Significance
The demonstration and clinical relevance of HIV-1–specific immune responses in exposed but uninfected seronegative individuals have been controversial. Studies seeking to detect these responses have generally been small in size and have varied in study population, methods of detection, and control groups. We conducted a large case–control immunology study of participants in the Preexposure Prophylaxis Initiative (iPrEx) chemoprophylaxis trial, selecting preinfection time points for those who became infected compared with persistently HIV-1–negative controls. We confirmed that HIV-1–specific responses are present in exposed uninfected individuals, sometimes at very high magnitude. HIV-1–specific responses also correlated with infection risk.
Keywords: HIV-1, T cell, designated HIV-1–exposed seronegative, preexposure chemoprophylaxis, vaccine
Abstract
HIV-1–specific T-cell responses in exposed seronegative subjects suggest that a viral breach of the exposure site is more common than current transmission rates would suggest and that host immunity can extinguish subsequent infection foci. The Preexposure Prophylaxis Initiative (iPrEx) chemoprophylaxis trial provided an opportunity to rigorously investigate these responses in a case–control immunology study; 84 preinfection peripheral blood mononuclear cell samples from individuals enrolled in the iPrEx trial who later seroconverted were matched with 480 samples from enrolled subjects who remained seronegative from both the placebo and active treatment arms. T-cell responses to HIV-1 Gag, Protease, Integrase, Reverse Transcriptase, Vif, and Nef antigens were quantified for all subjects in an IFN-γ enzyme-linked immunospot (ELISpot) assay. IFN-γ responses varied in magnitude and frequency across subjects. A positive response was more prevalent in those who remained persistently HIV-1–negative for Gag (P = 0.007), Integrase (P < 0.001), Vif (P < 0.001), and Nef (P < 0.001). When correlated with outcomes in the iPrEx trial, Vif- and Integrase-specific T-cell responses were associated with reduced HIV-1 infection risk [hazard ratio (HR) = 0.36, 95% confidence interval (95% CI) = 0.19–0.66 and HR = 0.52, 95% CI = 0.28–0.96, respectively]. Antigen-specific responses were independent of emtricitabine/tenofovir disoproxil fumarate use. IFN-γ secretion in the ELISpot was confirmed using multiparametric flow cytometry and largely attributed to effector memory CD4+ or CD8+ T cells. Our results show that HIV-1–specific T-cell immunity can be detected in exposed but uninfected individuals and that these T-cell responses can differentiate individuals according to infection outcomes.
HIV-1–specific immune responses have been documented in some HIV-1–exposed but uninfected individuals (1–11). These responses are thought to have been generated by exposure to HIV-1 or cross-reactivity to other antigens (12–14). Irrespective of how they are induced, the relationship of these immune responses to infection is controversial. Some groups have not reliably detected such responses, possibly because of the differences in strength or frequency in exposed uninfected subjects relative to those with a systemic infection (15–17). Thus, characterization of immune responses in exposed uninfected individuals and verification of how they relate to protection would require a well-designed study with clinical outcomes, similar to the immune correlates analysis for the RV144 vaccine trial (18–20). Such a study could reveal characteristics desirable for T-cell responses, which then could be used to inform the design of T-cell immunogens for a preventative HIV-1 vaccine (21).
Preexposure chemoprophylaxis studies, where there are long follow-up periods and infection outcomes, have the potential for discovery of naturally acquired or induced immunity related to HIV-1 infection risk. We designed a hypothesis-driven case–control immunology study nested within the global Preexposure Prophylaxis Initiative (iPrEx) chemoprophylaxis trial (22). Our study leveraged the rigorous design of the iPrEx trial to examine naturally acquired HIV-1–specific T-cell responses. The first objective of our study was to corroborate previous reports of HIV-1 antigen-specific IFN-γ responses in exposed but uninfected individuals. The second objective was to test the hypothesis that HIV-1–specific T-cell responses in persistently HIV-1–seronegative iPrEx participants [designated HIV-1–exposed seronegative (HESN)] differed from preinfection T-cell responses in those who eventually seroconverted [designated seroconverter–before infection (SC-BI)], thus relating such responses to infection risk.
Results
Distribution of HIV-1–Specific IFN-γ Responses Differentiates Cases from Controls.
Baseline demographic characteristics of SC-BI and HESN subjects were comparable (Table S1). The prevalence of self-reported sexual practices and characteristics considered to be risk factors for HIV-1 acquisition were higher for SC-BI subjects at study entry (Table S1).
Table S1.
Demographics of subjects included in the analysis
| Cases (n = 84) | Controls (n = 412) | |
| No. of controls contributing more than one sample* | NA | 61 |
| Age at enrollment, yr (SD) | 25 ± 6 | 27 ± 8 |
| Ethnicity† | ||
| African American | 4 (5%) | 21 (5%) |
| White | 6 (7%) | 41 (10%) |
| Mixed/other | 72 (86%) | 337 (82%) |
| Asian | 2 (2%) | 13 (3%) |
| City | ||
| Lima | 46 (55%) | 223 (54%) |
| Iquitos | 13 (15%) | 66 (16%) |
| Guayaquil | 18 (21%) | 84 (20%) |
| Sao Paulo | 1 (1%) | 6 (1%) |
| San Francisco | 1 (1%) | 5 (1%) |
| Boston | 1 (1%) | 5 (1%) |
| Cape Town | 2 (2%) | 12 (3%) |
| Chiang Mai | 2 (2%) | 11 (3%) |
| No. of partners in past 12 wk before baseline (median) | 22 | 17 |
| Transactional sex | 39 (46%) | 186 (45%) |
| ncRAI in past 12 wk before baseline | 72 (86%) | 262 (65%) |
| Known HIV-positive partner in 6 mo | 4 (5%) | 4 (1%) |
| Reported sexually transmitted infection in last 6 mo | 31 (37%) | 126 (31%) |
All numbers are reported as n except where noted. NA, not applicable.
The selection algorithm allowed for more than one time point from some individuals. This incidence density sampling approach allowed the conditional logistic regression to approximate the HRs associated with each covariate; 412 HESN subjects contributed 480 samples for the analysis.
Subjects were permitted to select more than one ethnicity.
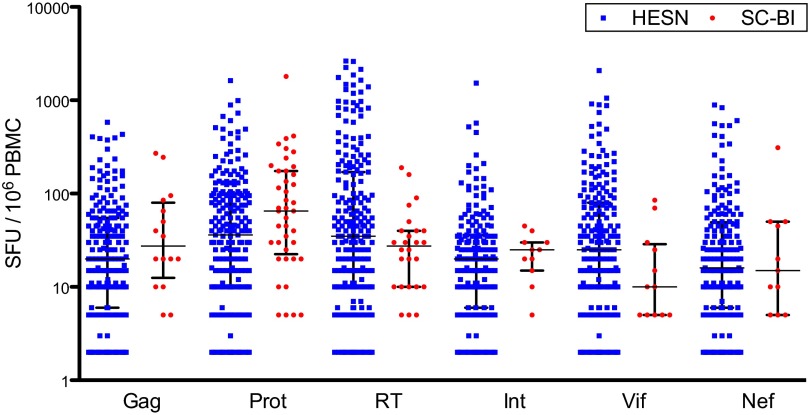
Antigen-specific responses [measured as spot-forming units (SFUs) in a standard enzyme-linked immunospot (ELISpot) assay] were skewed, with a large proportion of HESN and SC-BI subjects having no antigen-specific IFN-γ responses greater than two times the negative control (nonresponders). Thus, the SFU counts were analyzed with all values (combined) as well as with zero and nonzero SFU values independently (Table 1). HESN subjects had a greater proportion of responders compared with SC-BI subjects for the cumulative anti–HIV-1 response, but the difference did not reach statistical significance (Table 1 and Fig. S1). Among the individual antigens, responses were more prevalent in HESN subjects for Gag (P = 0.007), Integrase (P < 0.001), Vif (P < 0.001), and Nef (P < 0.001) antigens, whereas the differences did not reach significance for Reverse Transcriptase (RT) and Protease (Table 1 and Fig. S2). When summarizing the magnitude of responders only, HESN subjects had a greater cumulative anti–HIV-1 SFU count compared with SC-BI subjects (Fig. 1 and Table 1). A greater magnitude of SFU counts was also observed in responding HESN subjects among the individual antigen pools of RT, Integrase, Vif, and Nef, whereas the magnitude of both Gag and Protease SFU counts were numerically greater for SC-BI subjects (Fig. 2 and Table 1). When combining responders and nonresponders, the SFU count for HESN subjects was greater compared with for SC-BI subjects for Gag (P = 0.0161), RT (P = 0.022), Integrase (P = 0.007), Vif (P < 0.001), and Nef (P < 0.001) antigens but not Protease (Table 1). Anti–HIV-1 SFU counts for HESN and SC-BI subjects are represented in Fig. S3 as histograms inclusive of zero and nonzero values. A sample of imaged ELISpot wells from HESN and SC-BI subjects can be found in Figs. S4 and S5, respectively. Peripheral blood mononuclear cells (PBMCs) from a cohort of 30 unmatched low-risk healthy controls (LRHCs) were also tested in the ELISpot. The median (interquartile range) of the cumulative anti–HIV-1 IFN-γ response magnitude was 15 (10–25) SFUs (Fig. 1). We used a t test to compare cumulative SFUs in LRHCs with those in both HESN and SC-BI subjects among those who had a measureable response. We found a statistically significant difference between LRHCs and both HESN (P < 0.0001) and SC-BI (P < 0.0001) subjects. No LRHCs had an individual antigen response ≥55 SFUs (Fig. S3 B–G). Cumulative and antigen-specific anti–HIV-1 SFU counts for LRHC are represented in Fig. S3 A–G as histograms inclusive of zero and nonzero values. A sample of imaged ELISpot wells from LRHCs can be found in Fig. S6. Because of the unmatched nature of this control group and the low frequencies and magnitudes of LRHC responses relative to HESN and SC-BI responses, no additional analyses of LRHC responses were conducted.
Table 1.
Summary of SFUs for responders and nonresponders
| Antigen response/group | N | Frequency of response | Summary of SFU counts for responders only | Combined P value‡ | ||||
| Percentage | P value* | Mean (SFU per 106) | Median (SFU per 106) | IQR† (SFU per 106) | P value‡ | |||
| Cumulative anti–HIV-1 | ||||||||
| HESN | 480 | 71.9 | 0.721 | 336 | 120 | 40–325 | 0.610 | 0.158 |
| SC-BI | 84 | 63.1 | 172 | 70 | 35–180 | |||
| Gag | ||||||||
| HESN | 480 | 37.5 | 0.007 | 55 | 25 | 10–55 | 1 | 0.0161 |
| SC-BI | 84 | 19.0 | 62 | 28 | 15–75 | |||
| Protease | ||||||||
| HESN | 479 | 45.7 | 1 | 93 | 40 | 10–100 | 0.323 | 1 |
| SC-BI | 84 | 48.8 | 148 | 65 | 25–175 | |||
| Reverse Transcriptase | ||||||||
| HESN | 478 | 46.4 | 0.056 | 213 | 35 | 10–170 | 0.672 | 0.022 |
| SC-BI | 84 | 30.9 | 39 | 28 | 10–40 | |||
| Integrase | ||||||||
| HESN | 480 | 36.0 | <0.001 | 50 | 20 | 10–40 | 1 | 0.007 |
| SC-BI | 84 | 13.1 | 24 | 25 | 15–30 | |||
| Vif | ||||||||
| HESN | 480 | 42.5 | <0.001 | 92 | 30 | 10–80 | 0.128 | <0.001 |
| SC-BI | 84 | 14.3 | 23 | 10 | 5–28 | |||
| Nef | ||||||||
| HESN | 480 | 35.6 | <0.001 | 65 | 20 | 10–55 | 1 | <0.001 |
| SC-BI | 84 | 13.1 | 48 | 15 | 5–50 | |||
Frequency of response was based on the proportion of subjects with >0 SFUs after adjusting for background IFN-γ secretion (Methods). Response magnitude was summarized in responders only (>0 SFUs only) and combined (all data inclusive of 0 SFU values) because of the skewed nature of the SFU counts and large proportion of individuals lacking responses. IQR, interquartile range.
Reported P values were derived using a χ2 test on the proportion of nonresponders and accounted for multiple comparisons using a Bonferroni correction.
IQR represents the 25th to 75th percentiles.
Reported P values were derived using the Wilcoxon rank sum test and accounted for multiple comparisons using a Bonferroni correction.
Fig. S1.
Distribution of response frequency (>0 SFUs) for the cumulative anti–HIV-1 SFU count.
Fig. S2.
Distribution of response frequency (>0 SFUs) for the individual antigen anti–HIV-1 SFU count. Int, Integrase; Prot, Protease.
Fig. 1.
Distribution of cumulative anti–HIV-1 SFU counts in responders only. Cumulative nonzero SFU counts are shown with medians and interquartile ranges.
Fig. 2.
Distribution of anti–HIV-1 SFU counts for individual protein antigens in responders only. Nonzero SFU counts for individual antigens are shown with medians and interquartile ranges. Int, Integrase; Prot, Protease; RT, Reverse Transcriptase.
Fig. S3.
Histogram display of ELISpot SFU data. Cumulative anti–HIV-1 IFN-γ response for (A–G) HESN and SC-BI subjects and (A) LRHCs. Bins are customized to the range of SFUs observed for each antigen, except for the first four bins. The first four bins are consistent for all antigens and correspond to nonresponse (0), 1–10, 11–54, and 55–100 SFUs. Means are calculated inclusive of zero SFU values for nonresponders.
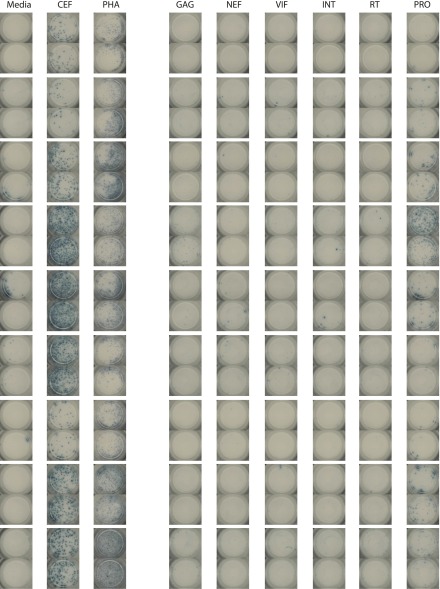
Fig. S4.
Example of control wells and HIV-1 antigen wells for nine HESN individuals. CEF, control peptide pool of cytomegalovirus (CMV), epstein-barr virus (EBV), and influenza peptides; Int, Integrase; PHA, phytohemagglutinin; Pro, Protease.
Fig. S5.
Example of control wells and HIV-1 antigen wells for nine SC-BI individuals. CEF, control peptide pool of cytomegalovirus (CMV), epstein-barr virus (EBV), and influenza peptides; Int, Integrase; PHA, phytohemagglutinin; Pro, Protease.
Fig. S6.
Example of control wells and HIV-1 antigen wells for nine LRHCs. CEF, control peptide pool of cytomegalovirus (CMV), epstein-barr virus (EBV), and influenza peptides; Int, Integrase; PHA, phytohemagglutinin; Pro, Protease.
HIV-1–Specific IFN-γ Responses Correlate with Infection Risk.
A multivariate conditional logistic regression model including all measured antigens was used as the primary analysis (because of the correlated nature of the responses) to elucidate the individual contributions of each antigen to HIV-1 infection risk (Table 2). Log10-transformed SFUs were used for all analyses to correct for the skewed distribution and wide range of SFU counts. Because we hypothesized that antigen responses arose from sexual exposure to HIV-1, reported noncondom receptive anal intercourse (ncRAI) was included in the model to address potential confounding between sexual practices, antigen response, and HIV-1 infection risk. Neither randomization to nor detectable levels of emtricitabine/tenofovir disoproxil fumarate were associated with antigen responses, and therefore, randomization was excluded from the model.
Table 2.
Association of antigen-specific T-cell responses with HIV-1 infection risk
| Antigen pool | Multivariate analysis | Multivariate analysis with backward selection | ||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Gag | 0.91 (0.57–1.45) | 0.711 | NR | NS |
| Protease | 2.5 (1.75–3.58) | <0.001 | 2.4 (1.69–3.41) | <0.001 |
| RT | 0.78 (0.52–1.18) | 0.238 | NR | NS |
| Integrase | 0.52 (0.28–0.96) | 0.038 | 0.49 (0.27–0.89) | 0.018 |
| Vif | 0.36 (0.19–0.66) | 0.001 | 0.33 (0.18–0.60) | <0.001 |
| Nef | 0.56 (0.30–1.04) | 0.065 | 0.52 (0.28–0.96) | 0.035 |
NR, not reported; NS, not significant; RT, Reverse Transcriptase.
Higher magnitudes of Vif and Integrase responses were associated with a statistically significant reduction in HIV-1 infection risk [hazard ratio (HR) = 0.36, 95% confidence interval (95% CI) = 0.19–0.66 and HR = 0.52, 95% CI = 0.28–0.96, respectively]. That is, for every 10-fold increase in the Vif and Integrase SFU count, the risk of HIV-1 infection decreased by 64% and 48%, respectively. In contrast, higher anti-Protease responses were associated with a statistically significant increase in infection risk (HR = 2.50, 95% CI = 1.75–3.58). Responses to the remaining antigens conferred a reduction in risk that did not reach statistical significance in the primary analysis (Table 2). The HR for Nef (HR = 0.52, 95% CI = 0.28–0.96) also reached statistical significance with backward selection of covariates (Table 2). We found similar results after limiting the analysis to include only those HESN subjects with HIV-1–negative antibody tests at subsequent protocol visits, arguing against the presence of an incubating or otherwise undetectable infection in those seronegatives with measureable antigen-specific IFN-γ responses. IFN-γ secretion in response to the positive control pool of cytomegalovirus (CMV), epstein-barr virus (EBV), and influenza peptides [CEF (pool of CMV, EBV, and influenza peptides)] was not associated with HIV-1 infection risk.
IFN-γ Responses Are Attributable to Effector Memory CD4+ and CD8+ T Cells.
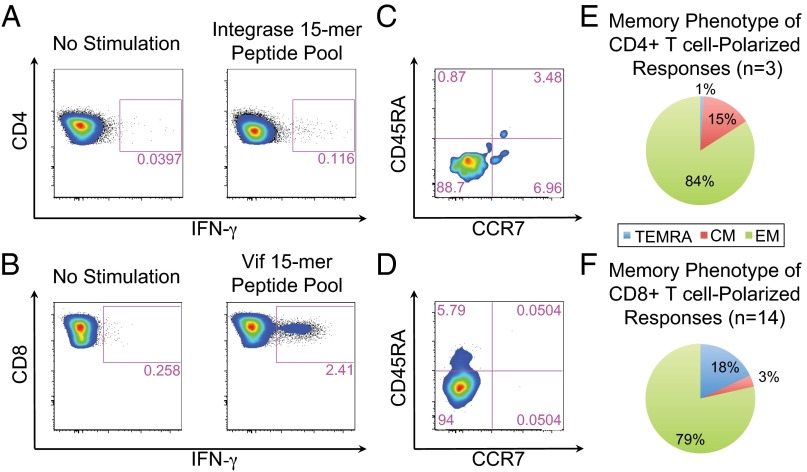
A subset of positive ELISpot responses (n = 17) was confirmed and characterized using multiparametric flow cytometry. Selection of samples was based on ELISpot reactivity (≥100 SFUs) and availability of cryopreserved PBMCs at the corresponding draw date. IFN-γ secretion was attributed to CD4+ or CD8+ T cells, with 14 responses (82%) attributed to a CD8+ dominant phenotype. An example of a CD4+ and CD8+ T-cell IFN-γ response from two different individuals can be found in Fig. 3 A and B; both responses were determined to be from effector memory cells (Fig. 3 C and D). For all of the antigens tested, IFN-γ responding cells were overwhelmingly of the effector memory phenotype (Fig. 3 E and F for CD4+ and CD8+, respectively). IFN-γ responses could be detected for each of the six peptide pools tested (Fig. S7).
Fig. 3.
Phenotypic analysis of IFN-γ–secreting cells. Examples of (A) a CD4+ Integrase-specific IFN-γ response and (B) a CD8+ Vif-specific IFN-γ response and (C and D, respectively) corresponding memory phenotyping. E and F summarize the memory phenotype of IFN-γ–secreting cells for all phenotypically characterized CD4+ and CD8+ responses, respectively. CM, central memory (CD45RA−CCR7+); EM, effector memory (CD45RA−CCR7−); TEMRA, terminally differentiated effector memory (CD45RA+CCR7−).
Fig. S7.
Examples of flow cytometry results for each antigen pool tested in the iPrEx Immunology Substudy. (A) HIV-1–specific antigen responses of CD3+ T cells. B depicts the proportions of CD4+ and CD8+ polarized responses from positive responses in HESN subjects tested in A.
Discussion
HIV-1–specific T-cell responses from exposed uninfected individuals have been previously reported (1–11, 23). Such responses have remained controversial, because some groups failed to observe such responses in exposed study subjects (16, 17). Overall, HESN studies have been limited in sample size and varied in testing protocols, survival bias, and other confounding factors that are difficult to control in cross-sectional studies. This study used a case–control design to address the limitations of previous reports, with a large subject number and comparable HIV-1 infection risk among those studied from a large controlled clinical trial. The data definitively show that HIV-1–specific T-cell responses exist in a subset of individuals at any given time in this exposed but uninfected population but that strong responses are infrequent, consistent with the variability in detection across previous HESN studies. The data also related HIV-1–specific T-cell responses to infection risk.
There was no detectable relationship of immune response to antiretroviral drug use, corroborating a recent report (24). Exposure data were not collected at the resolution necessary to estimate a temporal relationship between exposure and response strength. However, the low-response frequency for the individual protein antigens was consistent with the random association of exposure and PBMC draws. Nevertheless, the T-cell responses characterized in this study suggest that some HIV-1–exposed individuals encountered the virus or its proteins, triggering the observed immune response. This mechanism of inducing the observed responses is supported by the difference in response frequency and magnitude between LRHCs and both HESN and SC-BI subjects. Similar observations of T-cell responses in the absence of antibodies or detectable viremia have been documented in hepatitis C virus infection (25).
The role of T-cell responses in protection from HIV-1 infection has not been clearly established, but there is strong evidence of a T cell-mediated protective effect in nonclinical models of lentivirus infection. Protection has been observed in nonhuman primates with attenuated simian immunodeficiency virus (SIV) ΔNef vaccination, where a combination of T-cell and humoral immunity was induced (26, 27). Although a CMV-vectored vaccine that induced effector memory T-cell responses in Rhesus Macaques did not prevent SIV infection, the responses were associated with containment of SIV after mucosal exposure (28).
In humans, T-cell responses induced by the Ad5 Gag/Pol/Nef vaccine did not prevent HIV-1 infection in two separate studies of men who have sex with men and heterosexual men and women (29, 31). However, the vaccine constructs and elicited immunity differed from those observed in attenuated virus vaccination in nonhuman primates. The protein antigens in the Ad5 vaccine had been modified so as to abrogate their function (30), and they were based on laboratory-adapted HIV-1 isolates (31). Differences in immunodominance between naturally and vaccine-induced T-cell epitopes have been documented in other vaccine studies (32, 33). Thus, differences between naturally acquired and vaccine-induced HIV-1–specific immunity could explain the contradictory findings on the role of T cells in HIV-1 protection.
In this study, the association of Integrase-specific responses with reduced infection risk was modest but statistically significant. Responses directed against Integrase have been observed in previous HESN studies (9, 34). Integrase immunogens have also been included in recent vaccine studies along with Nef immunogens but not Vif in humans and nonhuman primate studies (31, 35).
Vif-specific IFN-γ responses were associated with the greatest reduction in infection risk relative to all others tested in this study. In the SIV model system, cellular immune responses to Vif have been used to track postchallenge viral replication (36), observed in SIV elite controllers (37), and associated with reduced SIV viral load and higher CD4+ T-cell counts postinfection (38). In humans, T-cell responses against Vif have been found in HIV-1 elite controllers (39) and HESN subjects (4). The accessory protein Vif is critical for HIV-1 replication, because it targets the intrinsic antiviral cytidine deaminase APOBEC (apolipoprotein B mRNA editing enzyme catalytic polypeptide-like enzyme) to the proteasome where it is degraded. Without Vif, HIV-1 is highly attenuated (40). Thus, the observed HR associated with a Vif response could indicate that the mechanistically related APOBEC cytidine deaminase in concert with T-cell responses or even alone provides the necessary reduction in R0, resulting in the control or clearance of infection foci. Supporting this notion are the colocation of T-cell epitopes with regions of the Vif protein associated with degradation of APOBEC3G/F, which was identified in a previous HESN study (4), and the relationship of APOBEC haplotypes to transmission (41).
Surprisingly, a T-cell response against Protease was associated with an increased risk of infection. Protease responses were not associated with emtricitabine/tenofovir disoproxil fumarate in this study and did not correlate with reported ncRAI. Given that the confirmation step of the study suggested a CD4+-polarized T-cell response from Protease (Fig. S7B) and that the dominant antigen in SC-BI subjects was Protease, one possible explanation for the association with increased risk could be an antigen-stimulated increase in target CD4+ T cells in the absence of CD8+ T cells or other counterbalancing immune mechanisms. Alternatively, a Protease-specific response could be an indication of an early, localized, or low-level infection without detectable viremia (eclipse phase) destined to become a productive systemic infection in the absence of protective immunity (non-Protease T-cell immunity or other mechanisms). Although highly sensitive clinical assays were used to verify the absence of HIV-1 viremia, early or otherwise undetectable infection could not be ruled out with the samples available for analysis. Irrespective of the mechanistic relationship of T-cell responses to subsequent infection outcomes, the proportion of HIV-1–exposed noninfected subjects who exhibited a detectable response suggests that virus–host interactions may be much more frequent than previously thought.
The frequency and magnitude of detectable T-cell responses in this study could be considered consistent with the definition of immune correlates of HIV-1 infection (18). The observed T-cell responses could also be a biomarker of exposure resulting from occult, controlled, abortive, or defective virus infection with a progressive infection determined by other contributing or coincident immune mechanisms. This study was not prospectively designed to address the potential contribution of preexisting cross-reactive IFN-γ responses or the effect of HIV-1 exposure on these responses (i.e., boosting of responses) (12, 13, 42). An expanded investigation would be needed to investigate intrinsic immune mechanisms (inclusive of APOBEC3D/F/G/H and their polymorphisms), conduct correlative virological studies, relate Protease-specific responses to the eclipse phase of infection, and test responses against protein antigens of HIV-1 not included in our analysis. We designed the nested case–control immunology study of the iPrEx trial to primarily investigate systemic T-cell responses in a large cohort of individuals. An extensive sample bank (large volume PBMC draws, tissues, and mucosal secretions) and systems biology experimental design were beyond the intended scope of this nested case–control study of the iPrEx trial.
In summary, our results conclusively show that HIV-1–specific T-cell immunity can be detected in a significant proportion of exposed but uninfected individuals. The mechanisms underlying the expansion of these T cells are unknown but may include cross-presentation of viral antigen, cross-reactivity from an unrelated antigen, or a transient, extinguished, or low-level HIV-1 infection. However, these T-cell responses can differentiate individuals according to infection outcomes, and certain T-cell response patterns correlate with infection risk. The results described here may also indicate that a renewed effort is warranted to more broadly investigate T cell-mediated mechanisms of infection resistance and the relationship to non-T cell-mediated mechanisms. Prospectively designed studies, such as of chemoprophylaxis or other large well-controlled HIV-1 prevention trials, of HESN subjects can afford the opportunity to probe such mechanisms with sufficient study sizes and sample repositories. Ultimately, a prospectively designed vaccine efficacy trial would be required to definitively establish protective mechanisms discovered in humans or nonhuman primates as correlates of protection (20).
Methods
Case–Control Study Design.
The iPrEx trial, inclusive of this immunology substudy, was approved by the University of California, San Francisco (UCSF) Committee on Human Research, the UCSF institutional review board. The iPrEx trial is registered with https://ClinicalTrials.gov (NCT00458393). For each case (SC-BI), up to six matched controls (HESN) were selected, with replacement from the same study site and comparable time on the study as cases, at the time of the cases’ first positive rapid tests (defined as a window of ≤84 d based on enrollment and drug dispensation date). The time on study match criteria was included so that controls would have similar exposure to drugs as cases at the point of assessment. Four controls were selected from the treatment arm: one with high sexual exposure, one with low sexual exposure, and two chosen at random. Two controls were selected from the placebo arm. Controls were matched to at least one case. The selection algorithm allowed for more than one time point from some individuals. This incidence density sampling approach allowed the conditional logistic regression to approximate the HRs associated with each covariate; 412 HESN subjects contributed 480 samples for the analysis. In both arms, high sexual exposure was defined as reporting ncRAI within 3 mo of the case or control specimen draw date. Low sexual exposure was defined as reporting no sexual partners within 3 mo of the case or control specimen. Absence of infection in controls was shown using rapid test HIV-1 antibody detection. For cases in whom HIV-1 antibodies were detected, HIV-1 RNA was also measured. Samples for unmatched LRHCs were obtained from a blood bank located in the San Francisco Bay area.
ELISpot Assay Methods.
IFN-γ ELISpot assays were conducted with pools of 15-mer peptides overlapping by 11 amino acids corresponding to HIV-1 consensus B sequences of Gag p24, Protease, Integrase, Reverse Transcriptase, Vif, and Nef that were obtained from the NIH AIDS Research and Reagent Program. Peptides were reconstituted with a minimum amount of DMSO, pooled, frozen at a concentration of 100 μg/mL, and used at a final concentration of 5 μg/mL. All peptides were filtered at 0.45 μm. Peptide pools were tested for reactivity in an ELISpot assay, with HIV-1–positive and LRHC samples as a quality control measure. PBMCs were plated at a concentration of 1 × 105 cells per well. No reassays were permitted, and all data were included in the analyses to ensure objectivity. The same analyst supervised or conducted all of the assay steps. Where possible, single lots or single manufacturers were used for reagents and labware.
Cytokine Flow Cytometry.
Cell preparation and antigen stimulation.
PBMCs were rapidly thawed in a 37 °C water bath and washed two times with cold R10 medium of RPMI, l-glutamine, Penicillin Streptomycin, and 10% (vol/vol) heat-inactivated filtered FBS. For in vitro stimulation, PBMCs were then resuspended in medium to 1 × 106 cells/mL and placed in 12 × 75-mm nonpyrogenic polypropylene tissue culture tubes (Becton Dickinson Falcon); 1 μg costimulatory anti-CD28 and anti-CD49d mAbs were added for every 1 × 106 cells. HIV-1 peptides were added at 5 μg/mL to culture tubes and incubated for 1 h at 37 °C at a 5° angle; 50 μL Brefeldin A at 5 μg/mL was added to each culture tube after 1 h, and tubes were then incubated overnight.
Immunofluorescence staining with antibodies.
After incubation, cells were washed at 1,800 rpm for 10 min with 5 °C 1× buffer of PBS, 0.05% sodium azide (NaN3), and 1% BSA. Excess volume was decanted, and cell surface stain was added at 75 μL per test. The cell pellet was resuspended in the cell surface stain and incubated at room temperature protected from light. PBMCs were stained with the following cell surface antibodies in the presence of human IgG (10 μg/mL; Sigma): Pacific Blue anti-CD3 (BioLegend), Brilliant Violet 605 anti-CD8 (clone RPA-T8; BioLegend), Brilliant Violet 650 anti-CD4 (clone OKT4; BioLegend), phycoerythrin/cyanine 7 anti-CD197 (CCR7; clone G043H7; BioLegend), FITC anti-CD45RA (clone HI100; BioLegend), and allophycocyanin/cyanine 7 anti-CD14 and anti-CD19 (clones HCD14 and HIB19, respectively; BioLegend). Amine Aqua (Life Technologies) was included in each stain to exclude dead cells. The cells were then fixed using 1× FACS Lyse Buffer (Becton Dickinson) in sterile water at room temperature. After 10 min, cells were washed, pelleted with buffer [PBS, 0.05% sodium azide (NaN3), 1% BSA], permeated with FACS Perm Buffer [25% (vol/vol) FACS lysing solution, 0.01% Tween-20 in sterile water], and then, repeated. Cells were washed, stained (75 μL per test) for intracytoplasmic IFN-γ with allophycocyanin anti–IFN-γ (clone B27; BD Pharmigen), and protected from light for 1 h. Cells were washed, pelleted, and fixed a final time in 2% paraformaldehyde in PBS.
Flow cytometric analysis of PBMCs.
Samples were analyzed on a Four-Laser LSR II Flow Cytometer (BD Biosciences). Data analysis was performed using FlowJo Version 9.6.1 software (TreeStar). To determine memory phenotypes of IFN-γ–producing HIV-1–specific cells, CCR7 and CD45RA expressions were assessed. Quadrant gates were created using the IFN-γ–negative cells and copied onto the IFN-γ–secreting cells. A minimum number of 50 events was required before the population could be considered and analyzed for memory phenotyping. A final event count range of 50–1,980 events was analyzed for the IFN-γ–negative and -positive cells.
Data Handling and Statistical Methods.
Data management.
ELISpot plates were counted using an automated plate reader (AID Diagnostika GmbH) and compiled in Microsoft Excel (version 14.2.1). The dataset was transferred to STATA SE 13.0. Samples were run in duplicate when possible, and results were averaged. In cases where duplicate results were not available (i.e., low cell numbers precluded running in duplicate), single results were used; only 3 singlicate data entries are reported for SC-BI subjects, and 29 singlicate data entries are reported for HESN subjects. For each result, two times the background IFN-γ (media) was subtracted. Background IFN-γ (median) was comparable for HESN and SC-BI at 1.5 and 5 spots per media-only well for both groups, respectively. Negative values were treated as nonresponses and set to zero. All statistical analyses were performed using STATA SE 13.0.
Pairwise analysis of response distribution.
The numbers of responders vs. nonresponders for cases and controls were analyzed using a χ2 test. Magnitude of each antigen response and overall response in responders were compared between cases and controls using the Wilcoxon rank sum test. Bonferroni adjustments were made to account for multiple comparisons.
Conditional logistic regression.
We related antigen responses to risk of HIV-1 seroconversion in analyses using conditional logistic regression. Multivariate conditional logistic regression was run including each HIV-1 antigen of interest on the log10 scale to relate antigen responses to risk of HIV-1 seroconversion. The model controlled for CEF and self-reported ncRAI to account for potential confounding. As a posthoc analysis, backward covariate selection eliminating all covariates that did not reach significance of P < 0.1 was used to better clarify parameter estimates.
Supplementary Material
Acknowledgments
We thank David Watkins, Frederick Hecht, Susan Buchbinder, and Teri Liegler for critically reviewing this manuscript. We thank Patricia Defechereux, Robert Hance, and Jeanny Lee for specimen distribution. We thank the Preexposure Prophylaxis Initiative (iPrEx) chemoprophylaxis trial participants for providing the samples. We also thank the iPrEx Study team and investigators Javier R. Lama, Juan Vicente Guanira, Martín Casapía, Orlando Montoya-Herrera, Susan Buchbinder, Valdilea G. Veloso, Kenneth H. Mayer, Suwat Chariyalertsak, Mauro Schechter, and Linda-Gail Bekker. We thank the NIH AIDS Reagent Program for providing the peptides. This work was partially supported by National Institutes of Health Grants AI62333, AI64002, AI087131, and RR024131; the Bill and Melinda Gates Foundation; the J. David Gladstone Institutes; the University of California, San Francisco (UCSF) Academic Senate Committee on Research (P.J.K.); the Peter and Shelagh Godsoe Family Foundation through the AIDS Research Institute at UCSF (P.J.K. and D.F.N.); a Howard Hughes Medical Institute Medical Fellowship (to R.G.C.); a fellowship from the American-Scandinavian Foundation (to E.M.E.); National Council for Scientific and Technological Development, Brazilian Ministry of Science and Technology/Coordination for the Improvement of Higher Education, Brazilian Ministry of Health Grant 056/2012 (to D.F.N.); and Fundação de Amparo a Pesquisa do Estado de São Paulo Grants 2010/05845-0 (to D.F.N. and E.G.K.) and 04/15856-9 (to E.G.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501443112/-/DCSupplemental.
References
- 1.Clerici M, et al. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J Infect Dis. 1992;165(6):1012–1019. doi: 10.1093/infdis/165.6.1012. [DOI] [PubMed] [Google Scholar]
- 2.Erickson AL, et al. Potentially exposed but uninfected individuals produce cytotoxic and polyfunctional human immunodeficiency virus type 1-specific CD8(+) T-cell responses which can be defined to the epitope level. Clin Vaccine Immunol. 2008;15(11):1745–1748. doi: 10.1128/CVI.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowke KR, et al. HIV-1-specific cellular immune responses among HIV-1-resistant sex workers. Immunol Cell Biol. 2000;78(6):586–595. doi: 10.1046/j.1440-1711.2000.00944.x. [DOI] [PubMed] [Google Scholar]
- 4.Kebba A, et al. Distinct patterns of peripheral HIV-1-specific interferon- gamma responses in exposed HIV-1-seronegative individuals. J Infect Dis. 2004;189(9):1705–1713. doi: 10.1086/383227. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn L, et al. T-helper cell responses to HIV envelope peptides in cord blood: Protection against intrapartum and breast-feeding transmission. AIDS. 2001;15(1):1–9. doi: 10.1097/00002030-200101050-00003. [DOI] [PubMed] [Google Scholar]
- 6.Legrand FA, et al. Strong HIV-1-specific T cell responses in HIV-1-exposed uninfected infants and neonates revealed after regulatory T cell removal. PLoS ONE. 2006;1:e102. doi: 10.1371/journal.pone.0000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowland-Jones SL, et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Invest. 1998;102(9):1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowland-Jones SL, et al. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet. 1993;341(8849):860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- 9.Holditch SJ, et al. Decay kinetics of HIV-1 specific T cell responses in vertically HIV-1 exposed seronegative infants. Front Immunol. 2011;2:94. doi: 10.3389/fimmu.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shacklett BL, et al. Dendritic cell amplification of HIV type 1-specific CD8+ T cell responses in exposed, seronegative heterosexual women. AIDS Res Hum Retroviruses. 2002;18(11):805–815. doi: 10.1089/08892220260139558. [DOI] [PubMed] [Google Scholar]
- 11.Kaul R, et al. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J Immunol. 2000;164(3):1602–1611. doi: 10.4049/jimmunol.164.3.1602. [DOI] [PubMed] [Google Scholar]
- 12.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38(2):373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trama AM, et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16(2):215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaul R, et al. Late seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J Clin Invest. 2001;107(3):341–349. doi: 10.1172/JCI10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young JM, Turpin JA, Musib R, Sharma OK. Outcomes of a National Institute of Allergy and Infectious Diseases Workshop on understanding HIV-exposed but seronegative individuals. AIDS Res Hum Retroviruses. 2011;27(7):737–743. doi: 10.1089/aid.2010.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Addo MM, et al. Lack of detectable HIV-1-specific CD8(+) T cell responses in Zambian HIV-1-exposed seronegative partners of HIV-1-positive individuals. J Infect Dis. 2011;203(2):258–262. doi: 10.1093/infdis/jiq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hladik F, et al. Most highly exposed seronegative men lack HIV-1-specific, IFN-gamma-secreting T cells. J Immunol. 2003;171(5):2671–2683. doi: 10.4049/jimmunol.171.5.2671. [DOI] [PubMed] [Google Scholar]
- 18.Qin L, Gilbert PB, Corey L, McElrath MJ, Self SG. A framework for assessing immunological correlates of protection in vaccine trials. J Infect Dis. 2007;196(9):1304–1312. doi: 10.1086/522428. [DOI] [PubMed] [Google Scholar]
- 19.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis. 2012;54(11):1615–1617. doi: 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lederman MM, et al. Determinants of protection among HIV‐exposed seronegative persons: An overview. J Infect Dis. 2010;202(Suppl 3):S333–S338. doi: 10.1086/655967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant RM, et al. iPrEx Study Team Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010;363(27):2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaul R, et al. HIV-1 Env-specific cytotoxic T-lymphocyte responses in exposed, uninfected Kenyan sex workers: A prospective analysis. AIDS. 2004;18(15):2087–2089. doi: 10.1097/00002030-200410210-00015. [DOI] [PubMed] [Google Scholar]
- 24.Pattacini L, et al. Partners PrEP Study Team Antiretroviral Pre-Exposure Prophylaxis Does Not Enhance Immune Responses to HIV in Exposed but Uninfected Persons. J Infect Dis. 2015;211(12):1943–1952. doi: 10.1093/infdis/jiu815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heller T, et al. Occupational exposure to hepatitis C virus: Early T-cell responses in the absence of seroconversion in a longitudinal cohort study. J Infect Dis. 2013;208(6):1020–1025. doi: 10.1093/infdis/jit270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258(5090):1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 27.Mori K, et al. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: Robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J Virol. 2001;75(9):4023–4028. doi: 10.1128/JVI.75.9.4023-4028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen SG, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502(7469):100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray GE, et al. HVTN 503/Phambili study team Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: A double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect Dis. 2011;11(7):507–515. doi: 10.1016/S1473-3099(11)70098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Priddy FH, et al. Merck V520-016 Study Group Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin Infect Dis. 2008;46(11):1769–1781. doi: 10.1086/587993. [DOI] [PubMed] [Google Scholar]
- 31.Buchbinder SP, et al. Step Study Protocol Team Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372(9653):1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betts MR, et al. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc Natl Acad Sci USA. 2005;102(12):4512–4517. doi: 10.1073/pnas.0408773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hertz T, et al. HIV-1 vaccine-induced T-cell responses cluster in epitope hotspots that differ from those induced in natural infection with HIV-1. PLoS Pathog. 2013;9(6):e1003404. doi: 10.1371/journal.ppat.1003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaul R, et al. CD8(+) lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J Clin Invest. 2001;107(10):1303–1310. doi: 10.1172/JCI12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15(3):293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473(7348):523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mudd PA, et al. Escape from CD8(+) T cell responses in Mamu-B*00801(+) macaques differentiates progressors from elite controllers. J Immunol. 2012;188(7):3364–3370. doi: 10.4049/jimmunol.1102470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martins MA, et al. T-cell correlates of vaccine efficacy after a heterologous simian immunodeficiency virus challenge. J Virol. 2010;84(9):4352–4365. doi: 10.1128/JVI.02365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarosso LF, et al. Unexpected diversity of cellular immune responses against Nef and Vif in HIV-1-infected patients who spontaneously control viral replication. PLoS ONE. 2010;5(7):e11436. doi: 10.1371/journal.pone.0011436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Refsland EW, et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10(11):e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie AJ, et al. Differences in HIV-specific T cell responses between HIV-exposed and -unexposed HIV-seronegative individuals. J Virol. 2011;85(7):3507–3516. doi: 10.1128/JVI.02444-10. [DOI] [PMC free article] [PubMed] [Google Scholar]