Abstract
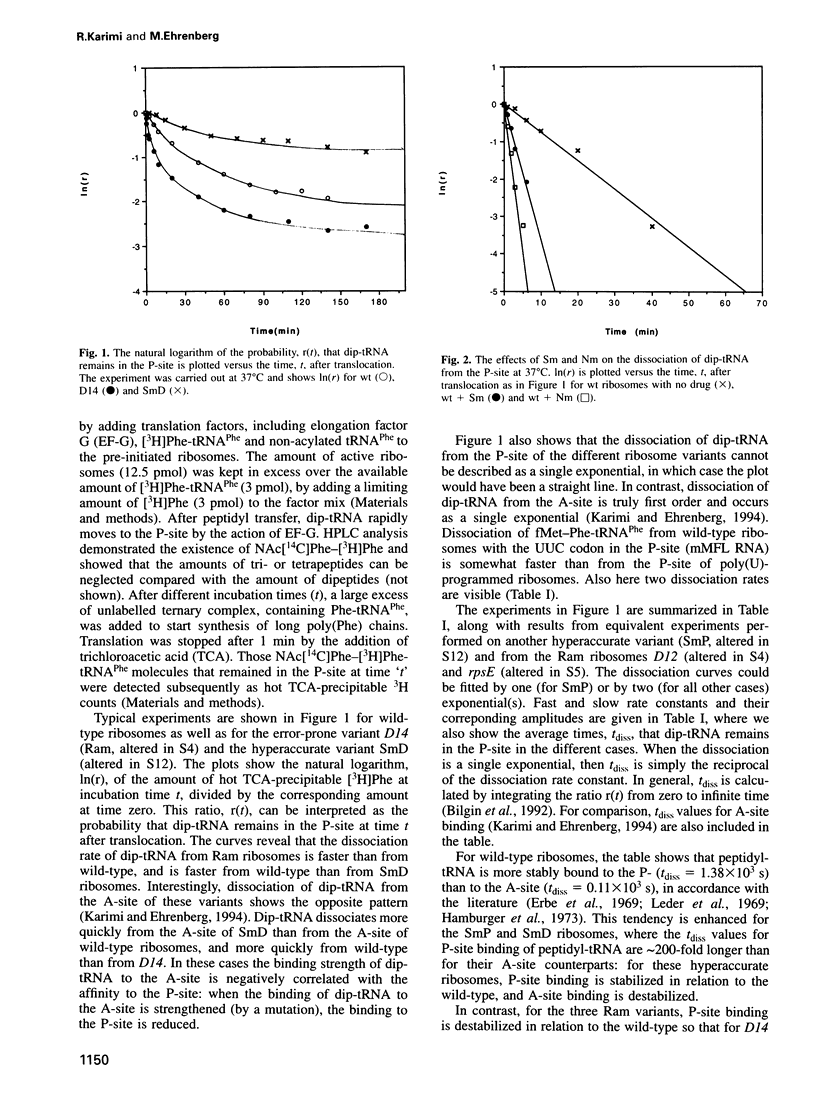
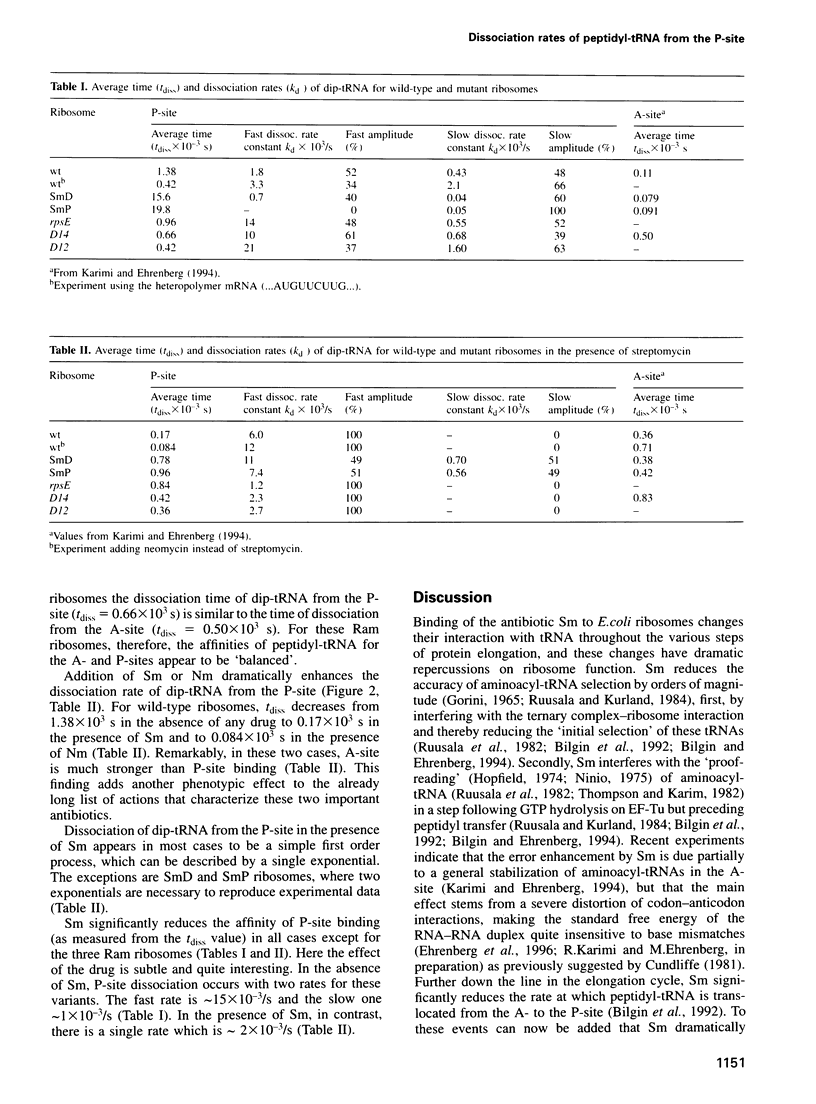
We studied the dissociation rates of peptidyl-tRNA from the P-site of poly(U)-programmed wild-type Escherichia coli ribosomes, hyperaccurate variants altered in S12 (SmD, SmP) and error-prone variants (Ram) altered in S4 or S5. The experiments were carried out in the presence and absence of streptomycin, and the effects of neomycin were tested in the wild-type ribosomes. Binding of peptidyl-tRNA to the P-site of wild-type ribosomes is much stronger than to their A-site. Addition of streptomycin dramatically reduces its affinity for the P-site. The S12 alternations make the P-site binding of peptidyl-tRNA much tighter, and the S4, S5 alterations make it weaker than in the case of the wild-type. We find that when binding of peptidyl-tRNA to the A-site is weak, then the affinity for the P-site is stronger, and vice versa. From these results, we formulate a hypothesis for the actions of streptomycin and neomycin based on deformations of the 16S rRNA tertiary structure. The results are also used to interpret some in vivo experiments on translational processivity.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson D. I., Andersson S. G., Kurland C. G. Functional interactions between mutated forms of ribosomal proteins S4, S5 and S12. Biochimie. 1986 May;68(5):705–713. doi: 10.1016/s0300-9084(86)80164-x. [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Kurland C. G. Ram ribosomes are defective proofreaders. Mol Gen Genet. 1983;191(3):378–381. doi: 10.1007/BF00425749. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Claesens F., Pahverk H., Ehrenberg M. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J Mol Biol. 1992 Apr 20;224(4):1011–1027. doi: 10.1016/0022-2836(92)90466-w. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Ehrenberg M. Mutations in 23 S ribosomal RNA perturb transfer RNA selection and can lead to streptomycin dependence. J Mol Biol. 1994 Jan 21;235(3):813–824. doi: 10.1006/jmbi.1994.1041. [DOI] [PubMed] [Google Scholar]
- Calogero R. A., Pon C. L., Canonaco M. A., Gualerzi C. O. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. B., Menninger J. R. Dissociation of peptidyl-tRNA from ribosomes is perturbed by streptomycin and by strA mutations. Mol Gen Genet. 1984;194(3):534–538. doi: 10.1007/BF00425571. [DOI] [PubMed] [Google Scholar]
- Davies J., Davis B. D. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J Biol Chem. 1968 Jun 25;243(12):3312–3316. [PubMed] [Google Scholar]
- Dong H., Kurland C. G. Ribosome mutants with altered accuracy translate with reduced processivity. J Mol Biol. 1995 May 5;248(3):551–561. doi: 10.1006/jmbi.1995.0242. [DOI] [PubMed] [Google Scholar]
- Erbe R. W., Nau M. M., Leder P. Translation and translocation of defined RNA messengers. J Mol Biol. 1969 Feb 14;39(3):441–460. doi: 10.1016/0022-2836(69)90137-5. [DOI] [PubMed] [Google Scholar]
- Hamburger A. D., Lapidot Y., De Groot N. Thermal stability of poly(U)-tRNA-ribosome complexes with Phe-tRNA Phe and peptidyl-tRNA Phe . Eur J Biochem. 1973 Feb 1;32(3):576–583. doi: 10.1111/j.1432-1033.1973.tb02644.x. [DOI] [PubMed] [Google Scholar]
- Hershey J. W., Thach R. E. Role of guanosine 5'-triphosphate in the initiation of Peptide synthesis, I. Synthesis of formylmethionyl-puromycin. Proc Natl Acad Sci U S A. 1967 Mar;57(3):759–766. doi: 10.1073/pnas.57.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc P. C., Kurland C. G. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc P. C. Rapid purification of highly active ribosomes from Escherichia coli. Anal Biochem. 1980 Jul 1;105(2):369–374. doi: 10.1016/0003-2697(80)90472-8. [DOI] [PubMed] [Google Scholar]
- Jørgensen F., Kurland C. G. Processivity errors of gene expression in Escherichia coli. J Mol Biol. 1990 Oct 20;215(4):511–521. doi: 10.1016/S0022-2836(05)80164-0. [DOI] [PubMed] [Google Scholar]
- Karimi R., Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994 Dec 1;226(2):355–360. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- Kaziro Y. The role of guanosine 5'-triphosphate in polypeptide chain elongation. Biochim Biophys Acta. 1978 Sep 21;505(1):95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- Kurland C. G., Ehrenberg M. Growth-optimizing accuracy of gene expression. Annu Rev Biophys Biophys Chem. 1987;16:291–317. doi: 10.1146/annurev.bb.16.060187.001451. [DOI] [PubMed] [Google Scholar]
- Kurland C. G., Ehrenberg M. Optimization of translation accuracy. Prog Nucleic Acid Res Mol Biol. 1984;31:191–219. doi: 10.1016/s0079-6603(08)60378-5. [DOI] [PubMed] [Google Scholar]
- Leder P., Bernardi A., Livingston D., Loyd B., Roufa D., Skogerson L. Protein biosynthesis: studies using synthetic and viral mRNAs. Cold Spring Harb Symp Quant Biol. 1969;34:411–417. doi: 10.1101/sqb.1969.034.01.047. [DOI] [PubMed] [Google Scholar]
- Menninger J. R., Caplan A. B., Gingrich P. K., Atherly A. G. Tests of the ribosome editor hypothesis. II. Relaxed (relA) and stringent (relA+) E. coli differ in rates of dissociation of peptidyl-tRNA from ribosomes. Mol Gen Genet. 1983;190(2):215–221. doi: 10.1007/BF00330642. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57(5):587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Olsson M. O., Isaksson L. A. Analysis of rpsD mutations in Escherichia coli. I. Comparison of mutants with various alterations in ribosomal protein S4. Mol Gen Genet. 1979 Feb 1;169(3):251–257. doi: 10.1007/BF00382271. [DOI] [PubMed] [Google Scholar]
- Piepersberg W., Böck A., Wittmann H. G. Effect of different mutations in ribosomal protein S5 of Escherichia coli on translational fidelity. Mol Gen Genet. 1975 Sep 29;140(2):91–100. doi: 10.1007/BF00329777. [DOI] [PubMed] [Google Scholar]
- Ruusala T., Andersson D., Ehrenberg M., Kurland C. G. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984 Nov;3(11):2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala T., Ehrenberg M., Kurland C. G. Is there proofreading during polypeptide synthesis? EMBO J. 1982;1(6):741–745. doi: 10.1002/j.1460-2075.1982.tb01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala T., Kurland C. G. Streptomycin preferentially perturbs ribosomal proofreading. Mol Gen Genet. 1984;198(2):100–104. doi: 10.1007/BF00328707. [DOI] [PubMed] [Google Scholar]
- Soffientini A., Lorenzetti R., Gastaldo L., Parlett J. H., Spurio R., La Teana A., Islam K. Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr Purif. 1994 Apr;5(2):118–124. doi: 10.1006/prep.1994.1018. [DOI] [PubMed] [Google Scholar]
- Thompson R. C., Karim A. M. The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP[gamma S]. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4922–4926. doi: 10.1073/pnas.79.16.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G., Jelenc P. C., Ehrenberg M., Kurland C. G. Rate of elongation of polyphenylalanine in vitro. Eur J Biochem. 1982 Feb;122(1):193–197. doi: 10.1111/j.1432-1033.1982.tb05866.x. [DOI] [PubMed] [Google Scholar]
- Weijland A., Parlato G., Parmeggiani A. Elongation factor Tu D138N, a mutant with modified substrate specificity, as a tool to study energy consumption in protein biosynthesis. Biochemistry. 1994 Sep 6;33(35):10711–10717. doi: 10.1021/bi00201a019. [DOI] [PubMed] [Google Scholar]
- Weijland A., Parmeggiani A. Toward a model for the interaction between elongation factor Tu and the ribosome. Science. 1993 Feb 26;259(5099):1311–1314. doi: 10.1126/science.8446899. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Young R., Dennis P. P., Nomura M. Role of ribosomal protein S12 in peptide chain elongation: analysis of pleiotropic, streptomycin-resistant mutants of Escherichia coli. J Bacteriol. 1977 Mar;129(3):1320–1329. doi: 10.1128/jb.129.3.1320-1329.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


