Abstract
The small molecule component of the chromoprotein enediyne antitumor antibiotics is biosynthesized through a convergent route, incorporating amino acid, polyketide and carbohydrate building blocks around a central enediyne hydrocarbon core. The naphthoic acid moiety of the enediyne neocarzinostatin plays key roles in the biological activity of the natural product by interacting with both the carrier protein and duplex DNA at the site of action. We have previously described the in vitro characterization of an S-adenosylmethionine-dependent O-methyltransferase (NcsB1) in the neocarzinostatin biosynthetic pathway (Luo, Y.; Lin, S.; Zhang, J.; Cooke, H. A.; Bruner, S. D. and Shen, B. (2008) J. Biol. Chem. 283, 14694–14702). Here we provide a structural basis for NcsB1 activity, illustrating that the enzyme shares an overall architecture with a large family of S-adenosylmethionine-dependent proteins. In addition, NcsB1 represents the first enzyme to be structurally characterized in the biosynthetic pathway of neocarzinostatin. By co-crystallizing the enzyme with various combinations of the cofactor and substrate analogs, details of the active site structure have been established. Changes in subdomain orientation were observed by comparing structures in the presence and absence of substrate, suggesting that reorientation of the enzyme is involved in binding the substrate. In addition, residues important for substrate discrimination were predicted and probed through site directed mutagenesis and in vitro biochemical characterization.
Enediyne antitumor antibiotics are structurally complex natural products possessing remarkable cytotoxicity (1). The nine-membered subfamily of enediynes are composed of a small molecule chromophore and an apoprotein, which sequesters and stabilizes the reactive enediyne and aids in delivery to target cells, leading to single and double stranded DNA cleavage (2). Biosynthetic gene clusters of multiple enediynes from actinomycetes have recently been sequenced and annotated, allowing detailed investigation into their complex biosynthesis (3–8). The pathways utilize diverse enzymology to construct the natural products, including the use of polyketide and nonribosomal peptide machinery. Neocarzinostatin (NCS, 1) is an archetypal example of the nine-membered enediynes, the studies of which have contributed to the general understanding of enediyne biosynthesis and mode of action (4, 9–12). The naphthoate moiety of NCS functions by both binding to the apoprotein and intercalating target DNA, thereby positioning the enediyne in the minor groove (9, 10, 13–15). Alterations of the peripheral moieties of NCS surrounding the enediyne core may be a way to control or mediate the reactivity and specificity of NCS (1).
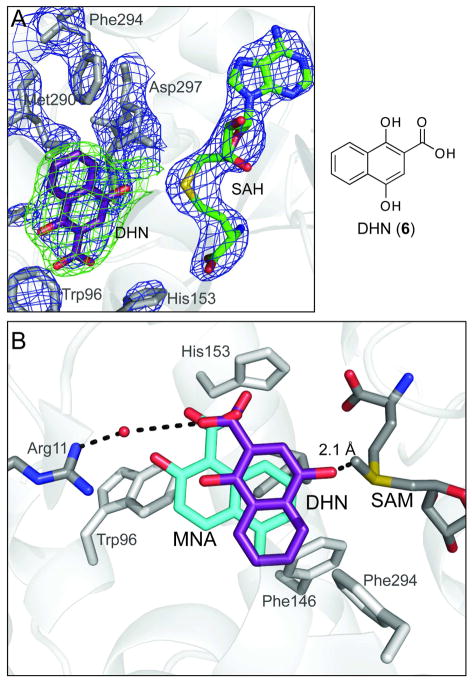
The recent cloning and sequencing of the NCS biosynthetic pathway from Streptomyces carzinostaticus revealed a convergent path with two polyketide synthases playing central roles in building the carbon framework (4, 12, 16). The scaffold for the naphthoate moiety in NCS is constructed by an iterative type I polyketide synthase via a mechanism common to the biosynthesis of diverse aromatic products (17–19). Four additional gene products decorate and couple the naphthoic acid to the enediyne core (Figure 1). The product of NcsB (2) is first hydroxylated at the C-7 position by a cytochrome P450 hydroxylase (NcsB3) to produce 3. The resulting hydroxyl group is methylated by NcsB1, producing the methyl ether 4 present in the natural product. This functionalized naphthoic acid is activated via adenylation, converted to the aryl-CoA 5 by the CoA-ligase NcsB2, and is finally attached to the enediyne core by NcsB4 (a putative acyltransferase) to produce the natural product (1). The assignment of the naphthoic acid biosynthetic pathway is based on bioinformatic analysis (4) and biochemical characterization of NcsB2 as a CoA-ligase (20) and NcsB1 as an S-adenosylmethionine (SAM)-dependent O-methyltransferase (O-MTase) (21). NcsB1 was shown to catalyze the third step in the biosynthesis of the naphthoate moiety of the NCS chromophore (3 to 4). Biochemical analysis also revealed a relaxed substrate flexibility towards substituted naphthoic acids, and preferred substrates were 3-hydroxy-2-naphthoic acids with the reactive hydroxyl group located at either the 5 or 7 position on the ring. NcsB1 also was demonstrated to methylate 1,4-dihydroxy-2-naphthoic acid (see Figure 5, 6) at the 4-hydroxyl group, which was surprising because of the large difference in the location of the reactive hydroxyl group from that on the natural substrate, necessitating a reorientation of the substrate in the active site. This observation led to the prediction of a model by which naphthoic acids flexibly bind the active site, and that the 1,2-hydroxy acid functionality was required for substrate binding and SAM-dependant methylation (21).
Figure 1.
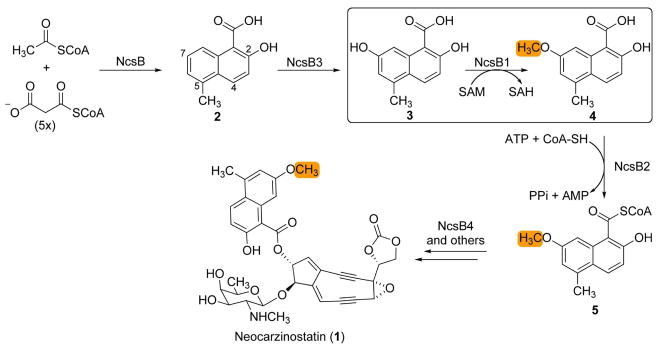
Biosynthesis of the naphthoic acid moiety and its attachment to the enediyne core, resulting in the natural product neocarzinostatin (1). The O-methyltransfer reaction catalyzed by NcsB1 is boxed (methyl group is highlighted in orange).
Figure 5.
Binding of alternate substrate, DHN (6). (A) Electron density map of NcsB1/SAH/6 active site, generated without 6. 2Fo – Fc map (blue, 1.5 σ) and Fo – Fc map (green, 3 σ) (B) Overlay of 2 and 6 in NcsB1 active site. Naphthoic acid 6 is hydrogen-bonded to Arg11 via a water and the 4-hydroxyl group is in sufficiently close proximity to SAM (2.1 Å, SAM from NcsB1/SAM/2 structure) for methyl transfer to occur.
There are ~160 classes of SAM-dependent MTases comprised of thousands of identified members. These functionally diverse enzymes methylate a wide range of substrates, including nucleic acids for regulation of gene expression, DNA repair or protection against restriction enzymes, proteins for repair or control of signal transduction pathways, hormones and neurotransmitters, and biosynthetic intermediates to produce secondary metabolites. Structurally characterized MTases can be segregated into five general classes based on specific structural features (22). Among the several examples of MTases that act on small molecules, catechol O-MTase isolated from rat liver was the first to be structurally characterized (23). This structure exemplifies the basic SAM-MTase core fold: a mixed α, β secondary structure with a β-sheet flanked on each side by three α-helices (24). A common feature of the active site of MTases is a glycine-rich region within the SAM binding pocket. Despite low overall sequence homology amongst family members, structurally characterized SAM-MTases share this common fold and the biologically active unit is a homodimer.
The role of NcsB1 was assigned in the enediyne neocarzinostatin biosynthetic pathway based on sequence homology to small molecule SAM dependent O-MTases including DnrK, an enzyme involved in the biosynthesis of the aromatic polyketide antibiotic daunorubicin from Streptomyces peucetius (44% sequence identity) (25). The X-ray crystal structure of DnrK revealed a homodimer with each monomer exhibiting the conserved small molecule MTase fold (26). RdmB from rhodomycin biosynthesis in Streptomyces purpurascens is also homologous to NcsB1, sharing 44% sequence identity and 58% similarity, but interestingly is a hydroxylase, not an O-MTase (27, 28). RdmB was used as a model for molecular replacement in solving the crystal structure of DnrK. In addition to DnrK, other bacterial small molecule O-MTases structures have recently been determined, including examples requiring a cation for catalysis. Cation-dependent O-MTases include CmcI from cephamycin biosynthesis (29), LiOMT from the pathogenic bacterium Leptospira interrogans (30), and BcOMT2 from Bacilus cereus (31). A limited number of cation-independent bacterial O-MTase structures have been structurally characterized and include DnrK and RebM from rebeccamycin biosynthesis (32).
Here we report the structural basis for NcsB1 activity in a variety of co-complexes with SAM or S-adenosyl-L-homocysteine (SAH) with or without 5-methyl-2-hydroxy naphthoic acid (2), product (4), or 1,4-dihydroxy-2-naphthoic acid (6), as an alternate substrate. The structures reveal that NcsB1 shares an overall architecture common to the large MTase family. The active site binding pocket is able to accommodate the natural substrate and structurally diverse analogs, allowing efficient methylation with distinct regiospecificity. The specificity determinants of the naphthoate binding pocket were probed using site-directed mutagenesis and alternate substrates. Based on the results, residues that affect the substrate specificity of the enzyme were identified.
MATERIALS AND METHODS
Protein Expression and Purification
NcsB1 was overproduced as an N-terminal His6-tagged fusion protein using the expression plasmid pBS5039 in E. coli BL21(DE3) cells as reported previously (21). Cells were grown in 1 L Luria-Bertani media at 37 °C, 150 RPM to an OD600 = 0.5–0.8. Overexpression was induced with 50 μM isopropyl β-D-thiogalactopyranoside at 18 °C for 16 hr. Cells were collected by centrifugation (20 min, 2000 g), resuspended in 25 mL of 20 mM Tris-HCl pH 7.5 and 500 mM NaCl, and flash frozen at −78 °C. Cell pellets were thawed and lysed by two passes through a French Press cell disruptor at 1000 psi. The lysate was then clarified by centrifugation (20 min, 10,000 g) and the protein was purified batchwise using Ni-NTA resin (Qiagen, Valencia, CA). NcsB1 used in biochemical assays was dialyzed into 20 mM Tris-HCl pH 7.5, 50 mM NaCl, and 1 mM β-mercaptoethanol, then concentrated using an Amicon Ultra-4 concentrator (10 kDa MW cutoff, GE Healthcare) and frozen with 40% supplemented glycerol at −25 °C. For crystallography, the Ni-NTA purified protein was dialyzed into 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM CaCl2, 1 mM β-mercaptoethanol) and concentrated to ~ 1 mL. The His6-tag was cleaved by incubation with the protease Factor Xa for 36 hr at 4 °C (monitored by SDS-PAGE). The protein solution was then diluted to 5 mL and purified on a HiTrap-Q ion exchange column (0–1 M NaCl gradient in 50 mM Tris-HCl pH 7.5 and 1 mM β-mercaptoethanol) followed by a Superdex 200 gel filtration column (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol). The purified protein was concentrated to ~10 mg/mL for crystallography (a total yield of 4 mg/L of cells). Protein concentration was determined using UV absorption at 280 nm and the Bradford assay.
Site-Directed Mutagenesis of ncsB1
The Quikchange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used to generate mutant constructs of NcsB1 using the following DNA oligonucleotide primers (mutant codon is underlined): Arg11Ala For: 5′ - GGC TGC ACA CAT CGG ATT GGC GGC GCT GGC CGA TCT GGC GAC - 3′ and Rev: 5′ - GTC GCC AGA TCG GCC AGC GCC GCC AAT CCG ATG TGT GCA GCC - 3′; Arg11Trp For: 5′ - GGC TGC ACA CAT CGG ATT GTG GGC GCT GGC CGA TCT GGC GAC - 3′ and Rev: 5′ - GTC GCC AGA TCG GCC AGC GCC CAC AAT CCG ATG TGT GCA GCC - 3′; Arg11Lys For: 5′ - GGC TGC ACA CAT CGG ATT GAA GGC GCT GGC CGA TCT GGC GAC - 3′ and Rev: 5′ - GTC GCC AGA TCG GCC AGC GCC TTC AAT CCG ATG TGT GCA GCC - 3′; Tyr293Ile For: 5′ - GCG CAT GCT CAC CAT CTT CGG AGG CAA GGA ACG C - 3′ and Rev: 5′-GCG TTC CTT GCC TCC GAA GAT GGT GAG CAT GCG C - 3′. The Quikchange protocol was followed as described with the addition of DMSO to the reaction to a final concentration of 5%. Mutations were confirmed by sequencing (Genewiz, S. Plainfield, NJ) and mutant constructs were transformed into E. coli BL21(DE3) cells, over-expressed and purified as described above.
Crystallization and X-ray Data Collection
Purified NcsB1 was crystallized with a substituted naphthoic acid and/or SAM/SAH by the hanging drop vapor diffusion method at 20 °C with ~4 M sodium formate (7 M stock solution from Hampton Research, see Table S1 for crystallization conditions). Stock solutions of the small molecules for co-crystallization were in 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol. Naphthoic acids were synthesized as described previously (20) or purchased from Aldrich. Typically, NcsB1 crystals appeared within a day and reached full size within one week. Mature crystals were transferred to a cryoprotectant solution (4.0 M sodium formate, 15% glycerol) and soaked briefly before flash freezing in liquid nitrogen. X-ray diffraction data was collected on the X25 beamline at the National Synchrotron Light Source at Brookhaven National Laboratories with an ADSC Q315 CCD X-ray detector. Diffraction intensities were indexed, integrated and scaled using HKL2000 (33) as summarized in Table 1. The crystals belonged to the space group P65 except for the NcsB1-SAH co-complex, which belonged to the C2221 space group.
Table 1.
Data collection and refinement statistics
| SAH | SAM and 2 | SAH and 4 | SAH and DHN (6) | |
|---|---|---|---|---|
| Space group | C2221 | P65 | P65 | P65 |
| Unit cell parameters (Å) | a=91.3, b=161.6, c=98.9 | a=b=109.1, c=206.9 | a=b=108.4, c=210.3 | a=b=108.0, c=211.9 |
|
| ||||
|
Data collection
| ||||
| Resolution (Å) | 25.0–2.08 (2.15–2.08) | 50–2.6 (2.69–2.60) | 30.0–2.69 (2.79–2.69) | 50.0–3.0 (3.1–3.0) |
| Wavelength (Å) | 1 | 1 | 1 | 1 |
| No. of reflections (measured/unique) | 345090/44038 | 967889/42568 | 658394/38115 | 35878/27798 |
| % Completeness* | 99.6 (99.6) | 99.8 (100.0) | 98.5 (100.0) | 99.8 (100.0) |
| Rmerge* | 0.052 (0.349) | 0.077 (0.441) | 0.094 (0.49) | 0.076 (0.523) |
| I/σ* | 32.2 (4.4) | 49.6 (6.4) | 24.0 (7.0) | 29.3 (4.5) |
| Redundancy* | 7.8 (7.4) | 22.7 (22.9) | 17.3 (17.8) | 12.6 (12.8) |
|
| ||||
|
Refinement
| ||||
| No. of reflections (total/test) | 41442/2071 | 41357/2064 | 37165/3704 | 26923/2698 |
| Rwork | 0.240 | 0.206 | 0.212 | 0.217 |
| Rtest | 0.272 | 0.227 | 0.247 | 0.245 |
| No. amino acids (chains A/B) | 325/303 | 325/325 | 328/328 | 328/328 |
| No. atoms | ||||
| Protein (chains A/B) | 2379/2193 | 2379/2379 | 2406/2406 | 2406/2406 |
| Ligands (cofactor/substrate/glycerol) | 52/-/6 | 54/30/12 | 52/34/12 | 50/30/12 |
| Water molecules | 305 | 249 | 163 | 95 |
| Mean B (protein/SAM/4/water) (Å2) | 46.0/36.0/-/50.1 | 53.0/42.3/51.6/55.6 | 71.4/59.6/81.4/74.3 | 69.9/59.9/57.2/65.4 |
| R. m. s. d. from ideal geometry | ||||
| r.m.s.d bonds (Å) | 0.006 | 0.006 | 0.006 | 0.0013 |
| r.m.s.d angles (°) | 1.2 | 1.3 | 1.2 | 1.7 |
| Ramachandran plot by PROCHECK (%) | ||||
| Core region | 90.6 | 93.2 | 91.7 | 88.8 |
| Allowed region | 9.0 | 6.2 | 7.9 | 10.4 |
| Generously allowed | 0.2 | 0.5 | 0.2 | 0.7 |
| Disallowed | 0.2 | 0.0 | 0.2 | 0.0 |
|
| ||||
| PDB code | 3I53 | 3I5U | 3I58 | 3I64 |
overall (highest resolution shell)
Structure Determination
An initial solution for the NcsB1/SAM/2 ternary complex was obtained using the molecular replacement program PHASER (34), part of the CCP4 suite (35). A polyalanine dimer model of DnrK from Streptomyces peucetius (PDB ID code 1TW2) truncated to residues 15–350 was used as the search model. Manual building of the NcsB1 model was performed using the program COOT (36) and the structure was refined using CNS (37, 38). This solution was used to solve phases in subsequent datasets in the P65 space group using molecular replacement. For the NcsB1-SAH co-complex (C2221 space group), a partial solution (chain A and the N-terminus of chain B) was found using PHASER. The remaining C-terminus of chain B was placed into the model using the program MOLREP (39). All final structures were subjected to multiple rounds of building and refinement until the R values converged. Non-crystallographic restraints were used for all P65 space group structures until final stages of refinement. Ligands were fit into the electron density maps and the PRODRG server (40) was used for generating topology and parameter files; SAM and SAH coordinates were obtained from the HICup server (41) and were subsequently altered to fit electron density. The program PyMOL (Delano Scientific, San Carlos, CA) was used to generate graphic images. RMSD values were calculated via structural comparison using TopMatch (42, 43).
The NcsB1/SAH structure (C2221 space group) was solved at a resolution of 2.08 Å and refined to an Rwork and Rfree of 24.0 and 27.2% (Table 1). The asymmetric unit consisted of two monomers. For chain A, there was sufficient electron density to build in residues 5–330 and the bound SAH molecule. For chain B, the final model consisted of residues 9–150, 159–274, 282–297 and 302–330, as insufficient electron density was available to build the remaining 29 residues. Electron density for the SAH molecule was well defined, albeit with higher B factors for the SAH molecule in chain B. In addition, the dimer contained one glycerol molecule. The NcsB1 co-complex with SAM and the substrate analog (2) was solved at a resolution of 2.60 Å with Rwork and Rfree of 20.6 and 22.7%, respectively. For both chains, N-terminal residues 1–4 could not be built, nor could residues 279–281, though sufficient electron density was available for both SAM and 2 to be built into each active site. NcsB1 with SAH and product (4) diffracted to 2.69 Å and was refined to an Rwork and Rfree of 21.2 and 24.7%, respectively. All residues of both chains could be built in as well as SAH, the product in the active site, and four glycerol molecules.
The final structure was a co-complex of NcsB1 with SAH and an alternate substrate, 1,4-dihydroxy-naphthoic acid (6). The crystal diffracted to 3.0 Å, likely a result of reduced crystal quality as 10 mM β-mercaptoethanol was required for crystal growth because of problematic oxidation of the substrate. The final structure refined to an Rwork and Rfree of 21.7 and 24.5% and included residues 4–332, SAH and 6 in the final model.
Kinetics of NcsB1 Mutants
To determine the kinetic parameters of all mutant constructs with the natural substrate, the previously reported biochemical assay was used with minor modifications (21). Reactions were run at 25 °C and contained 10 μM enzyme, 2.5 mM SAM, 100 mM phosphate buffer at pH 6.5 and varying concentrations of 3 ranging from 50 μM to 2.5 mM for a total volume of 50 μL. Reactions were quenched with trifluoroacetic acid to a final concentration of 16% after 15 minutes, except for WT and Arg11Ala, which were quenched at 10 min. Samples were centrifuged at 14,000 g for 2 minutes and the supernatant was analyzed by HPLC at 340 nm (eluent A: water with 0.1% trifluoroacetic acid; eluent B: acetonitrile; gradient: 0–12 min at 10–60% acetonitrile, 12–17 min at 0% at a flow rate of 1 mL/min on a VYDAC C18 Protein and Peptide column, 4.6 × 250 mm). To obtain kinetic parameters, initial rates were plotted against substrate concentration and were fitted to the Michaelis-Menten equation using KaleidaGraph (Synergy Software, Reading, PA).
RESULTS AND DISCUSSION
Crystallization and X-ray Structure Determination
The 34.5 kDa/332 residue NcsB1 was co-crystallized with various combinations of SAH, SAM, deshydroxy-substrate analog 2 and the methylated product 4. The analog 2 was used in crystallization experiments due to the oxidative instability of the substrate (3) over extended time periods. Two crystal forms were generated depending on the nature of the bound small molecules. When co-crystallized with a substrate analog/product and SAH or SAM, the co-complex crystallized in the P65 space group. In the absence of naphthoic acid, distinct crystals belonging to the C2221 space group formed. The ternary complex of NcsB1/SAM/2 was the initial structure solved using molecular replacement with a polyalanine dimer model of DnrK (44% identity) as the search model. This structure was then used as a search model for the alternate co-complexes. Solution of the NscB1/SAH complex (C2221 space group) by molecular replacement was not straightforward, suggesting an alternate overall structure. The final structural model was obtained by performing rotation and translation searches separately on the N- and C-terminal domains. While a 0.6 Å RMSDCα was observed amongst the P65 structures, the NcsB1/SAH structure differed significantly from the others by an RMSDCα of 1.8 Å. In addition, the two subunits of the NcsB1/SAH structure were quite distinct, with an RMSDCα of 2.8 Å between the homodimer chains.
Overview of the Structures
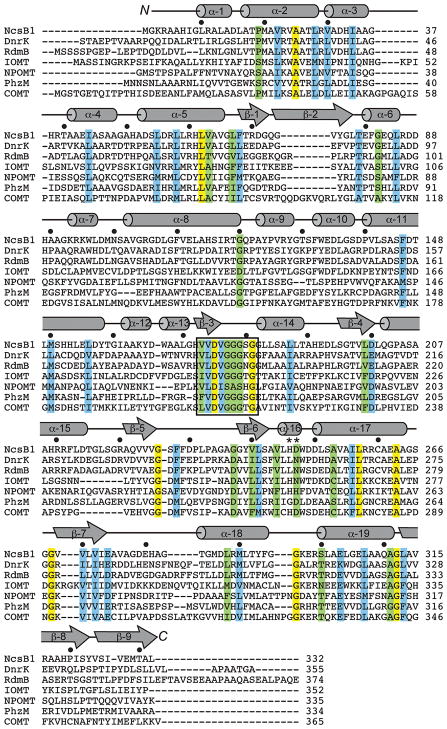
The resulting dimer models of NcsB1 display a high degree of structural similarity with DnrK (RMSDCα = 1.8 Å). In the initial NcsB1/SAM/2 structure, and others in the P65 space group, the two monomeric subunits exhibit a high degree of symmetry (Figure 2A). Each subunit of the NcsB1 dimer is made up of three domains totaling nineteen α-helices and eight β-sheets (Figure 2B). The N-terminal domain is largely α-helical, with just two β-strands (β1 and β2), and constitutes most of the residues involved in the homodimer interface (~5300 Å2 total surface). Of note, α5 (residues 52–65) has significant interactions with α18 (residues 284–292) from the C-terminal domain of the paired dimer. The dimer interface is made up of primarily hydrophobic residues, and a dimeric biological unit was supported by size-exclusion chromatography (Figure S1). A middle domain acts as a hinge-like region between the two larger terminal domains and is made up of α-helices 9–11. The C-terminal domain exhibits a Rossman-like fold (β3–9 flanked by α12–19), which is conserved in structural homologs and makes up the majority of the SAM binding site (24). Based on structural alignment using the DALI server, NcsB1 shares the highest structural similarity with SAM-dependent enzymes that act on small molecules (Figure 3, Table S2) (44). Homologs include DnrK (an O-MTase) and RdmB (a hydroxylase), isoflavone-O-MTase from alfalfa (45), a putative O-MTase from Nostoc punctiforme, a phenazine-specific N-MTase from Pseudomonas aeruginosa (46) and caffeic acid 3-O-MTase from alfalfa (47). Among the structures, an indicative glycine-rich region of the SAM binding pocket is conserved. These six structural homologs demonstrate the ability of the conserved scaffold to catalyze a variety of transformations.
Figure 2.
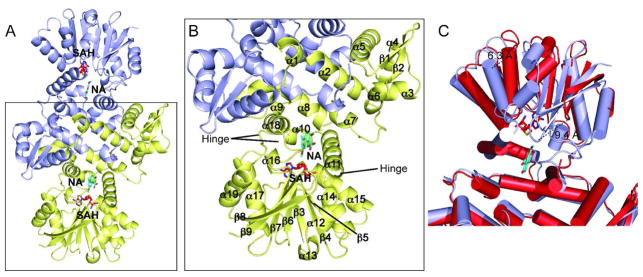
Cartoon representations of NcsB1. (A) Dimer of NcsB1 with the active site region indicated. Chain A is colored light blue and chain B yellow. Ligands are depicted in stick format with SAH in red and naphthoic acid 4 in cyan. B) Close up view of monomer with secondary structural elements and hinge regions labeled. C) Overlay of NcsB1/SAH/4 monomer (light blue) and NcsB1/SAH monomer (red).
Figure 3.
Sequence alignment of NcsB1 structural homologs as determined by DALI structural alignment server. Abbreviations used with accession codes from the RCSB Protein Data Bank in parentheses: DnrK, S. peucetius O-MTase (Q06528); RdmB, S. purpurascens hydroxylase (Q54527); IOMT, alfalfa O-MTase (O24529); NPOMT, N. punctiforme putative O-MTase (ZP_00112478); PhzM, P. aeruginosa N-MTase (Q9HWH2); COMT, alfalfa O-MTase (P28002). Completely conserved residues are yellow, highly conserved residues are blue, and conserved residues are green. Catalytic residues are indicated by an asterisk and conserved glycine region is boxed.
In the NcsB1/SAH co-complex structure, the C-terminal domain of chain B is rotated by ~20° compared to NcsB1/SAH/2, displacing α13 by 6.8 Å and β8/β9 by 9.4 Å (Figure 2C). This movement results in a more open conformation of the active site, possibly functioning to allow the entrance of substrate. A similar domain displacement was seen in the DnrK ternary complex where two crystals with different space groups were also observed (26). Both DnrK crystals were bound to SAH and product, but the B-factors were higher in one of the two subunits, suggesting lower occupancy of the ligands. One of the crystal structures exhibited a significant difference between the two monomers, with an RMSDCα of 1.3 Å. Likewise, the RdmB structures exhibit a domain movement upon substrate binding, though in this co-complex structure, both domains were displaced (27). The NcsB1/SAH co-complex described here exhibits a larger difference, likely due to complete lack of substrate analog or product in the crystallization conditions. In addition, SAH bound in the open subunit (chain B) has higher relative B-factors. Several regions in chain B showed weak or no electron density, possibly indicating areas of flexibility on which the domain can twist. These hinge regions include part of α11 (residues 150–159), the loop region between β7 and α18 (residues 274–282) and the loop between α18 and α19 (residues 297–301) (Figure 2B).
SAM/SAH Binding Pocket
The SAM/SAH binding pocket is located at the C-terminus of the β-strands comprising part of the Rossman-like fold. Electron density for SAM or SAH is clearly defined in all four co-complex structures and the SAM binding interactions are largely conserved among small molecule MTases. The adenine ring is involved in a hydrogen-bonding interaction with Ser227, a π interaction with Phe228 and the binding pocket is lined with additional aromatic and/or hydrophobic residues, including Trp133, Trp248, Phe229 and Leu201 (Figure 4A). The ribosyl moiety is anchored by two hydrogen bonds between Asp200 and Ser143 and the homocysteine portion of SAM forms hydrogen bonding interactions with the sidechains of Asp175, Ser242, His153 and the backbone carbonyl of Gly177. The LDXGXGXG motif indicative of SAM-utilizing proteins is found in NcsB1 as VDVGGGSG and is located between β3 and α14. We previously reported that NcsB1 alkylates the substrate 3 with a variety of SAM analogs, including S-ethyl and S-n-propyl, to the corresponding 7-alkyl ether with reasonable efficiency (21). The nearest side chain to the methyl-group on SAM is the sterically small Ala243 (3.5 Å distance). Based on the large size and flexibility of the substrate binding pocket, it is not surprising that NcsB1 could accommodate larger SAM analogs.
Figure 4.
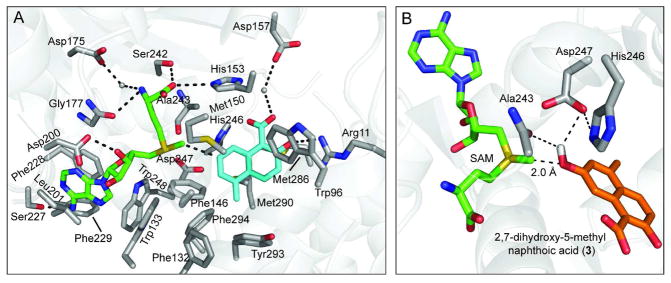
Active site of NcsB1/SAM/2. (A) Substrate and SAM binding pocket. SAH is shown in green and naphthoic acid 2 in cyan; residues in the substrate and SAM binding pockets are shown in grey with pertinent hydrogen-bonds indicated. (B) Close up of active site in NcsB1/SAM/2 structure with 7-hydroxy group modeled onto 2, with the likely catalytic residues shown in grey. SAM is shown in green and the naphthoic acid 3 in orange. The methyl group on SAM is 2.0 Å from the hydroxyl group on 3.
Naphthoic Acid Binding Pocket
The naphthoic acid binding site is located at the juncture of helices from all three subdomains, including α7, α10, α11, α16, and α18, and the pocket is lined primarily with hydrophobic/aromatic residues (Figure 4A). Three methionine sidechain thioethers (Met150, 286 and 290) are present on either side of the naphthoate ring forming van der Waals interactions with the substrate. The carboxylate and 2-hydroxyl group of the substrate interacts with Arg11 directly and with Asp157 through an ordered water. This interaction represents the only hydrogen bonds between the enzyme and substrate and appears to anchor the naphthoic acid into the active site adjacent to the bound SAM. In order to probe the importance of Arg11 in enzyme activity, site-directed mutagenesis was used to alter this site and the activity was evaluated. This residue was mutated to Ala and Lys to assess the requirement of this hydrogen-bonding interaction for binding naphthoic acids. Unexpectedly, the Arg11Ala mutant still effectively methylated naphthoic acid 3 (Table 2), although the results showed a doubling of KM that was countered by a similar increase in kcat. The Arg11Lys mutant had 110% of the catalytic efficiency as compared to the WT. This mutant likewise showed an increased rate of turnover. This observation suggests that the specific hydrogen bond from Arg11 is not entirely crucial for substrate turnover and can be compensated for by a similar interaction with Lys or by increased overall catalytic turnover. In addition, simple benzoic acids lacking a hydroxyl group adjacent to the aryl acid (for example 3-hydroxy benzoic acid, see Figure S4) were methylated by NcsB1, although at a significantly decreased rate, further suggesting that the 1,2-hydroxy acid is not absolutely required for enzyme-catalyzed chemistry.
Table 2.
Kinetics of NcsB1 and mutant constructs
| Protein/substrate | KM (μM) | kcat (min−1) | kcat/KM (rel to WT/3) |
|---|---|---|---|
| WT/3 (18) | 206 ± 49 | 0.69 ± 0.05 | 1 |
| Tyr293Ile/3 | 649 ± 9 | 0.76 ± 0.04 | 0.35 |
| Arg11Trp/3 | 400 ± 68 | 0.60 ± 0.03 | 0.45 |
| Arg11Ala/3 | 419 ± 61 | 1.24 ± 0.06 | 0.88 |
| Arg11Lys/3 | 319 ± 40 | 1.18 ± 0.05 | 1.10 |
NcsB1 Residues Involved in Methyltransferase Chemistry
Roles for residues in the active site relevant for catalysis could include activation of the phenol for nucleophilic attack via acid/base chemistry. Histidine residues specifically have been implicated to play this role in RebM, a carbohydrate O-MTase in rebeccamycin biosynthesis, where mutation of two histidine residues in the substrate binding pocket to alanines led to a complete loss of activity, while single mutations showed a marked decrease in activity (32). In NcsB1, three residues are in close proximity to the substrate phenol; the diad of His246 and Asp247 side chains and the backbone carbonyl of Ala243 may function to aid in activation and/or proton shuttling (Figure 4B).
Binding of 1,4-Dihydroxy-2-Naphthoic Acid in Active Site of NcsB1
Previous exploration of substrate specificity of NcsB1 suggested a large degree of flexibility for naphthoate binding (21). A model was proposed that involved anchoring of the substrate by the 1,2-hydroxy acid motif and subsequent methylation of exposed phenols in proximity to bound SAM. In order to probe this hypothesis, the structure of NcsB1 bound to 1,4-dihydroxy-2-naphthoic acid (DHN, 6) was determined. Methylation of this substrate would necessitate binding in a conformation distinct from the natural substrate while retaining the interaction with the enzyme through the 1,2-hydroxy acid. In the co-crystal structure, the proximity and orientation of 6 to SAH and important hydrogen bonding partners were readily apparent (Figure 5A). The substrate is bound in an appropriate position for regioselective methylation by SAM with an alternate orientation of the bound naphthoic acid. The methyl group on SAM is ~2.1 Å away from the 5-hydroxyl group of 6 and the carboxylate is hydrogen bonded to Arg11 via an ordered water (Figure 5B).
Probing the Determinants of the Substrate Specificity for NcsB1
The biosynthetic gene cluster for the related enediyne C-1027 contains a gene (sgcD4) with high homology to ncsB1 (56% identity, 67% similarity) and the enzyme is proposed to methylate a benzoxazolinate (Figure S3) (6, 48). Of nine residues in NcsB1 identified to play a role in substrate binding, four have identical counterparts in SgcD4 (Trp96, Phe146, Met286, and Met290), two are similar (Met150NcsB1 is a Leu in SgcD4 and Phe294NcsB1 is a Tyr) and two residues are distinct (Arg11NcsB1 is a Trp and Tyr293NcsB1 is an Ile). In order to probe the substrate specificity determinants of NcsB1, two rational mutations were made on the residues that significantly differed structurally between NcsB1 and SgcD4: Tyr293Ile and Arg11Trp. These two additional mutant constructs were assayed against the natural substrate (3) (Table 2). Both mutants methylated 3, though showed significant increases in KM compared to the wild-type enzyme. A larger decrease in catalytic efficiency was seen with the Tyr293Ile mutant, which had a KM approximately three times higher than wild-type. The KM was doubled for the Arg11Trp. The ability of Arg11 mutants, especially the Trp and Ala mutants, to methylate naphthoic acid suggests that the aromatic binding pocket of NcsB1 is substantially more crucial to binding substrate than the hydrogen bond from Arg11. The reduced aromatic character of the Tyr293Ile mutant supports this theory. We next tested a simple substrate analog of the benzoxazolinate, 2,5-dihydroxybenzoic acid. In this simple model compound, methylation occurs at the 5-position, although with significantly lower efficiency (Figure S3). 2,5-Dihydroxybenzoic acid was assayed with the panel of mutants in Table 2, and indeed alternate specificity was observed as compared to wild-type NcsB1 (Table S3). For example, although the relative measured activities were low, contrary to the wild-type enzyme, the Arg11Trp mutant has higher relative activity toward 2,5-dihydroxybenzoic acid and the Arg11Lys was less efficient. These results implicate Arg11 in substrate discrimination for these related methyltransferases.
In summary, we have solved the crystal structures of NcsB1 in two conformations with SAH/SAM bound, with and without a substrate analog or product. These structures revealed a large displacement of the C-terminal domain, a movement that likely opens up the active site for naphthoate binding. Additionally, the ternary complex structure of 1,4-dihydroxy naphthoic acid and SAH bound to NcsB1 was solved and showed a rotation of this alternate substrate in the binding pocket, allowing for methylation of the hydroxyl group at the 4 position. These results led us to probe substrate binding using active site mutants, demonstrating altered substrate specificity and revealing the importance of key residues in substrate binding.
Supplementary Material
Acknowledgments
This work was supported in part by funds from the Damon Runyon Cancer Research Fund (to S.D.B) and NIH grants CA78747 and CA113297 (to B.S.).
We thank Tim Montavon for critical reading of the manuscript and members of the Bruner and Shen groups for helpful discussions.
ABBREVIATIONS
- NCS
neocarzinostatin
- SAH
S-adenosyl-L-homocysteine
- SAM
S-adenosyl-L-methionine
- CoA
coenzyme A
- PPi
pyrophosphate
- MTase
methyltransferase
- DHN
1,4-dihydroxy-2-naphthoic acid
- RMSDCα
root mean squared difference at α-carbons
Footnotes
Coordinates have been deposited within the Protein Data Bank (PDB codes 3I53, 3I5U, 3I58, 3I64)
Supporting Information Available
Crystallization conditions (Table S1), determination of biologically active unit of NcsB1 (Figure S1), table containing results from DALI structural homolog search (Table S2), electron density maps (Figure S2), alignment of NcsB1 with SgcD4 (Figure S3), and detailed methods and results for benzoic acid substrates (Figure S4, Table S3) are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shen B, Liu W, Nonaka K. Enediyne natural products: biosynthesis and prospect towards engineering novel antitumor agents. Curr Med Chem. 2003;10:2317–2325. doi: 10.2174/0929867033456701. [DOI] [PubMed] [Google Scholar]
- 2.Maeda H, Aikawa S, Yamashita A. Subcellular fate of protein antibiotic neocarzinostatin in culture of a lymphoid cell line from Burkitt’s lymphoma. Cancer Res. 1975;35:554–559. [PubMed] [Google Scholar]
- 3.Liu W, Ahlert J, Gao Q, Wendt-Pienkowski E, Shen B, Thorson JS. Rapid PCR amplification of minimal enediyne polyketide synthase cassettes leads to a predictive familial classification model. Proc Natl Acad Sci USA. 2003;100:11959–11963. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 7.Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 9.Povirk LF, Dattagupta N, Warf BC, Goldberg IH. Neocarzinostatin chromophore binds to deoxyribonucleic acid by intercalation. Biochemistry. 1981;20:4007–4014. doi: 10.1021/bi00517a009. [DOI] [PubMed] [Google Scholar]
- 10.Lee SH, Goldberg IH. Sequence-specific, strand-selective, and directional binding of neocarzinostatin chromophore to oligodeoxyribonucleotides. Biochemistry. 1989;28:1019–1026. doi: 10.1021/bi00429a015. [DOI] [PubMed] [Google Scholar]
- 11.Kappen LS, Ellenberger TE, Goldberg IH. Mechanism and base specificity of DNA breakage in intact cells by neocarzinostatin. Biochemistry. 1987;26:384–390. doi: 10.1021/bi00376a008. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc Natl Acad Sci USA. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbaniak MD, Bingham JP, Hartley JA, Woolfson DN, Caddick S. Design and synthesis of a nitrogen mustard derivative stabilized by apo-neocarzinostatin. J Med Chem. 2004;47:4710–4715. doi: 10.1021/jm040790d. [DOI] [PubMed] [Google Scholar]
- 14.Caddick S, Muskett FW, Stoneman RG, Woolfson DN. Synthetic ligands for apo-neocarzinostatin. J Am Chem Soc. 2006;128:4204–4205. doi: 10.1021/ja058419i. [DOI] [PubMed] [Google Scholar]
- 15.Baker JR, Woolfson DN, Muskett FW, Stoneman RG, Urbaniak MD, Caddick S. Protein-small molecule interactions in neocarzinostatin, the prototypical enediyne chromoprotein antibiotic. Chem Bio Chem. 2007;8:704–717. doi: 10.1002/cbic.200600534. [DOI] [PubMed] [Google Scholar]
- 16.Sthapit B, Oh TJ, Lamichhane R, Liou K, Lee HC, Kim CG, Sohng JK. Neocarzinostatin naphthoate synthase: an unique iterative type I PKS from neocarzinostatin producer Streptomyces carzinostaticus. FEBS Lett. 2004;566:201–206. doi: 10.1016/j.febslet.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 17.Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tu57 and production of new antibiotics. Chem Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- 18.Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem Biol. 2006;13:575–585. doi: 10.1016/j.chembiol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Q, He Q, Ding W, Tang M, Kang Q, Yu Y, Deng W, Zhang Q, Fang J, Tang G, Liu W. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem Biol. 2008;15:693–705. doi: 10.1016/j.chembiol.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Cooke HA, Zhang J, Griffin MA, Nonaka K, Van Lanen SG, Shen B, Bruner SD. Characterization of NcsB2 as a promiscuous naphthoic acid/coenzyme A ligase integral to the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin. J Am Chem Soc. 2007;129:7728–7729. doi: 10.1021/ja071886a. [DOI] [PubMed] [Google Scholar]
- 21.Luo Y, Lin S, Zhang J, Cooke HA, Bruner SD, Shen B. Regiospecific O-methylation of naphthoic acids catalyzed by NcsB1, an O-methyltransferase involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin. J Biol Chem. 2008;283:14694–14702. doi: 10.1074/jbc.M802206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 24.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 25.Madduri K, Torti F, Colombo AL, Hutchinson CR. Cloning and sequencing of a gene encoding carminomycin 4-O-methyltransferase from Streptomyces peucetius and its expression in Escherichia coli. J Bacteriol. 1993;175:3900–3904. doi: 10.1128/jb.175.12.3900-3904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansson A, Koskiniemi H, Mantsala P, Niemi J, Schneider G. Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products. J Biol Chem. 2004;279:41149–41156. doi: 10.1074/jbc.M407081200. [DOI] [PubMed] [Google Scholar]
- 27.Jansson A, Koskiniemi H, Erola A, Wang J, Mantsala P, Schneider G, Niemi J. Aclacinomycin 10-hydroxylase is a novel substrate-assisted hydroxylase requiring S-adenosyl-L-methionine as cofactor. J Biol Chem. 2005;280:3636–3644. doi: 10.1074/jbc.M412095200. [DOI] [PubMed] [Google Scholar]
- 28.Jansson A, Niemi J, Lindqvist Y, Mantsala P, Schneider G. Crystal structure of aclacinomycin-10-hydroxylase, a S-adenosyl-L-methionine-dependent methyltransferase homolog involved in anthracycline biosynthesis in Streptomyces purpurascens. J Mol Biol. 2003;334:269–280. doi: 10.1016/j.jmb.2003.09.061. [DOI] [PubMed] [Google Scholar]
- 29.Oster LM, Lester DR, Terwisscha van Scheltinga A, Svenda M, van Lun M, Genereux C, Andersson I. Insights into cephamycin biosynthesis: the crystal structure of CmcI from Streptomyces clavuligerus. J Mol Biol. 2006;358:546–558. doi: 10.1016/j.jmb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Hou X, Wang Y, Zhou Z, Bao S, Lin Y, Gong W. Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans. J Struct Biol. 2007;159:523–528. doi: 10.1016/j.jsb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Cho JH, Park Y, Ahn JH, Lim Y, Rhee S. Structural and functional insights into O-methyltransferase from Bacillus cereus. J Mol Biol. 2008;382:987–997. doi: 10.1016/j.jmb.2008.07.080. [DOI] [PubMed] [Google Scholar]
- 32.Singh S, McCoy JG, Zhang C, Bingman CA, Phillips GN, Jr, Thorson JS. Structure and mechanism of the rebeccamycin sugar 4′-O-methyltransferase RebM. J Biol Chem. 2008;283:22628–22636. doi: 10.1074/jbc.M800503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr, A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey S. The CCP4 Suite - Programs for Protein Crystallography. Acta Crystallogr Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Proc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 40.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 41.Kleywegt GJ. Crystallographic refinement of ligand complexes. Acta Crystallogr Sect D: Biol Crystallogr. 2007;63:94–100. doi: 10.1107/S0907444906022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sippl MJ, Wiederstein M. A note on difficult structure alignment problems. Bioinformatics. 2008;24:426–427. doi: 10.1093/bioinformatics/btm622. [DOI] [PubMed] [Google Scholar]
- 43.Sippl MJ. On distance and similarity in fold space. Bioinformatics. 2008;24:872–873. doi: 10.1093/bioinformatics/btn040. [DOI] [PubMed] [Google Scholar]
- 44.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol. 2001;8:271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- 46.Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zubieta C, Kota P, Ferrer JL, Dixon RA, Noel JP. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002;14:1265–1277. doi: 10.1105/tpc.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Lanen SG, Lin S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027 involves a new branching point in chorismate metabolism. Proc Natl Acad Sci USA. 2008;105:494–499. doi: 10.1073/pnas.0708750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.