figure 7A–B). The predominant autoantibody isotype in response to infection was IgG2a, and IgG2b and IgG3 were also positive to a lesser extent (Figure 7C), suggesting that infection promote the production of pathogenic autoantibodies to nuclear antigens. Furthermore, mice infected with curli-competent wild type bacteria had significantly stronger ANA staining (Figure 7D). Interestingly, sera from mice infected with S. Typhimurium showed both nuclear and cytoplasmic staining, while mice infected with E. coli showed nuclear staining only, suggesting that different bacteria may diversify the autoantibody repertoire. After sacrificing the animals, we analyzed blood, spleens, and livers for colony forming units (CFUs) and found no evidence of persistent infection at the time of euthanasia (data not shown). Together our results indicate that the exposure to curli or infection with biofilm-competent bacteria promotes the development of autoantibodies in susceptible mice.
Figure 7. Infection with curli-producing bacteria accelerates autoimmunity in lupus-prone mice.
Anti-dsDNA and anti-chromatin autoantibody ELISAs from NZBxW/F1 mice injected with live E. coli or S. Typhimurium strains. (A) Wild type E. coli or its csgA mutant was injected i.p. into NZBxW/F1 mice at 105 CFU for four times. (B) Live virulent S. Typhimurium or its isogenic csgBA mutant was injected i.p. into NZBxW/F1 mice at 105 CFU for four times. (C) Autoantibody isotypes. The dotted horizontal line indicates cutoff for positivity, calculated as two standard deviations above the average of sera from naïve C57BL/6 mice. *, P < 0.05. (D) Two representative ANA stainings from each experimental group are shown. Magnified insets at the right corner of two pictures.
DISCUSSION
Studies using amyloid specific dyes demonstrated that up to 40 % of the bacterial biofilms contain amyloids (Larsen et al., 2007). However, it has been a challenge to identify novel bacterial amyloid proteins using basic amino acid alignment searches since these proteins do not share any sequence homology but they have a common quaternary structure. Although curli fibers were initially thought to be encoded by only the members of the Enterobactericeae family, gene homologs encoding curli were recently determined also in four phyla, Bacteroidetes, Proteobacteria, Firmicutes, and Thermodesulfobacteria (Dueholm et al., 2012; Hufnagel et al., 2013). Among these, Bacteriodetes, Firmicutes and Proteobacteria are the major phyla found in the gastrointestinal tract, but more work is needed to determine the abundance of amyloids in the gastrointestinal tract (Gill et al., 2006; Hooper et al., 2002; Turnbaugh et al., 2007). Besides curli fibers, amyloids are produced also by Staphylococcus aureus and Mycobacterium tuberculosis (Alteri et al., 2007; Chapman et al., 2002; Hufnagel et al., 2013; Schwartz et al., 2012).
Although it was known that amyloids and DNA are incorporated into the biofilm ECM(Whitchurch et al., 2002), little is known about their interactions. Here, we demonstrate that DNA released during biofilm formation is incorporated into curli fibers of S. Typhimurium and accelerates the fiber polymerization process. Since we found that only approximately 10% of the bacteria die in the biofilm population, the high number of live bacteria with a red DNA corona around them point to an active nucleic acid extrusion mechanism during S. Typhimurium biofilm development. Furthermore, the hollow areas lacking green bacteria but showing only the red amyloid stain suggest that in order to incorporate into curli fibers, eDNA must be present during the initial stages of amyloid production and polymerization.
Our results using eukaryotic DNA from salmon sperm also show that eukaryotic DNA can be incorporated into bacterial amyloid fibers, which suggest that host DNA, released in the vicinity of a bacterial biofilm possibly during tissue damage, may be incorporated into the ECM through this mechanism. Moreover, neutrophils extrude large amounts of DNA called the neutrophil extracellular traps (NETs) as a mechanism to kill bacteria (Brinkmann et al., 2004). Recent studies have shown that P. aeruginosa can incorporate neutrophil-derived DNA into its ECM in order to form a thicker biofilm (Walker et al., 2005). Together with our results, these findings suggest that bacteria may take advantage of eukaryotic DNA in the vicinity to enhance its ECM.
DCs are the sentinels of the immune system, which recognize pathogen associated molecular patterns (PAMPs) (Iwasaki and Medzhitov, 2004) and damage associated molecular patterns (DAMPs) (Gallo and Gallucci, 2013) during infection or tissue damage, and initiate the adaptive immune response (Merad et al., 2013). We found that DCs are highly activated by biofilms and by curli-DNA composites in particular. We have also reported that the amyloid component of curli fibers stimulates the macrophages through the TLR2-TLR1 complex (Rapsinski et al., 2013; Tukel et al., 2010). These results indicate that curli fibers are PAMPs, able to stimulate the innate immune system. Although studies on eDNA and its sensors are limited, bacterial dsDNA can stimulate TLR9 (Hemmi et al., 2000) and cytosolic STING-dependent DNA sensors resulting in a Type I interferon response (Ishikawa et al., 2009). The fact that curli-DNA composites are more immunostimulatory than curli or DNA alone suggests that different immune receptors might synergize to recognize these novel molecular patterns in bacterial biofilms.
As many other PAMPs, curli-DNA composites strongly induce the expression of Type I IFNs and the IFN stimulated response in DCs in vitro and in vivo. A Type I IFN Signature is up-regulated in PBMCs of SLE patients (Baechler et al., 2003; Bennett et al., 2003; Crow et al., 2003; Feng et al., 2006; Han et al., 2003) and in DCs from lupus prone mice (Sriram et al., 2012), and administration of exogenous Type I IFNs accelerates autoimmunity in lupus prone mice (Mathian et al., 2005). Therefore, the results that curli-DNA composites accelerate the onset of autoimmunity in lupus-prone mice can be partially explained by the strong induction of Type I IFNs in DCs as well as by the polyclonal activation of the adaptive immune system, as curli up-regulated activation markers in splenic T and B cells.
SLE manifests with the production of antinuclear antibodies, including those directed against DNA, ribonucleoprotein complex (RNP), and nucleosomes (Tan, 2012). Our findings that naturally occurring bacterial curli-DNA composites, which can contain both bacterial and eukaryotic DNA, efficiently trigger Type I IFN stimulation and autoantibody production points to a unique bacterial stimulator of autoimmunity. The fact that eukaryotic DNA complexes with the bacterial amyloid curli brings up the possibility of the exposure of the lupus-prone immune system to autoAgs by the curli-DNA composites in presence of the adjuvant effects caused by TLR triggering and Type I IFN stimulation. Self-Ags can be released by NETs or by necrotic cells during tissue damage induced by bacterial infections. Moreover, defects in phagocytosis and clearance of apoptotic cells are associated with lupus (Shigekazu et al., 2010) and may allow self DNA from post-apoptotic cells to be incorporated into biofilms and become immunogenic.
SLE has a multifactorial pathogenesis in which the genetic make-up and environmental triggers are considered the major players (Liu and Davidson, 2012). Among the environmental triggers, infections have been proposed as either initiators of autoimmunity or triggers of flares, the sudden increase in disease severity (Petri, 1998). Several studies have tested this hypothesis, providing conflicting results. For example lupus-prone MRL/lpr mice bred in germ-free conditions develop lupus-like disease (Maldonado et al., 1999), suggesting that genetics trump environmental factors. However, the fact that SLE has a concordance of 30–40% in identical twins (Connolly and Hakonarson, 2012), clearly indicate that the environmental triggers are required in human disease. Consistent with the idea that bacterial infections contribute to lupus pathogenesis, we found that infection with biofilm-competent, curli-producing bacteria exacerbates autoimmunity in lupus-prone mice. These results are possibly due to the direct exposure to curli itself but also to the fact that curli-expressing bacteria are better protected from the immune response – thereby increasing exposure and causing a greater immune activation.
How bacterial infections contribute to lupus pathogenesis and flares remain inconclusive. Our study suggest that biofilm-derived curli-DNA composites can accelerate autoimmunity through the induction of Type I IFNs, the activation of DCs, and the polyclonal stimulation of T and B cells. The fact that also the commensal strain of E. coli could accelerate autoimmunity, and that Salmonella and E.coli induced different patterns of autoantibodies, highlights the need to address how the interaction with pathogens and microbiota affects autoimmunity in genetically susceptible individuals.
Our results that curli-DNA composites can trigger autoimmunity even in non-autoimmune prone mice raise the question of why autoimmunity is not more frequent in the human population that is often exposed to biofilm-forming bacteria. A working hypothesis is that in non-lupus-prone individuals, or murine strains, natural infections do not normally result in the systemic release of curli-DNA composites in amount equivalent to those we injected in this study; while in susceptible individuals, genetically determined dysfunctions may increase the systemic exposure to curli-DNA composites during chronic infections. The more frequent outcome of Salmonella infection in bacteremia and complications in soft tissues in SLE patients (Lim et al., 2001; Pablos et al., 1994; Tsao et al., 2002) and the recent identification of increased levels of circulating endotoxin in SLE patients (Shi et al., 2014), support this hypothesis. In addition, since inflammatory cytokines can decrease intestinal mucosal barrier function (Turner, 2009), the high levels of cytokines present in pre-disease stages of lupus (Connolly and Hakonarson, 2012; Sriram et al., 2012), might compromise mucosal barrier function and allow the biofilm material produced by microbiota at mucosal sites to access the immune system, act as danger signals and trigger lupus onset.
Our study highlights a role of bacterial amyloid-DNA composites in stimulating the innate and the adaptive immune system, and suggests that chronic biofilm-producing bacterial infections may represent an important environmental contributor to SLE pathogenesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains
S. Typhimurium strain IR715 is a fully virulent, nalidixic acid-resistant strain derived from the ATCC strain 14028 (Stojiljkovic et al., 1995). An unmarked deletion of csgBA mutant of S. Typhiumurium strain 14028 (Nishimori et al., 2012), S. Typhimurium EHW26, a fliCfljB mutant, and S. Typhimurium RPW3 that contains a mutation in msbB were previously described (Raffatellu et al., 2005). E. coli Nissle 1917 was first described by Alfred Nissle (Nissle, 1957).
Acceleration of lupus onset
NZBxW/F1 mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). C57BL/6 (B6), congenic Sle1,2,3 and 129/SvJ mice were purchased from Jackson Laboratories and bred and maintained in our colonies in accordance with the guidelines of the Institutional Animal Care and Use Committees of Temple University, a member of American Association for the Accreditation of Laboratory Animal Care-accredited facilities. 6-week-old female NZBxW/F1 mice and B6 and 129 mice were injected i.p. with PBS, or 50µg curli-DNA composites or BSA in 0.5 ml of PBS either once a week, or three times a week. Serum was collected to monitor autoantibody production.
Lupus and Infections
E. coli and S. Typhimurium were grown in biofilm-promoting conditions. Live wild type E. coli or its csgA mutant was injected i.p. into 8 weeks old female NZBxW/F1 mice at 105 CFU per injection once a week for four weeks (4 injections total). Live S. Typhimurium or its isogenic csgBA mutant was injected i.p. into age-matched female NZBxW/F1 mice at 104 CFU per injection about once every two weeks for 8 weeks (4 injections total). Mice were allowed to recover from infection for 4 and 8 weeks after the last injection (for S. Typhimurium and E. coli respectively).
The rest of the Methods are in Supplemental Procedures.
Supplementary Material
HIGHLIGHTS.
Bacterial amyloid curli and DNA composites form within bacterial biofilms.
DNA accelerates the polymerization of bacterial amyloid curli.
Curli-DNA composites induce autoantibodies and Type I interferon.
Infections with amyloid-expressing bacteria trigger autoimmunity.
ACKNOWLEDGEMENTS
We would like to thank Drs. A.J. Baumler, P.L. Cohen, P. J. Piggot and P. Matzinger for reading the manuscript and providing constructive critiques.
Work in C.T.’s laboratory was supported by the NIH, NIAID 1R03AI107434 and 1R21AI105370. Work in S.G.’s laboratory was supported by the NIH, NIAID RO1-AI076423, the FCCC-Temple University Nodal grant and the Lupus Research Institute, Innovative Research Grant. RC was supported by NIH, NIAMS RO1-AR061569. PMG was partially supported by the Lupus Foundation’s Goldie Simon Preceptorship Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Co-first authors PMG and GJR contributed equally to this work. GJR performed the experiments related to the initial discovery of nucleic acids in curli fibers. GJR also performed biofilm experiments, amyloid polymerization experiments as well as confocal microscopy. PMG performed the experiments of activation of dendritic cells in vitro, in vivo treatment of mice, autoantibody ELISAs, ANA assay. PMG and RPW conducted the experiments for the in vivo acceleration of lupus with infections. US helped with the in vitro dendritic cell stimulations experiments. GOO purified curli fibers for in vivo and in vitro experiments and helped perform ELISAs. MG constructed the E. coli csgA mutant. BB optimized the confocal imaging of the S. Typhimurium biofilms. RC helped with the ANA staining experiments and provided a human disease perspective. SG and CT designed and oversaw all experimental work, wrote the manuscript, which was further edited by all authors. All authors approved the final manuscript.
REFERENCES
- Alteri CJ, Xicohtencatl-Cortes J, Hess S, Caballero-Olin G, Giron JA, Friedman RL. Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci U S A. 2007;104:5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Vargas A, Gomez-Martin D, Merayo-Chalico J, Ponce-de-Leon A, Alcocer-Varela J. Risk Factors for Drug-resistant Bloodstream Infections in Patients with Systemic Lupus Erythematosus. J Rheumatol. 2014 doi: 10.3899/jrheum.131261. [DOI] [PubMed] [Google Scholar]
- Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS. 2013;(Suppl):1–51. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- Bollinger RR, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the normal human large bowel: fact rather than fiction. Gut. 2007;56:1481–1482. [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Bygrave AE, Rose KL, Cortes-Hernandez J, Warren J, Rigby RJ, Cook HT, Walport MJ, Vyse TJ, Botto M. Spontaneous autoimmunity in 129 and C57BL/6 mice-implications for autoimmunity described in gene-targeted mice. PLoS Biol. 2004;2:E243. doi: 10.1371/journal.pbio.0020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes R, Dumitru C, Ravetch JV. Uncoupling of immune complex formation and kidney damage in autoimmune glomerulonephritis. Science. 1998;279:1052–1054. doi: 10.1126/science.279.5353.1052. [DOI] [PubMed] [Google Scholar]
- Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JJ, Hakonarson H. Role of cytokines in systemic lupus erythematosus: recent progress from GWAS and sequencing. J Biomed Biotechnol. 2012;2012:798924. doi: 10.1155/2012/798924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Reis P, Nativ S, Isgro J, Rodrigues T, Yildirim-Toruner C, Starr A, Saiman L, Imundo L, Eichenfield A. Major infections in a cohort of 120 patients with juvenile-onset systemic lupus erythematosus. Clin Immunol. 2013;149:442–449. doi: 10.1016/j.clim.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–490. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- Desai DD, Krishnan MR, Swindle JT, Marion TN. Antigen-specific induction of antibodies against native mammalian DNA in nonautoimmune mice. J Immunol. 1993;151:1614–1626. [PubMed] [Google Scholar]
- Di Domizio J, Dorta-Estremera S, Gagea M, Ganguly D, Meller S, Li P, Zhao B, Tan FK, Bi L, Gilliet M, et al. Nucleic acid-containing amyloid fibrils potently induce type I interferon and stimulate systemic autoimmunity. Proc Natl Acad Sci U S A. 2012a;109:14550–14555. doi: 10.1073/pnas.1206923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domizio J, Zhang R, Stagg LJ, Gagea M, Zhuo M, Ladbury JE, Cao W. Binding with nucleic acids or glycosaminoglycans converts soluble protein oligomers to amyloid. J Biol Chem. 2012b;287:736–747. doi: 10.1074/jbc.M111.238477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueholm MS, Albertsen M, Otzen D, Nielsen PH. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One. 2012;7:e51274. doi: 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M, Fukuyama H, McGaha TL. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE. The Journal of …. 2006 doi: 10.1084/jem.20052438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon KB, Stone VV. Type I interferon and systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31:803–812. doi: 10.1089/jir.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS, Dong X, Chen W, Kim MH, Weng HH, et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- Fessler BJ. Infectious diseases in systemic lupus erythematosus: risk factors, management and prophylaxis. Best Pract Res Clin Rheumatol. 2002;16:281–291. doi: 10.1053/berh.2001.0226. [DOI] [PubMed] [Google Scholar]
- Fournie GJ, Lambert PH, Meischer PA. Release of DNA in circulating blood and induction of anti-DNA antibodies after injection of bacterial lipopolysaccharides. J Exp Med. 1974;140:1189–1206. doi: 10.1084/jem.140.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo PM, Gallucci S. The dendritic cell response to classic, emerging, and homeostatic danger signals. Implications for autoimmunity. Front Immunol. 2013;4:138. doi: 10.3389/fimmu.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S, Lolkema M, Matzinger P. Natural adjuvants: endogenous activators of dendritic cells. Nat Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- Gerona JG, Navarra SV. Salmonella infections in patients with systemic lupus erythematosus: a case series. Int J Rheum Dis. 2009;12:319–323. doi: 10.1111/j.1756-185X.2009.01440.x. [DOI] [PubMed] [Google Scholar]
- Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GM, Chen SL, Shen N, Ye S, Bao CD, Gu YY. Analysis of gene expression profiles in human systemic lupus erythematosus using oligonucleotide microarray. Genes Immun. 2003;4:177–186. doi: 10.1038/sj.gene.6363966. [DOI] [PubMed] [Google Scholar]
- Hang L, Slack JH, Amundson C, Izui S, Theofilopoulos AN, Dixon FJ. Induction of murine autoimmune disease by chronic polyclonal B cell activation. J Exp Med. 1983;157:874–883. doi: 10.1084/jem.157.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hoebe K, Janssen EM, Kim SO, Alexopoulou L. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nature …. 2003 doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Hufnagel DA, Tukel C, Chapman MR. Disease to dirt: the biology of microbial amyloids. PLoS Pathog. 2013;9:e1003740. doi: 10.1371/journal.ppat.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S, Figueiredo J, Khare S, Nunes J, et al. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol. 2003;48:1357–1376. doi: 10.1046/j.1365-2958.2003.03507.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Jimenez JS. Protein-DNA interaction at the origin of neurological diseases: a hypothesis. J Alzheimers Dis. 2010;22:375–391. doi: 10.3233/JAD-2010-100189. [DOI] [PubMed] [Google Scholar]
- Johnson L, Horsman SR, Charron-Mazenod L, Turnbull AL, Mulcahy H, Surette MG, Lewenza S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013;13:115. doi: 10.1186/1471-2180-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen TN, Thurman J, Izui S, Falta MT, Metzger TE, Flannery SA, Kappler J, Marrack P, Kotzin BL. Genetic susceptibility to polyI:C-induced IFNalpha/beta-dependent accelerated disease in lupus-prone mice. Genes Immun. 2006;7:555–567. doi: 10.1038/sj.gene.6364329. [DOI] [PubMed] [Google Scholar]
- Kai-Larsen Y, Luthje P, Chromek M, Peters V, Wang X, Holm A, Kadas L, Hedlund KO, Johansson J, Chapman MR, et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010;6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Su B, Nestle FO, Zal T, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- Lim E, Koh WH, Loh SF, Lam MS, Howe HS. Non-thyphoidal salmonellosis in patients with systemic lupus erythematosus. A study of fifty patients and a review of the literature. Lupus. 2001;10:87–92. doi: 10.1191/096120301675973164. [DOI] [PubMed] [Google Scholar]
- Liu Z, Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 2012;18:871–882. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Bahrami B, Macfarlane GT. Mucosal biofilm communities in the human intestinal tract. Adv Appl Microbiol. 2011;75:111–143. doi: 10.1016/B978-0-12-387046-9.00005-0. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- Maldonado MA, Kakkanaiah V, MacDonald GC, Chen F, Reap EA, Balish E, Farkas WR, Jennette JC, Madaio MP, Kotzin BL, et al. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. Journal of immunology (Baltimore, Md : 1950) 1999;162:6322–6330. [PubMed] [Google Scholar]
- Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black × New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174:2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L. Genetics of SLE: evidence from mouse models. Nat Rev Rheumatol. 2010;6:348–357. doi: 10.1038/nrrheum.2010.63. [DOI] [PubMed] [Google Scholar]
- Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, Wakeland EK. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki H, Higuchi K, Nakakuki K, Takeda T. Kinetic analysis of amyloid fibril polymerization in vitro. Lab Invest. 1991;65:104–110. [PubMed] [Google Scholar]
- Nishimori JH, Newman TN, Oppong GO, Rapsinski GJ, Yen JH, Biesecker SG, Wilson RP, Butler BP, Winter MG, Tsolis RM, et al. Microbial amyloids induce interleukin 17A (IL-17A) and IL-22 responses via Toll-like receptor 2 activation in the intestinal mucosa. Infect Immun. 2012;80:4398–4408. doi: 10.1128/IAI.00911-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissle A. [Sanitation of intestinal flora as a prophylactic measure] Medizinische. 1957;10:82–85. [PubMed] [Google Scholar]
- O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- Pablos JL, Aragon A, Gomez-Reino JJ. Salmonellosis and systemic lupus erythematosus. Report of ten cases. Br J Rheumatol. 1994;33:129–132. doi: 10.1093/rheumatology/33.2.129. [DOI] [PubMed] [Google Scholar]
- Petri M. Infection in systemic lupus erythematosus. Rheum Dis Clin North Am. 1998;24:423–456. doi: 10.1016/s0889-857x(05)70016-8. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Baumler AJ. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun. 2005;73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsinski GJ, Newman TN, Oppong GO, van Putten JP, Tukel C. CD14 protein acts as an adaptor molecule for the immune recognition of Salmonella curli fibers. J Biol Chem. 2013;288:14178–14188. doi: 10.1074/jbc.M112.447060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel J. Protein folding: The dark side of proteins. Nature. 2010;464:828–829. doi: 10.1038/464828a. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012;8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Zhang Z, Yu AM, Wang W, Wei Z, Akhter E, Maurer K, Costa Reis P, Song L, Petri M, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One. 2014;9:e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigekazu N, Rikinari H, Kohki K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Sriram U, Varghese L, Bennett HL, Jog NR, Shivers DK, Ning Y, Behrens EM, Caricchio R, Gallucci S. Myeloid dendritic cells from B6.NZM Sle1/Sle2/Sle3 lupus-prone mice express an IFN signature that precedes disease onset. J Immunol. 2012;189:80–91. doi: 10.4049/jimmunol.1101686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Tan EM. Autoantibodies, autoimmune disease, and the birth of immune diagnostics. The Journal of clinical investigation. 2012;122:3835–3836. doi: 10.1172/JCI66510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao CH, Chen CY, Ou LS, Huang JL. Risk factors of mortality for salmonella infection in systemic lupus erythematosus. J Rheumatol. 2002;29:1214–1218. [PubMed] [Google Scholar]
- Tukel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Baumler AJ. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol. 2010;12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, et al. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- Tukel C, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Baumler AJ. Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe. 2009;6:45–53. doi: 10.1016/j.chom.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- von Rosenvinge EC, O’May GA, Macfarlane S, Macfarlane GT, Shirtliff ME. Microbial biofilms and gastrointestinal diseases. Pathog Dis. 2013;67:25–38. doi: 10.1111/2049-632X.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, Nick JA. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun. 2005;73:3693–3701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.