Significance
Proteolytic enzymes are inhibited in vivo by protein inhibitors. Such inhibitors are used by symbiotic bacteria in our gut to protect themselves from digestive peptidases. This is the case for Escherichia coli, which has acquired a large, multidomain inhibitor of broad inhibitory spectrum [Escherichia coli α2-macroglobulin (ECAM)]. We studied ECAM and found it is cleaved by host peptidases, which triggers large conformational rearrangement of the inhibitor—shown by protein crystallography and electron microscopy reconstructions—as well as covalent binding of the peptidase. The latter is blocked similarly to a mouse by a snap trap, which prevents damage to the bacterial envelope. Prey peptidases, however, can still be active in the digestion of intake proteins.
Keywords: protein inhibitor, gut microbiome, conformational rearrangement, X-ray crystal structure, cryo-electron microscopy
Abstract
The survival of commensal bacteria requires them to evade host peptidases. Gram-negative bacteria from the human gut microbiome encode a relative of the human endopeptidase inhibitor, α2-macroglobulin (α2M). Escherichia coli α2M (ECAM) is a ∼180-kDa multidomain membrane-anchored pan-peptidase inhibitor, which is cleaved by host endopeptidases in an accessible bait region. Structural studies by electron microscopy and crystallography reveal that this cleavage causes major structural rearrangement of more than half the 13-domain structure from a native to a compact induced form. It also exposes a reactive thioester bond, which covalently traps the peptidase. Subsequently, peptidase-laden ECAM is shed from the membrane and may dimerize. Trapped peptidases are still active except against very large substrates, so inhibition potentially prevents damage of large cell envelope components, but not host digestion. Mechanistically, these results document a novel monomeric “snap trap.”
The human microbiome plays a crucial role in host health and disease (1). Successful commensalism requires microorganisms to neutralize damaging host factors, but the mechanisms to maintain symbiosis are only poorly understood (2). In particular, their habitat is rich in host proteolytic enzymes, which are generally held in check by protein inhibitors (3). Several Gram-negative proteobacteria, including human pathogens, contain genes similar to the widespread metazoan α2-macroglobulins (α2Ms) (4). These are large, multidomain glycoproteins that uniquely function as broad-spectrum endopeptidase inhibitors and mostly contain a reactive β-cysteinyl-γ-glutamyl thioester bond (5). The potential bacterial α2Ms (bα2Ms) occur in two independent forms: one provided with a thioester bond (represented by Escherichia coli α2M; ECAM) and cotranscribed with penicillin-binding protein 1C, and the other lacking a thioester bond and transcribed from an operon further encoding other proteins (represented by E. coli YfaS).
In humans, α2M (hα2M) circulates mostly in blood plasma as an abundant “native” ∼720-kDa tetramer. After cleavage in a “bait region” (6), the tetramer closes under large conformational rearrangement to yield an “induced” form, which encages the peptidase following an irreversible “Venus flytrap” mechanism (5, 7, 8). Inside the cage, within a large “central prey chamber,” peptidases still cleave small-to-medium substrates (<10–20 kDa) (9), which enter the tetramer through any of 12 entrances (8), but not large substrates. In some cases, prey lysines may be covalently bound through the thioester bond of mammalian α2Ms. However, other α2M-family inhibitors such as ovostatins lack thioester bonds and only encage, but they are as efficient inhibitors as hα2M (10). Induction of tetrameric hα2M exposes C-terminal receptor-binding domains (RBDs), which are bound by specific cell-surface receptors. This exposure triggers receptor-mediated endocytosis and clearance of the inhibitor and its prey from the circulation (11). For successful encaging, at least two protomers are required to wrap around a standard-size endopeptidase (12), but the detailed molecular mechanism of tetrameric α2M inhibition is unknown, as only the molecular structure of induced hα2M is available (8). Little is also known about the physiology and function of bα2Ms, as only a YfaS-ortholog from Pseudomonas aeruginosa and ECAM have been partially studied to date (13–16). The crystal structure of native α2M from Salmonella enterica (SEAM) is available (16), but its working mechanism is also unknown so far.
To shed light on the structure and function of α2Ms, we studied ECAM functionally, biophysically, and structurally by X-ray crystallography and cryoelectron microscopy (cryo-EM). We found that cleavage at the bait region of ECAM triggers major conformational rearrangement and covalent binding of the prey peptidase after a monomeric snap-trap mechanism, which differs from the encaging Venus flytrap mechanism of tetrameric hα2M.
Results and Discussion
Native and Induced Forms of ECAM.
In thioester proteins in general, the reactive thioester bond is protected in native forms to prevent precocious opening (17). In hα2M, treatment with small nucleophiles such as methylamine (MA) opens the thioester bond and rearranges the tetramer. This rearrangement is equivalent to the status induced by prey peptidases (18), after which the thioester loop is exposed on the inner protein surface of the cage and becomes accessible to surface lysines of the prey (7, 8). Native and peptidase- or MA-induced hα2M differ in their biophysical properties (19). In contrast, recombinant native ECAM (nECAM) and MA-treated ECAM (MA-ECAM) were equivalent in size-exclusion chromatography (SEC), native PAGE, thermofluor assays, and circular dichroism spectroscopy (CD), and they were both monomeric (SI Appendix, Fig. S1 A–D). This indicates that MA treatment of nECAM, which opens the thioester bond, as shown by the emergence of free cysteines (SI Appendix, Table S2), does not produce an induced species. This, in turn, is consistent with the structural equivalence of native and MA-treated SEAM (16). Induced ECAM (iECAM) was obtained only by incubation with endopeptidases such as the physiologically relevant mammalian host digestive enzyme trypsin (20). Unlike nECAM, iECAM formed monomers and dimers, both diverging from nECAM in SEC, native PAGE, thermofluor assays, and CD, which is consistent with conformational rearrangement on induction (SI Appendix, Fig. S1 A–D and J). MA-ECAM, in turn, was transformed into an induced form similar to iECAM after cleavage in the bait region. Other endopeptidases capable of ECAM induction were thermolysin, chymotrypsin, and subtilisin (SI Appendix, Fig. S1E).
Analysis of trypsin-induced iECAM by denaturing SDS/PAGE and N-terminal Edman degradation showed that induction entailed cleavage at R946–F947 (numbering follows UniProt P76578), which falls into the “bait-region domain” (BRD; see Fig. 1A for domain organization and acronyms) of the inhibitor. Further cleavages contributed to a complex band pattern (SI Appendix, Fig. S1F), which occasionally gave rise to high-molecular-mass products of proteinase-generated fragments, as previously described for hα2M (7). To simplify the picture, we produced an ECAM mutant (TEV-ECAM), in which bait-region positions A943–G951 had been replaced with a recognition sequence for tobacco-etch virus (TEV) peptidase. This protein was cleaved by TEV peptidase at a single site (SI Appendix, Fig. S2A) and gave rise to an induced form (TEV-iECAM; SI Appendix, Fig. S2 B and C). We also observed cleavage in the bait region of ECAM by ulilysin, chymotrypsin, pancreatic elastase, subtilisin, and thermolysin (SI Appendix, Fig. S2F). In all cases, this cleavage was efficient and showed that the bait region contains accessible recognition sites for peptidases with distinct substrate specificities, despite being shorter than in hα2M [∼25 residues, Q934–G958 (see following) vs. 39 residues (see ref. 6)]. These results indicate that ECAM is a pan-proteinase target protein and that cleavage in the bait region is required and suffices to generate iECAM.
Fig. 1.
Monomeric iECAM structure. (A) Domain organization of ECAM with spanning residues (see also SI Appendix, Fig. S7). A, flexible segment; BRD, bait-region domain; CUB, domain type first described in proteins C1r/C1s, Uegf, and Bmp1; L, linker; MG0–MG7, macroglobulin-type domains 0 to 7; NIE, N-terminal domain of induced ECAM; RBD, receptor-binding domain; SP, signal peptide; TED, thioester domain. The bait region and the thioester segment are depicted as green and purple handles, respectively. Red arrows pinpoint the primary trypsin cleavage sites for induction (R946) and solubilization/dimerization (R162). Domain naming is based on hα2M, which lacks A, MG0, and NIE (8). (B) Scheme of iECAM in front view. (C) Ribbon-plot of monomeric iECAM in front, lateral, and back views. In B and C, the domain colors are as in A, and the visible polypeptide chain ends upstream (U) and downstream (D) of the cleaved bait region, as well as the thioester (cyan ellipse; blown up in the inset in C), are indicated.
Induced ECAM Is Released as Monomers and Dimers.
Among the trypsin cleavage sites of ECAM was also R162–D163, which falls between the first two N-terminal domains: macroglobulin-type domain 0 (MG0) and the N-terminal domain of induced ECAM (NIE) (Fig. 1A). This linker was likewise targeted by pancreatic elastase and thermolysin (SI Appendix, Fig. S1 G and H). As the N-terminal flexible segment A (Fig. 1A) of ECAM is anchored to the periplasmic side of the inner membrane through a “lipobox”-mediated lipidic linkage of the N-terminal cysteine residue of the secreted protein (C18) through posttranslational modification, cleavage at MG0-NIE removes the membrane anchor, thus yielding soluble iECAM. This strongly suggests shedding is a relevant step of the working mechanism of ECAM after induction. It is noteworthy that this cleavage was also responsible for freshly purified monomeric iECAM being slowly transformed by trypsin to a noncovalent dimer. Evidence for this came from ECAM mutant R162G, which was not cleaved by trypsin at MG0-NIE and remained largely monomeric even after extended incubation periods (SI Appendix, Fig. S1G). In addition, TEV peptidase, which transformed native TEV-ECAM into an induced form (see earlier text), did not cleave at MG0-NIE, nor did it form dimers (SI Appendix, Fig. S2B). Extended dimerization was mainly observed at high trypsin:nECAM ratios or at prolonged incubation times (SI Appendix, Fig. S1I). Dimers of iECAM were stable and separable from monomers, did not revert to monomers under any condition assayed, and were unaffected by high salt, detergents, or reducing agents. In addition, dimers were conformationally equivalent to iECAM monomers in CD and thermofluor assays (SI Appendix, Fig. S1 C and D), as well as in cryo-EM reconstructions (see following), which supports that once induction has occurred on monomeric ECAM, dimerization just entails association of two preformed moieties. We conclude that shedding under removal of the first ∼140 residues of secreted ECAM (segment A and domain MG0; Fig. 1A) after its induction is required for dimerization by trypsin and ulilysin. However, other peptidases targeting the MG0-NIE linker such as pancreatic elastase and thermolysin did not significantly form dimers (SI Appendix, Fig. S1G). Thus, iECAM dimerization is restricted to treatment with particular peptidases, possibly depending on their size and shape.
ECAM Inhibits Cleavage of Very Large Substrates and Cell Wall Components.
We assessed the inhibitory activity of ECAM in vitro against a cohort of model serine- and metallopeptidases of differing specificity in the presence of a wide range of substrates (SI Appendix, Supplementary Experimental Procedures [SEP] section [§] 1.2). We did not detect inhibition against low- or medium-molecular-mass substrates. In contrast, proteolytic activity was inhibited for trypsin against thyroglobulin (660 kDa) and aldolase (160 kDa), for chymotrypsin against thyroglobulin, and for subtilisin against thyroglobulin and fumarase (200 kDa), indistinguishable by both monomeric and dimeric iECAM (SI Appendix, Fig. S3 A–C). These experiments were complemented with assays against cell envelope extracts prepared from E. coli K12 cells (SI Appendix, SEP §1.3), which are rapidly processed by endopeptidases that separate the outer membrane and the peptidoglycan. These experiments revealed that digestion by trypsin, chymotrypsin, and subtilisin was inhibited by ECAM in a concentration-dependent manner (SI Appendix, Fig. S3D). In addition, we also found that ECAM is a cell wall protector in vivo, as its absence results in diminished cell viability in the presence of host peptidases (SI Appendix, Supplementary Results and Discussion [SRD] §2.1). Accordingly, ECAM inhibits proteolysis of large globular proteins and proteins embedded in the cell envelope, but not of isolated peptides and medium-to-large-sized proteins.
ECAM Is a Pan-Peptidase Covalent Inhibitor.
We also found that the inhibitory mechanism of ECAM further requires covalent bonding of prey peptidases by means of an intact thioester bond targeted by a lysine from the prey. Covalent linkage was shown in a zymogram, which yielded ECAM cleavage fragments showing tryptic activity against casein, and similar for monomeric and dimeric trypsin-induced iECAM (SI Appendix, Fig. S2D). In addition, purification of trypsin-treated monomeric and dimeric iECAM by SEC revealed the presence of the peptidase in the elution peaks of the latter, as shown by activity against small fluorogenic trypsin substrates and peptide-mass fingerprinting. Evidence for the involvement of the thioester bond came from the finding that iECAM showed free cysteines compared with trypsin-untreated nECAM (SI Appendix, Table S2); that is, the bond had been broken during induction. To verify that lysines were the targets of the thioester bond, we probed lysine-methylated TEV peptidase and found that, in contrast to the untreated protein, the enzyme was not bound by TEV-ECAM on cleavage induction (SI Appendix, Fig. S2C).
In contrast, incubation of MA-ECAM, with a broken thioester bond, with trypsin underwent cleavage in the bait region and the MG0-NIE linker (SI Appendix, Fig. S1J) and was induced, but it did not bind the peptidase, as shown by the lack of peptidolytic activity of SEC-purified peptidase-induced MA-ECAM and the absence of cross-linked peptidases in denaturing SDS/PAGE (SI Appendix, Fig. S1J). In addition, peptidase activity against cell envelope extracts was inhibited by nECAM (see earlier text), but not by MA-ECAM (SI Appendix, Fig. S3D).
Finally, binding of trypsin through ECAM was semiquantitatively assessed by using fluorogenic methylcoumarin-labeled peptidase. This revealed that one peptidase molecule was covalently bound by between three and four iECAM molecules on average; that is, thioester-mediated prey binding is relatively inefficient because a surface lysine needs to be close to the thioester bond to be bound when the bait region is cleaved. This contrasts with the high efficacy of bait-region cleavage (see earlier text) and indicates that iECAM can be either bound or unbound to the peptidase.
Crystal Structure of Trypsin-Induced iECAM.
To shed light on the structural basis of the molecular mechanism of ECAM, we crystallized and solved the structure of trypsin-induced iECAM by four-wavelength anomalous diffraction with a selenomethionine derivative of the protein and a dataset to high resolution from wild-type protein (SI Appendix, SEP §1.10–1.11 and Tables S3 and S4). Although the peptidase was inside the crystals, as revealed by fluorescence microscopy (SI Appendix, Fig. S4), we could not localize it because of the low binding efficiency of iECAM (see previous section) and the intrinsic disorder in its overall conformation. The iECAM oligomeric structure in the crystals is a dimer formed by a crystallographic dyad (for a detailed description of the dimer, see SI Appendix, SRD §2.2 and Fig. S5). The iECAM monomer structure includes fragment P166–P1653, which is organized in 12 domains and a linker region (NIE to RBD; Fig. 1 A and B). The molecule is arranged as an elliptical grommet with a ∼105-Å major axis and a ∼60-Å minor axis (Fig. 1C, Center). A large hook protrudes ∼80 Å from one of its major axis vertices and is inclined ∼30° toward the center of the ellipse. We distinguish between a front convex face (Fig. 1C, Left; reference orientation hereafter) and a back concave face (Fig. 1C, Right). The polypeptide starts at the bottom with domain NIE, which features one of the ellipse major-axis ends. Thereafter, six MG domains (MG1–MG6) are arranged as a one-and-a-half-turn superhelix (MG-superhelix) around a central lumen ∼20 Å in diameter (“entrance 1”) in such a way that domains MG5 and MG6 are, respectively, aligned and in contact with MG1 and MG2. Perpendicularly attached to MG3 and MG6, domain MG7 features the opposite end of the ellipse and leads to the hook, which includes a domain type first described in proteins C1r/C1s, Uegf, and Bmp1 (CUB); a thioester domain (TED); and RBD. Overall, iECAM includes six structurally different domain types: MGs, NIE, CUB, TED, RBD, and BRD (SI Appendix, Fig. S6 A–E).
MG domains are fibronectin-type-III-like β-sandwiches comprising a three- and a four-stranded antiparallel β-sheet, whose planes are rotated away by ∼40° (SI Appendix, Fig. S6A; for assignment of secondary structure elements, see SI Appendix, Fig. S7). Into this basic scaffold, additional elements are inserted, which cause the eight MG domains (including MG0, see following) to span between 78 and 128 residues and vary in domain length (along the sheets; SI Appendix, Fig. S6 A, F, and G) between ∼30 Å (MG1) and ∼50 Å (MG7). Domain NIE is a variant of an MG domain, into which an extra short strand has been inserted between NIE-β6 and NIE-β7, which interacts with NIE-β1 (SI Appendix, Fig. S6E). This entails that although the four-stranded β-sheet overlaps with that of MG1 (SI Appendix, Fig. S6I), the three-stranded back sheet is rotated and translated, thus causing the planes of the two NIE sheets to intersect at an angle of ∼70° on the right lateral face while the opposite lateral face opens. In addition, two helices are inserted in the segment-connecting strands β3 and β4 of domain NIE (NIE-β3→β4).
The CUB domain is a β-sandwich of two parallel four-stranded antiparallel β-sheets (I and II), which is unrelated to the MG fold (Fig. S6D). A short helix is inserted at CUB-β6→β7, as is domain TED at CUB-β3→β4. The TED domain, in turn, is a sixfold α/α-toroid made up by six α-hairpins that resides on the outer surface of CUB sheet I and whose central axis is rotated ∼45° away from the sheet planes of the CUB β-sandwich. The arrangement of the α-hairpins is clockwise when viewed from the entry surface of the toroid (Fig. S6C). The thioester segment is a 15-atom thiolactone ring composed of four residues: C1187–L1188–E1189–Q1190. It is located at the beginning of the first toroid helix, TED-α2, on the domain entry face, and compatible with an induced peptidase-bound inhibitor, the thioester bond is broken (Fig. 1C, Right). This segment is shielded by TED-α4→α5. However, although the side chain of C1187 is surrounded by the side chains of E1189, L1242, and W1243, Q1190 points to the bulk solvent, which is consistent with a disordered trypsin molecule bound to its side chain in the crystal structure.
The C-terminal domain of ECAM, RBD (name based on its structural similarity with hα2M RBD, see ref. 8), occupies a key position in iECAM and interacts with TED, CUB, and MG7 (Fig. 1C). In addition, it stabilizes the hook structure protruding from the MG-superhelix by interacting with MG2 and MG3. RBD has a complex topology (SI Appendix, Fig. S6 B and H) and consists of a central MG core expanded to a six-stranded front and a five-stranded back β-sheet, whose planes are rotated away by ∼40°, as in MG domains.
The BRD is inserted at MG6-β3→β4, spans 66 residues (S901–N966), and is folded irregularly. It plays an important role not only in triggering the conformational rearrangement when cleaved but also in the stability of nECAM. A mutant in which BRD had been replaced by three glycines (protein ECAMΔBRD; SI Appendix, Table S1) yielded properly folded protein but was completely digested under conditions that only produced stable induced protein for the wild-type. BRD is defined for S901–G938 (upstream of the cleavage site) and G949–N966 (downstream of the cleavage site) in the crystal structure as a result of trypsin cleavage after R946. The upstream segment of BRD is freely accessible: it lines part of the concave surface of the monomer and contains two helices. It interacts with MG2, the segment linking MG2 and MG3, MG6, and RBD. After the second helix, the BRD chain runs in extended conformation along the inner MG-superhelix surface, and between Q921 and G938, the polypeptide is trapped between MG5, L, NIE, MG4, and MG1, with BRD segment A926–I931 performing a β-ribbon interaction with MG1-β1. The last upstream-segment residue defined in the structure, G938, emerges on the lower left outer surface of the monomer (“U” in Fig. 1C). The downstream segment of BRD, in turn, is defined from G949 onward (“D” in Fig. 1C), at the interface between MG2 and CUB. It encompasses a short helix, BRD-α3, and runs upward, mainly interacting with MG2, MG7, CUB, RBD, and the MG7-CUB and CUB-RBD linkers before rejoining MG6.
Single-Particle Cryo-EM and Homology Modeling of Native ECAM.
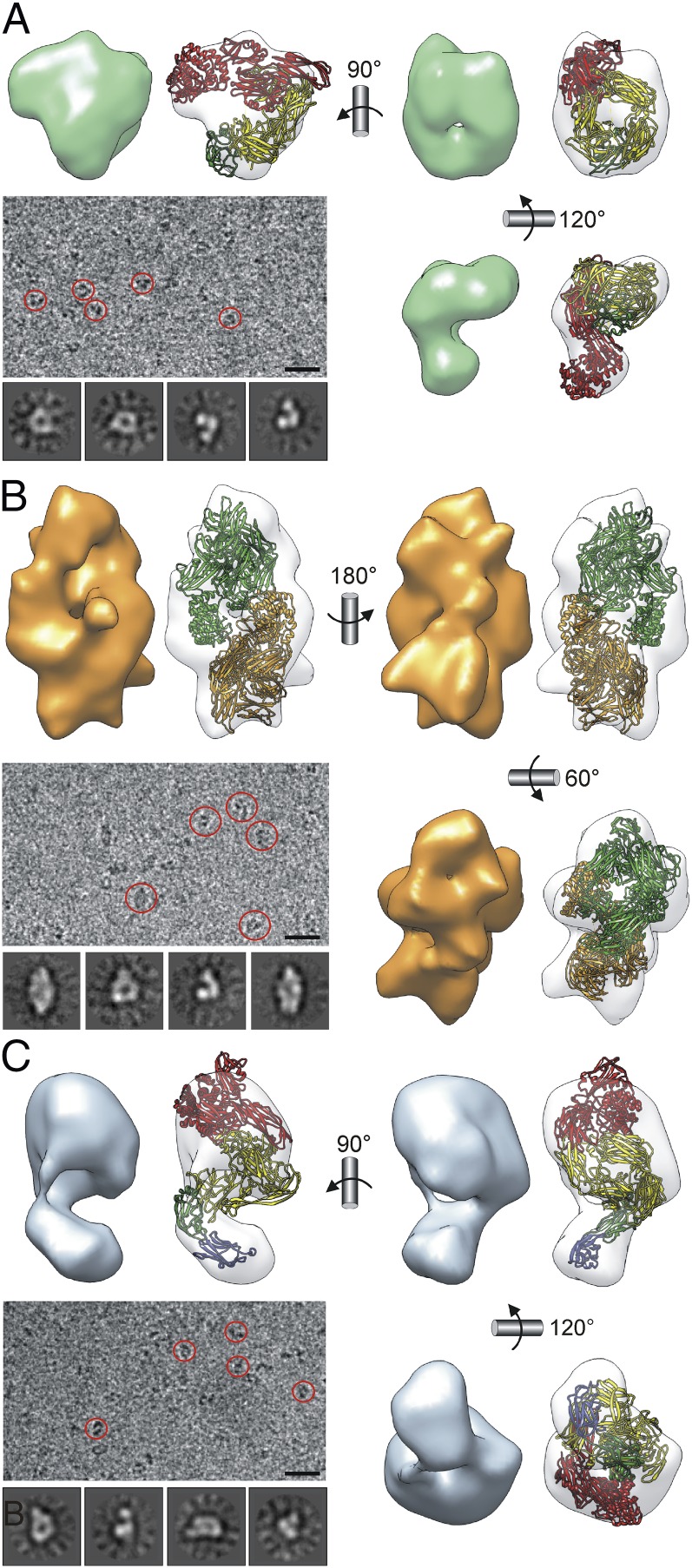
To complement the aforementioned crystal structure of iECAM, monomeric and dimeric trypsin-induced iECAM were further analyzed by 3D cryo-EM reconstructions of single particles (Fig. 2 A and B). Therefore, purified proteins were applied to carbon-coated grids, blotted, and plunged into liquid ethane. Images were recorded on a CCD camera at low-dose conditions, with a 200-kV electron microscope equipped with a field emission gun. Images were classified using a reference-free clustering approach to select homogeneous populations of 18,346 and 33,536 particles for monomeric iECAM and dimeric iECAM, respectively, which were used for reconstruction. The final resolution of the models was estimated to be, respectively, 17 and 14 Å (SI Appendix, Fig. S8). The crystallographic coordinates of monomeric and dimeric iECAM (SI Appendix, SRD §2.2 and Fig. S5) were adequately fitted into the corresponding cryo-EM maps, and the concordance of both structures is clear with the exception of the slight deviation from the C2 symmetry of the dimeric iECAM cryo-EM map (Fig. 2 A and B). Although dimerization surfaces are probably flexible in vivo, such dimerization does not lead to major conformational rearrangement of a monomer once induced. Fostered by the agreement between the cryo-EM and X-ray structures, we further obtained a cryo-EM reconstruction for nECAM to 16 Å on the basis of 46,842 particles (Fig. 2C), as crystallization of the full-length protein produced only poorly diffracting crystals. To get additional insight into the structure of nECAM at atomic level, we assayed several constructs and managed to solve the crystal structure of three of them, respectively spanning domains MG0-NIE-MG1, NIE-MG1, and MG7-CUB(TED)-RBD alias nECAMΔN.
Fig. 2.
ECAM single-particle cryo-EM reconstructions. (A) Monomeric iECAM. (B) Dimeric iECAM. (C) Monomeric nECAM. In each case, the cryo-EM map (solid surface) and the fitting of the respective atomic models (semitransparent surfaces) are shown in three views. Representative cryo-EM field views are also depicted (selected particles are encircled). (Scale bar, 300 Å.) In addition, four representative averaged classes are shown. Monomeric ribbon models are depicted in four colors (MG7-CUB[TED]-RBD in red, MG1–MG6 in yellow, NIE in green, and MG0 in blue). In the dimeric model, whole monomers are in green and orange. Left orientation of A and C as in Fig. 3D. Best fitting of dimeric iECAM was achieved by rotating one molecule 19° away from the C2-symmetric position.
The first structure was solved by multiwavelength anomalous dispersion (MAD)/multiple isomorphous replacement, including anomalous signal with data from a three-wavelength MAD experiment (peak, inflection point, and high-energy remote) performed with a selenomethionine-derivatized protein crystal and a dataset obtained from wild-type protein. The other two structures were solved by single isomorphous replacement including anomalous signal by measuring a selenomethionine derivative of the protein at the absorption peak wavelength and a dataset from wild-type protein to higher resolution (SI Appendix, Tables S3 and S4 and Fig. S9). According to these crystal structures and that of native SEAM (16), we constructed a composite homology model of nECAM for which its cryo-EM map at 16 Å resolution was used as a constraint (Fig. 2C). This model supported the major conformational rearrangement that occurs on induction, as anticipated by the biophysical studies presented earlier. The structure of native SEAM was essentially used to position the respective ECAM domains, with the exception of MG0 and NIE, and to confirm that the entire isolated four-domain structure of nECAMΔN was in a native conformation (SI Appendix, SEP §1.13).
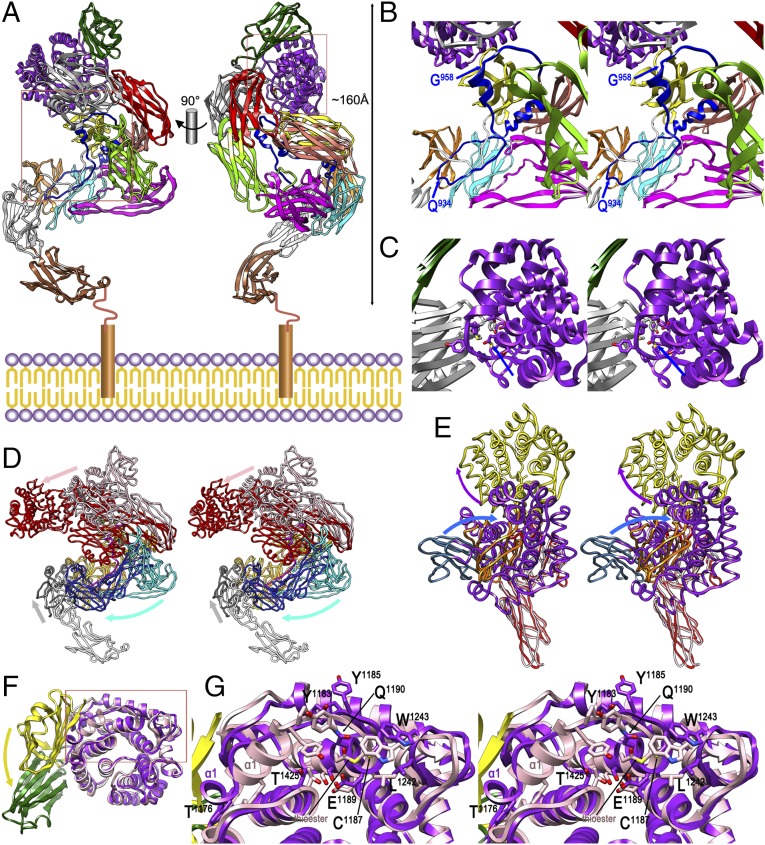
Similar to SEAM, the nECAM model (MG0–RBD; K57–P1653) reveals an elongated helicoidal structure of ∼160 Å maximal length (Fig. 3A). MG0, at the N terminus of ECAM and projecting away from NIE, probably faces the inner membrane in vivo and is flexibly linked with NIE, so it is easily removed after induction (see earlier text). nECAM contains a central MG-superhelix as in iECAM, which, however, is distorted and lacks a central “entrance 1” (SI Appendix, SRD §2.2). In addition, segments MG7-CUB(TED)-RBD and MG0 protrude, respectively, from opposite ends of the MG ellipsoid in opposite directions. According to this model, BRD would be flexible and line the inner surface of the superhelix, with three segments in helical conformation, as in iECAM (Fig. 3B). BRD would interact with linker L and domains MG1, MG2, and MG4-MG6, and the bait region, flexible and freely accessible for prey peptidases, would span segment Q934–G958. This is consistent with the cleavage sites detected for several model peptidases, including trypsin (after R946). Overall, given the shape of nECAM and the 40 missing residues at the N terminus leading to the membrane anchor, which are predicted to be flexible, this bait region could potentially cover up to 170–210 Å of the width of the periplasmic space above the inner membrane. As the total width of the periplasm in E. coli is ∼210 Å (21), ECAM would thus protect the entire periplasm, including the lipoproteins anchored to the periplasmic side of the outer membrane, against intruding endopeptidases (SI Appendix, SRD §2.1). In addition, the thioester region at the beginning of TED-α2 is buried in our nECAM model and faces the outer surface of the six-stranded front sheet of RBD (Fig. 3C). The thioester bond itself is intact, as revealed by the experimental nECAMΔN structure, and protected by residues from TED (T1425, E1189, T1191, L1242, W1243, Y1185, and Y1183) and, in particular, RBD-β3 and RBD-β6→β7 (Y1635, M1634, and L1546). This explains why an ECAM mutant lacking RBD (protein ECAMΔRBD; SI Appendix, Table S1) encoded a well-folded protein that nevertheless did not form a thioester bond. This indicated that RBD has a relevant functional role in thioester integrity. This role differs from metazoan α2Ms, in which RBD targets cell surface receptors before endocytosis (11).
Fig. 3.
Native ECAM and transition to induced ECAM. (A) Composite homology model of full-length nECAM anchored to the inner membrane in two orthogonal views, with domains colored as in Fig. 1A. (B) Close-up in cross-eye stereo of the left rectangle of A. The suggested limits of the (modeled) bait region within BRD, based on potential accessibility, are pinpointed by blue arrows. (C) Close-up in stereo of the right rectangle of A, showing the protected intact thioester (blue arrow; see also G). (D) Structures of nECAM (as in A) and iECAM in stereo after optimal superposition of MG1-L-MG2 and MG5-MG6 (both in orange for iECAM, in yellow for nECAM). Diverging segments are MG7-CUB(TED)-RBD (iECAM, red; nECAM, pink), MG3–MG4 (iECAM, dark blue; nECAM, cyan), and MG0-NIE [iECAM (NIE only), gray; nECAM, white]. Arrows pinpoint the overall displacements on induction of the three groups in the color of the respective nECAM segments. (E) Detail in stereo of the experimental crystal structures of nECAMΔN and iECAM showing MG7 and RBD (both in red for iECAM, in pink for nECAM), CUB (iECAM, orange; nECAM, blue), and TED (iECAM, yellow; nECAM, purple). Arrows pinpoint the overall relative displacements of CUB and TED on induction in the color of the respective nECAM domains. (F) Detail of CUB-TED of the nECAMΔN (yellow-pink) and iECAM (green-purple) crystal structures after optimal superposition of the TED domains only. The CUB domain undergoes a relative 90° rotation on induction (yellow arrow). (G) Close-up of the rectangle of F in stereo showing the thioester region. Some residues of iECAM are labeled for reference, as is the intact thioester bond of nECAM and helix TED-α1 for both structures.
Structure-Derived Snap-Trap Mechanism of Induction.
Comparison of iECAM and nECAM reveals the detailed mechanism of ECAM induction mediated by cleavage in the bait region (Movie S1). This process yields a more compact structure (Fig. 3D), which is consistent with higher electrophoretic mobility, similar to what happens with hα2M (18), and to the aforementioned differences in biophysical assays. Superposition shows that the structures only coincide on the bilayered side of the MG-superhelix (MG1-L-MG2 and MG5-MG6), and, partially, at BRD (up to Y932 and from H964 onward). On induction, MG3 and MG4 are flipped inward toward MG6 as a rigid body because of a ∼90° rotation around the anchor point of MG3 with MG2 and a concomitant translation downward of up to ∼50 Å (for MG4; see Fig. 3D for spatial orientation hereafter). The new position of MG4 forces NIE to be moved outward along the outer surface of the four-stranded sheet of MG1. This movement traps the segment of the bait region upstream of the cleavage site after a ∼180° rotation downward around G933 (see earlier text). The bait region is undefined from Q939 to G948 in iECAM, and the distance between the flanking residues is too great to be covered by the 10 missing residues (66 Å).
In contrast, in hα2M, the corresponding distance easily accommodates the missing residues (SI Appendix, SRD §2.3 and Fig. S10 for a detailed comparison between iECAM and hα2M). This explains why in ECAM, cleavage in the bait region must occur to yield the induced form, whereas in hα2M, the induced form is compatible with an intact bait region and thus can be obtained by MA treatment (8). The displacement of MG3 is also concomitant with MG7 and RBD becoming rotated as a rigid body by ∼25° downward, so RBD is displaced by ∼25Å toward newly positioned MG3. Rearrangement of MG7 and RBD also causes CUB and TED to move downward and outward, the former being rotated by ∼25° and displaced by ∼35 Å, and the latter becoming rotated and translated by ∼45 Å. When comparing these two domains only, CUB is rotated by 90° with respect to TED around the domain interface because of the presence of residues that favor such hinge motions, P1442–G1443 and P1169–P1170 (Fig. 3 E–G). Rotating away CUB causes loops TED-α3→α4 (G1210–D1216) and TED-β2→α5 (F1240–E1249) to be displaced to the right, which further causes TED-α1 and TED-α1→α2 (P1169–L1188) to undergo major rearrangement. In particular, TED-α1 (I1173–A1182) becomes unwound for its last six residues in iECAM. This causes displacement of segment Y1183–G1186, which acts as a protective lid of the thioester bond in nECAM. In this way, the thioester becomes exposed and solvent-accessible in iECAM, so it can be targeted by prey surface lysines (Fig. 3G). Most noteworthy, the initial movement of the mechanism, that of MG3 relative to MG2, is blocked in nECAM by the BRD segment after the bait region, which passes above the MG2–MG3 linker (Fig. 3B). On cleavage in the bait region, this constraint is released, and the segment downstream of the cleavage site becomes rotated by ∼50° around N963 toward and above MG2, and approaches the outer surface of CUB in its induced position.
Conclusions
Taken together, these results, together with the functional characterization in vitro and partially in vivo, indicate that ECAM works as an irreversible monomeric snap trap. This snap trap definitively differs from the tetrameric Venus flytrap of mammalian α2Ms. Monomeric nECAM represents the baited and set trap, with a spring-loaded bar (the hidden thioester) and a trip (BRD segment after the bait region) to release it. When the bait region is cleaved, induction occurs under large conformational rearrangement and exposure of a hidden thioester bond, which is analogous to setting off the trap through the rapid swing-down of the spring-loaded bar. However, only if the thioester bond is targeted by a surface lysine of the prey peptidase to yield a covalent bond is the prey trapped by the released bar. In any case, the trap would remain irreversibly inactivated, either with or without a trapped peptidase. In contrast to a true snap trap, however, the prey peptidase is not disabled by ECAM, but merely restricted in its radius of action and substrate size.
Materials and Methods
Wild-type ECAM and its variants were produced, purified, and assayed for activity following standard techniques. Proteins were studied for their 3D structure through X-ray crystallography and cryo-EM. A detailed description of the experimental procedures is provided in the SI Appendix. The latter also includes four supplementary tables, 10 supplementary figures, the Acknowledgments, and SI Appendix, SRD.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4ZIU, 4ZJH, 4ZJG, and 4ZIQ). The cryo-electron microscopy reconstructions of nECAM have been deposited with the EMDataBank (EMDataBank codes EMD-3016, EMD-3017, and EMD-3018).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506538112/-/DCSupplemental.
References
- 1.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Kantyka T, Rawlings ND, Potempa J. Prokaryote-derived protein inhibitors of peptidases: A sketchy occurrence and mostly unknown function. Biochimie. 2010;92(11):1644–1656. doi: 10.1016/j.biochi.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budd A, Blandin S, Levashina EA, Gibson TJ. Bacterial α2-macroglobulins: Colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 2004;5(6):R38. doi: 10.1186/gb-2004-5-6-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett AJ, Starkey PM. The interaction of α2-macroglobulin with proteinases. Characteristics and specificity of the reaction, and a hypothesis concerning its molecular mechanism. Biochem J. 1973;133(4):709–724. doi: 10.1042/bj1330709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sottrup-Jensen L, Sand O, Kristensen L, Fey GH. The α-macroglobulin bait region. Sequence diversity and localization of cleavage sites for proteinases in five mammalian α-macroglobulins. J Biol Chem. 1989;264(27):15781–15789. [PubMed] [Google Scholar]
- 7.Sottrup-Jensen L. α-macroglobulins: Structure, shape, and mechanism of proteinase complex formation. J Biol Chem. 1989;264(20):11539–11542. [PubMed] [Google Scholar]
- 8.Marrero A, et al. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew Chem Int Ed Engl. 2012;51(14):3340–3344. doi: 10.1002/anie.201108015. [DOI] [PubMed] [Google Scholar]
- 9.Bieth JG, Tourbez-Perrin M, Pochon F. Inhibition of α 2-macroglobulin-bound trypsin by soybean trypsin inhibitor. J Biol Chem. 1981;256(15):7954–7957. [PubMed] [Google Scholar]
- 10.Nagase H, Harris ED., Jr Ovostatin: A novel proteinase inhibitor from chicken egg white. II. Mechanism of inhibition studied with collagenase and thermolysin. J Biol Chem. 1983;258(12):7490–7498. [PubMed] [Google Scholar]
- 11.Strickland DK, et al. Sequence identity between the α2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J Biol Chem. 1990;265(29):17401–17404. [PubMed] [Google Scholar]
- 12.Feldman SR, Gonias SL, Pizzo SV. Model of α2-macroglobulin structure and function. Proc Natl Acad Sci USA. 1985;82(17):5700–5704. doi: 10.1073/pnas.82.17.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neves D, et al. Conformational states of a bacterial α2-macroglobulin resemble those of human complement C3. PLoS ONE. 2012;7(4):e35384. doi: 10.1371/journal.pone.0035384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan N, Gettins PGW. α-Macroglobulins are present in some gram-negative bacteria: Characterization of the α2-macroglobulin from Escherichia coli. J Biol Chem. 2008;283(42):28747–28756. doi: 10.1074/jbc.M803127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robert-Genthon M, et al. Unique features of a Pseudomonas aeruginosa α2-macroglobulin homolog. MBio. 2013;4(4):e00309–e00313. doi: 10.1128/mBio.00309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong SG, Dessen A. Structure of a bacterial α2-macroglobulin reveals mimicry of eukaryotic innate immunity. Nat Commun. 2014;5:4917. doi: 10.1038/ncomms5917. [DOI] [PubMed] [Google Scholar]
- 17.Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Mol Immunol. 2004;40(12):903–908. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Van Leuven F, Cassiman JJ, Van den Berghe H. Functional modifications of α 2-macroglobulin by primary amines. I. Characterization of α2 M after derivatization by methylamine and by factor XIII. J Biol Chem. 1981;256(17):9016–9022. [PubMed] [Google Scholar]
- 19.Barrett AJ, Brown MA, Sayers CA. The electrophoretically ‘slow’ and ‘fast’ forms of the α2-macroglobulin molecule. Biochem J. 1979;181(2):401–418. doi: 10.1042/bj1810401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Merwe JP, Mol GJ. Levels of trypsin and α-chymotrypsin in feces from patients with Crohn’s disease. Digestion. 1982;24(1):1–4. doi: 10.1159/000198767. [DOI] [PubMed] [Google Scholar]
- 21.Matias VRF, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185(20):6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.