Significance
Transcriptional regulation of the rDNA cluster is a tightly regulated process owing to the cellular demand for ribosomal subunits. Dysregulation of ribosomal biogenesis is associated with genomic instability and tumor progression. Nucleolar-remodeling complex (NoRC) has emerged as a key complex in the maintenance of rDNA repression. However, the mechanisms that regulate the association of NoRC to rDNA gene loci are yet to be understood. We unravel the role for a BANP, E5R, and Nac1 (BEN) domain-containing transcription repressor, BEND3, in regulating rDNA repression. BEND3 modulates the stability of a NoRC component, TTF-1–interacting protein 5 (Tip5), by controlling the USP21-mediated deubiquitination of Tip5. These results provide crucial insights into the role of a transcription factor in controlling rDNA gene expression by controlling the recruitment and activity of chromatin-modifying complex.
Keywords: BEND3, NoRC, Tip5, transcription repression, rDNA
Abstract
Ribosome biogenesis dictates the translational capacity of cells. Several mechanisms establish and maintain transcriptional output from eukaryotic ribosomal DNA (rDNA) loci. rDNA silencing is one such mechanism that ensures the inactivity and hence the maintenance of a silenced state of a subset of rRNA gene copies. Whereas oncogenic agents stimulate rRNA gene transcription, tumor suppressors decrease rRNA gene transcription. We demonstrate in mammalian cells that BANP, E5R, and Nac1 (BEN) domain 3 (BEND3), a quadruple BEN domain-containing protein, localizes in nucleoli and binds to ribosomal RNA gene promoters to help repress rRNA genes. Loss of BEND3 increases histone H3K4 trimethylation and, correspondingly, decreases rDNA promoter DNA methylation, consistent with a role for BEND3 in rDNA silencing. BEND3 associates with the nucleolar-remodeling complex (NoRC), and SUMOylated BEND3 stabilizes NoRC component TTF-1–interacting protein 5 via association with ubiquitin specific protease 21 (USP21) debiquitinase. Our results provide mechanistic insights into how the novel rDNA transcription repressor BEND3 acts together with NoRC to actively coordinate the establishment of rDNA silencing.
Transcriptional regulation of ribosomal RNA (rRNA) genes is crucial for normal cell growth and proliferation because it dictates ribosome biogenesis in eukaryotic cells (1). rRNA synthesis and processing occurs in nucleoli, nuclear compartments that form at clusters of repetitive rDNA in which rRNA genes are found in tandem arrays. rRNA genes encode a precursor transcript (45S prerRNA) that is processed and modified to yield 18S, 5.8S, and 28S rRNA. However, more than half of the rDNA repeats are rendered transcriptionally silent by epigenetic mechanisms (2–6). The nucleolar remodeling complex (NoRC), a member of the ISWI family of the ATP-dependent chromatin-remodeling complex consisting of TTF-1–interacting protein 5 (Tip5) and sucrose nonfermenting 2 homolog (Snf2h), contributes to the silent state of rRNA genes (7, 8). NoRC targets histone-modifying enzymes and DNA methyltransferase to rDNA to establish a heterochromatic state that inhibits transcription activation (7, 9, 10).
BANP, E5R, and Nac1 domain 3 (BEND3) is a quadruple BEN domain-containing protein that associates with heterochromatin, and its overexpression causes extensive heterochromatinization (11). Using an artificial in vivo gene locus reporter assay, we previously demonstrated that BEND3 can efficiently repress transcription. However, the role of BEND3 in the physiological context, and the endogenous target genes whose expression is modulated by BEND3, remained to be determined.
The BEN domain is an α-helical module found in many proteins in metazoans including BANP/SMAR1, NAC1, BEND3, and BEND5 and is also present in several viral genes (12). The BEN domain is a conserved DNA-binding domain, and BEN domain-containing transcription factors mediate chromatin organization and transcription (11, 13). Several of the BEN-superfamily proteins are known transcriptional repressors, including SMAR1, NAC1, and BEND3, -5, and -6 (11, 13, 14). BEND3 is a key factor that mediates the switch from constitutive to facultative heterochromatin in embryonic stem cells (15). BEND6 acts as a corepressor of notch transcription factor (16). BEND5 has been found to express in neurons and function as a sequence-specific transcription repressor that regulates neurogenesis (17).
In this paper, we demonstrate that BEND3 localizes to nucleoli, binds to rDNA promoter elements, and interacts with NoRC. Our results show that BEND3 contributes to silencing of rDNA transcription and that the cooperativity between SUMOylated BEND3 and NoRC is essential for repression of rRNA gene transcription. Furthermore, BEND3 stabilizes members of the NoRC via its association with the ubiquitin specific protease 21 (USP21) deubiquitinase.
Results
BEND3 Localizes to Nucleoli and Associates with rDNA Promoters.
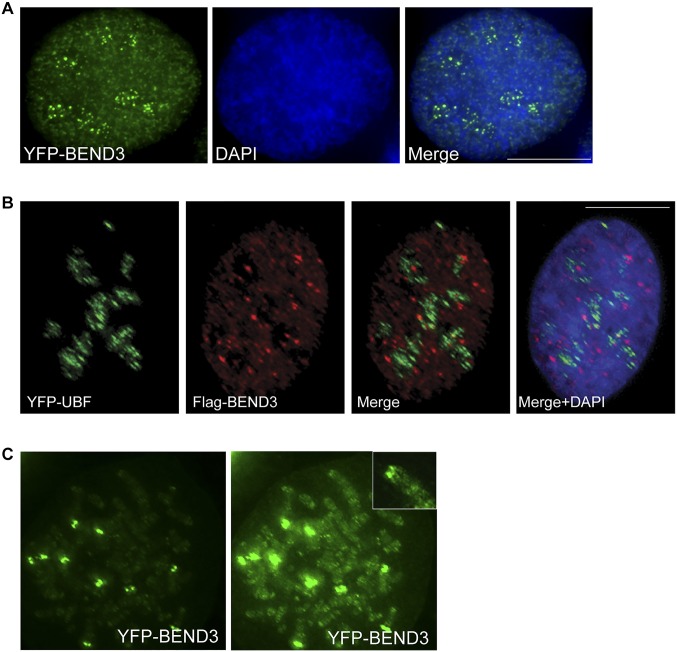
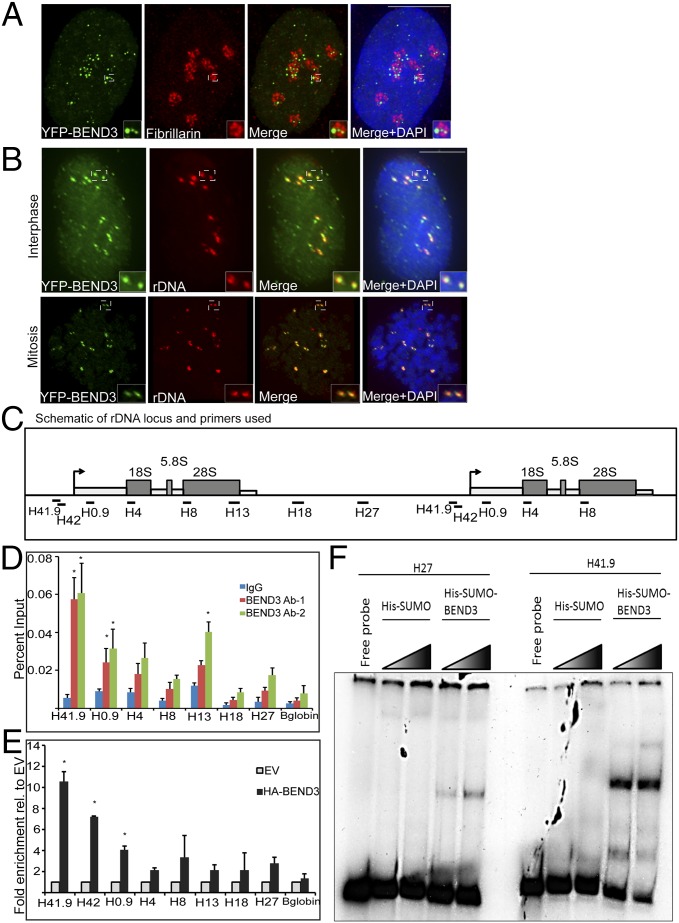
BEND3 is a nuclear protein that associates with the HP1-containing heterochromatin loci (11). Examination of the localization of BEND3 in human cells showed multiple punctate foci of YFP-BEND3 appearing closer to or within DAPI-less regions (Fig. S1A). Immunolocalization of the nucleolar marker fibrillarin and UBF confirmed the distribution of several punctate foci of BEND3 within the nucleoli during interphase (Fig. 1A and Fig. S1B). BEND3 also associates with telomeric heterochromatic regions during mitosis (Fig. S1C). The rDNA clusters are distributed close to telomeres in the acrocentric chromosomes 13, 14, 15, 21, and 22 (6). Ribosomal DNA FISH using a probe complementary to the rDNA repeat unit in cells expressing YFP-BEND3 revealed that YFP-BEND3 colocalized with most of the rDNA foci during interphase as well as mitosis (Fig. 1B). Furthermore, chromatin immunoprecipitation (ChIP) experiments using two different BEND3 antibodies (BEND3 Ab-1 and Ab-2) confirmed the association of BEND3 at the rDNA promoter (as seen with primer set H41.9), with a gradual decrease in enrichment across the transcribed and intergenic spacer (IGS) regions (Fig. 1 C and D and Table S1). Similar to the endogenous BEND3, we found that exogenously expressed HA-BEND3 associated primarily at the rDNA promoter as evidenced by its 7- to 10-fold enrichment relative to the empty vector when H41.9 or H42 primer sets were used (Fig. 1E).
Fig. S1.
BEND3 localizes to the nucleoli. (A) U2OS cells transfected with YFP-BEND3 (green). (B) YFP-UBF and FLAG-BEND3 distribution in human U2OS cells. (Scale bar: 10 µm.) (C) YFP-BEND3 distribution on mitotic chromosomes. Note the localization of BEND3 at telomeric sites of five pairs of acrocentric chromosomes, reminiscent of the rDNA sites on chromosomes 13, 14, 15, 21, and 22. Higher exposure shows weak signal of BEND3 at other telomeric sites. (Inset) Highly overexposed image.
Fig. 1.
BEND3 localizes to rDNA. (A) Immunostaining of the nucleolar marker Fibrillarin (red) in YFP-BEND3 transfected cells. (B) rDNA FISH analysis (red) in YFP-BEND3 transfected U2OS cells during interphase and mitosis. DNA is counterstained with DAPI. (Scale bar: 10 µm.) (C) A schematic of 1.5 units of the human rDNA repeat and primers used for ChIP analysis. H41.9 and H42 are at the promoter; H4, H8, and H13 span the coding region whereas H18 and H27 are at the IGS region. (D) ChIP analysis showing BEND3 occupancy at the rDNA locus. Results are plotted as percentage of input. (E) ChIP analysis showing HA-BEND3 occupancy at the rDNA locus. HA-ChIP was performed in cells stably expressing either an empty vector (EV) or HA-tagged BEND3. Results are plotted as percentage of input values normalized to EV control. Error bars represent SD; n = 3. *P value < 0.05. (F) Mobility shift assay showing His-SUMO-BEND3 binding to rDNA promoter (H41.9) and IGS (H27) sequences in vitro.
Table S1.
Primers
| Name | Forward | Reverse | Coordinate |
| H41.9 | CCGTGGGTTGTCTTCTGACT | AAGCGAAACCGTGAGTCG | 41907–42035 |
| H 0.9 | AACGGTGGTGTGTCGTTC | TCTCGTCTCGTCTCACTCAA | 852–977 |
| H4 | CGACGACCCATTCGAACGTCT | CTCTCCGGAATCGAACCCTGA | 3990–4092 |
| H8 | AGTCGGGTTGCTTGGGAATGC | CCCTTACGGTACTTGTTGACT | 8204–8300 |
| H13 | ACCTGGCGCTAAACCATTCGT | GGACAAACCCTTGTGTCGAGG | 12855–12970 |
| H18 | GTTGACGTACAGGGTGGACTG | GGAAGTTGTCTTCACGCCTGA | 18155–18280 |
| H27 | CCTTCCACGAGAGTGAGAAGCG | CTCGACCTCCCGAAATCGTACA | 27366–27478 |
| H42 | AGAGGGGCTGCGTTTTCGGCC | CGAGACAGATCCGGCTGGCAG | 41982–42075 |
| Meth | GTATATCTTTCGCTCCGAGTCG | ACAGGTCGCCAGAGGACAG | −47 to +28 |
| 45S | GCCTTCTCTAGCGATCTGAGAG | CCATAACGGAGGCAGAGACA | 1406–1487 |
| 45S-1 | GAACGGTGGTGTGTCGTT | GCGTCTCGTCTCGTCTCACT | 851–980 |
| 45S-2 | CTCCGTTATGGTAGCGCTGC | GCGGAACCCTCGCTTCTC | 1477–1572 |
| 47S | GTCAGGCGTTCTCGTCTC | GCACGACGTCACCACAT | 308–442 |
| c-Myc | CCACAGCAAACCTCCTCACAG | GCAGGATAGTCCTTCCGAGTG | |
| BEND3 | Taqman assay (Invitrogen) | Assay no. Hs01028508_s1 |
Next, we tested if BEND3 could bind rDNA directly. To address this, we performed mobility shift assays using bacterially expressed His-SUMO-BEND3. To test if BEND3 recognizes the rDNA promoter sequence-specifically, we used the same primer sets from our ChIP analysis to generate probes corresponding to various rDNA sequences. As shown in Fig. 1F, incubation of rDNA promoter probe (H41.9) showed a dramatic shift in mobility, indicating strong BEND3 binding to rDNA promoter sequences in samples from His-SUMO-BEND3 lysate but not His-SUMO lysate. BEND3 displayed a weak shift when a probe from an IGS region (H27) was used compared with the promoter region. These data strongly corroborate our ChIP results showing that BEND3 binds to the rDNA promoter region in a sequence-dependent manner. Taken together, our data clearly demonstrate that BEND3 localizes to nucleoli and binds to rDNA promoters with high sequence specificity.
BEND3 Represses rDNA Transcription.
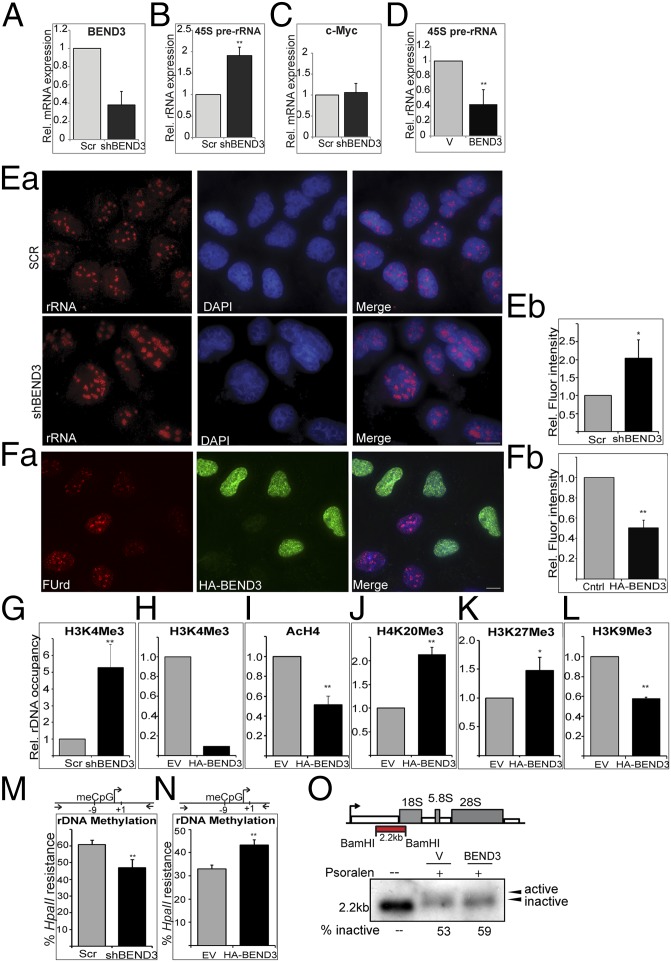
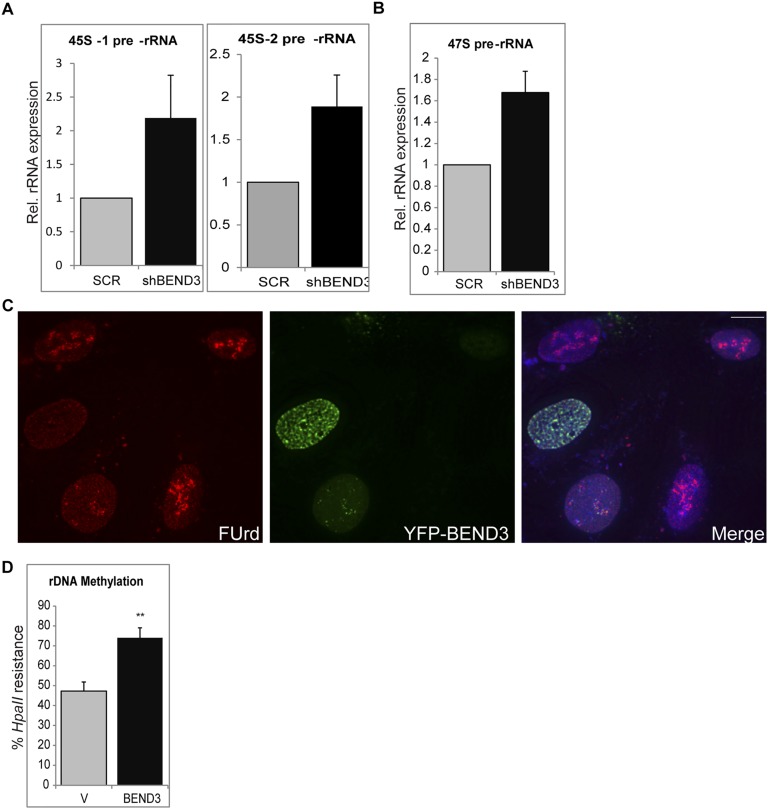
Association of BEND3 with rDNA promoter sequences raises the possibility that BEND3 regulates transcription of rDNA. We generated cell lines stably expressing an shRNA construct against BEND3 (shBEND3) or a Scramble (Scr) shRNA as a control (Fig. 2A). Knockdown of BEND3 resulted in a significant increase in the abundance of the 45S prerRNA transcript as well as the unprocessed 47S prerRNA, suggesting that BEND3 is a repressor of rDNA transcription (Fig. 2B and Fig. S2 A and B). The levels of c-Myc transcript (shown as control) did not change upon BEND3 knockdown (Fig. 2C), indicating that BEND3 specifically regulates rDNA transcription. We also observed that cells exogenously expressing BEND3 showed a marked decrease in the pre-rRNA levels compared with the control cells (Fig. 2D). These results were corroborated with results obtained through RNA-FISH where the level of rRNA signals in BEND3-shRNA–treated cells was markedly increased compared with control cells (Fig. 2 Ea and Eb).
Fig. 2.
BEND3 represses rDNA transcription. (A) Validation of BEND3 knockdown by qRT-PCR using a highly specific probe-based Taqman Gene Expression Assay. Note greater than 60% reduction in BEND3 mRNA levels in cells stably expressing an shRNA against human BEND3 (shBEND3). (B) qRT-PCR analysis of 45S pre-rRNA transcript in control (Scr) and shBEND3-expressing cells. c-Myc is used a negative control in C. (D) Relative expression of 45S pre-rRNA transcript in cells transfected with vector (V) or HA-BEND3 (BEND3). Error bars represent SD; n = 3. **P value < 0.01. (Ea) RNA FISH analysis using a probe complementary to rRNA in U2OS cells stably expressing scrambled (Scr) shRNA or shRNA against BEND3 (shBEND3). (Eb) Quantification of the data from Ea. (Fa) Fluorouridine labeling in HA-BEND3–expressing cells. (Fb) Quantification of the data from Fa. (G) ChIP analysis using H3K4me3 Ab at the rDNA promoter in shBEND3-expressing cells and Scr control. (H) H3K4Me3 Ab ChIP in cells stably expressing HA-BEND3 relative to EV control (data are representative of two independent experiments). (I) ChIP using antibody against pan H4 acetylation (AcH4) at rDNA promoters in cells stably expressing HA-BEND3 compared with empty vector (EV) control. (J) H4K20me3 Ab. (K) H3K27me3 Ab. (L) H3K9me3 Ab ChIP in HA-BEND3 or EV stable cells. (M and N) Methylation-sensitive restriction analysis used to measure rDNA promoter methylation levels in BEND3-depleted cells (M) or in HA-BEND3–expressing cells (N). meCpG levels were measured by digestion with HpaII followed by qPCR of the indicated promoter region (schematic). Error bars represent SD; n = 3. *P < 0.05, **P value < 0.01. (O) Active and inactive gene fraction by psoralen cross-linking in vector or BEND3-overexpressing cells.
Fig. S2.
(A) 45S prerRNA levels in BEND3-depleted cells using two different primer sets. (B) 47S prerRNA levels in BEND3-depleted cells. (C). Fluorouridine labeling in YFP-BEND3–expressing cells. (D) rDNA promoter methylation in HEK 293T cells transfected with YFP-BEND3.
We investigated if BEND3 modulates rRNA levels by affecting rRNA synthesis or processing. We employed a metabolic labeling approach using Fluorouridine (FUrd) in vivo pulse labeling. FUrd is incorporated into nascent RNA and can then be visualized using a BrdU antibody. As seen in Fig. 2F, cells that express HA-BEND3 (green) showed a dramatic decrease in nucleolar FUrd signal (red), suggesting a decreased rate of rRNA synthesis in these cells compared with neighboring cells that did not express HA-BEND3. Similar results were also observed in cells transfected with YFP-BEND3 (Fig. S2C), suggesting that BEND3 can efficiently reduce rRNA synthesis/transcription.
Because we observed a significant increase in the expression of pre-rRNA in BEND3-depleted cells, we examined the status of various chromatin modifications at rDNA loci. ChIP revealed that BEND3-depleted cells showed significant increase in the H3K4 trimethylation levels at the rDNA promoter, suggesting that transcription at rDNA loci was elevated in the absence of BEND3 (Fig. 2G). In contrast, cells expressing HA-BEND3 showed a dramatic decrease in H3K4 trimethylation and H4 acetylation, as well as an increase in repressive chromatin marks such as H4K20 trimethylation and H3K27 trimethylation. Interestingly, we observed a decrease in H3K9 trimethylation levels in BEND3-expressing cells (Fig. 2 H–L). Our results suggest that BEND3 levels influence the chromatin state at rDNA loci by modulating histone posttranslational modifications, thereby influencing rRNA transcription.
Silencing at rDNA loci has been closely linked to methylation of rDNA promoters, and this has been implicated in impaired assembly of transcription machinery at the rDNA promoter (18). We examined whether BEND3 is required for the maintenance of CpG methylation at rDNA promoters. The extent of CpG methylation at rDNA promoters was assessed by the enzyme HpaII that cleaves only at unmethylated sites. Depletion of BEND3 decreased rDNA promoter methylation, consistent with the increased rDNA transcriptional output observed under these conditions (Fig. 2M). In contrast, overexpression of BEND3 caused an increase in promoter DNA methylation in multiple cell lines (U2OS, Fig. 2N; 293T, Fig. S2D), indicative of its role in establishing de novo DNA methylation. Altogether, our results conclusively demonstrate that BEND3 is a transcriptional repressor that modulates the chromatin state to induce and maintain transcriptional repression at the rDNA locus.
Next, we used the psoralen cross-linking assay to determine the status of transcribed/transcribable active versus inactive rRNA genes in BEND3 overexpressed cells (19). Following psoralen cross-linking, active and inactive rRNA genes can be distinguished by Southern hybridization owing to their differential mobility on agarose gels. We observed that overexpression of BEND3 increased the fraction of inactive rRNA genes (Fig. 2O), further supporting our observations that BEND3 plays a key role in repressing rDNA transcription.
BEND3 Associates with NoRC to Establish rDNA Silencing.
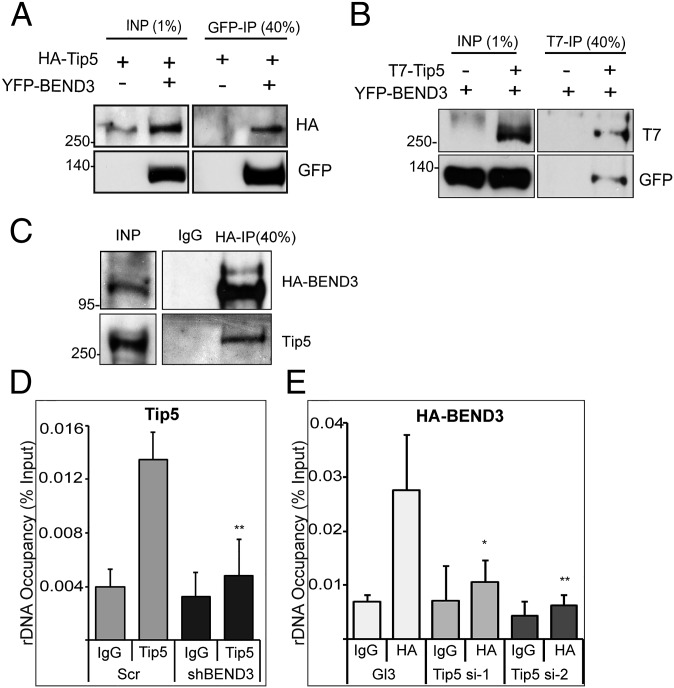
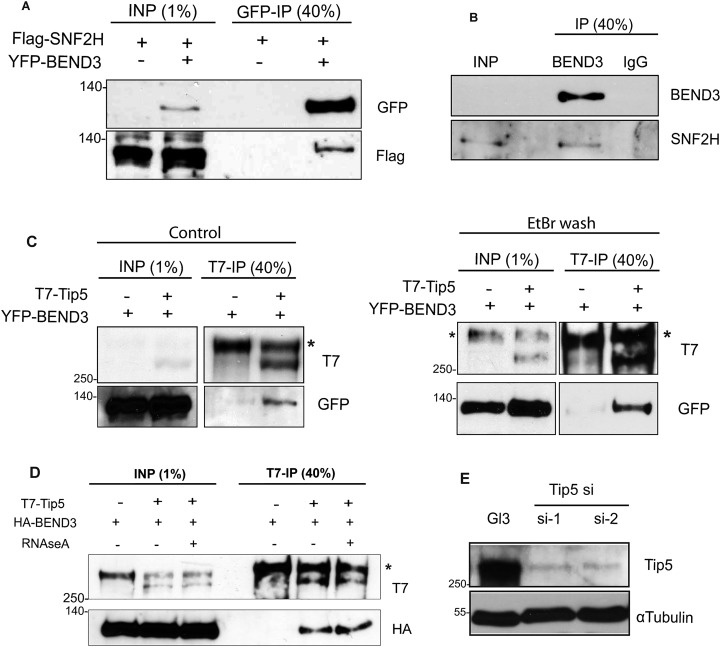
NoRC, consisting of Tip5 and Snf2h, mediates silencing of rDNA loci by altering the chromatin structure at rDNA promoters (7–10). Because both BEND3 and NoRC alter chromatin structure, and are required for rDNA silencing, we examined whether BEND3 associates with NoRC. Immunoprecipitation (IP) carried out in lysates from cells cotransfected with YFP-BEND3 and HA-Tip5 showed interaction between Tip5 and BEND3 (Fig. 3A). Similarly, reciprocal IP using T7 antibody in cells expressing YFP-BEND3 and T7-Tip5 confirmed interaction between BEND3 and Tip5 (Fig. 3B). IP carried out in lysates from cells transiently transfected with Flag-Snf2h along with YFP-BEND3 demonstrated the interaction of BEND3 with Snf2h (Fig. S3A). We further confirmed the association of BEND3 with endogenous Tip5 in U2OS cell lines stably expressing HA-BEND3 (Fig. 3C). Furthermore, IP from human U2OS nuclear extracts using BEND3 antibody showed a robust interaction of BEND3 with endogenous Snf2h (Fig. S3B). Because NoRC-associated RNA (pRNA) is crucial for targeting NoRC to chromatin, and is also required for rDNA silencing (20), we next examined if the interaction of BEND3 with NoRC is dependent on RNA. We cotransfected cells with YFP-BEND3 and T7-Tip5 and examined the dependence of their interaction on RNA by conducting ethidium bromide (EtBr) washes, a treatment that destabilizes protein–nucleic acid associations by intercalating with DNA or RNA (21). We did not observe any change in the interaction between Tip5 and BEND3 under these conditions (Fig. S3C). Similarly, RNaseA treatment did not affect the association of Tip5 and BEND3, suggesting that this complex does not require an RNA component for its stability (Fig. S3D).
Fig. 3.
BEND3 associates with NoRC. (A) BEND3 interacts with Tip5. Immunoprecipitation was performed using GFP antibody in cells expressing HA-Tip5 in the presence or absence of YFP-BEND3. (B) Reciprocal immunoprecipitation using T7 antibody was performed in cells expressing YFP-BEND3 with or without T7-Tip5. (C) Immunoprecipitation using HA antibody in cells stably expressing HA-BEND3. Note that BEND3 associates with endogenous NoRC (Tip5). (D) Tip5 occupancy at rDNA promoters in control (Scr) and BEND3-depleted cells (shBEND3). (E) HA-BEND3 occupancy at rDNA promoters in control (Gl3) and Tip5-depleted cells (si-1, si-2). ChIP data are represented as percentage of input. Error bars represent SD; n = 3. *P < 0.05, **P value < 0.01.
Fig. S3.
(A) BEND3 interacts with SNF2H. Immunoprecipitation using GFP antibody in cells expressing YFP-BEND3 with or without Flag-SNF2H shows SNF2H being co-immunoprecipitated along with BEND3 as seen in the immunoblots using antibodies against Flag. (B) Immunoprecipitation of endogenous BEND3 in U2OS cells co-immunoprecipitates endogenous SNF2H as detected in the immunoblots using antibodies against BEND3 or SNF2H. (C and D) BEND3-Tip5 interaction is not RNA-dependent. BEND3-Tip5 co-immunoprecition in the presence of EtBr (C) or RNase (D). (E) Depletion of Tip5 using two different siRNA oligonucleotides.
Finally, we examined if BEND3 and NoRC show any functional cooperativity in vivo to establish rDNA silencing. Because BEND3 and NoRC were observed in one complex, we analyzed if BEND3 dictates the binding of NoRC to rDNA promoters or vice versa. A significant reduction in the amount of Tip5 binding to rDNA was observed upon BEND3 depletion (Fig. 3D). Similarly, BEND3 ChIP in cells lacking Tip5 showed reduction in the association of BEND3 at the rDNA promoters (Fig. 3E and Fig. S3E). These results demonstrate that BEND3 and NoRC cooperate to establish rDNA silencing. Similar cooperativity among NoRC components has previously been reported (21).
SUMOylated BEND3 Stabilizes NoRC.
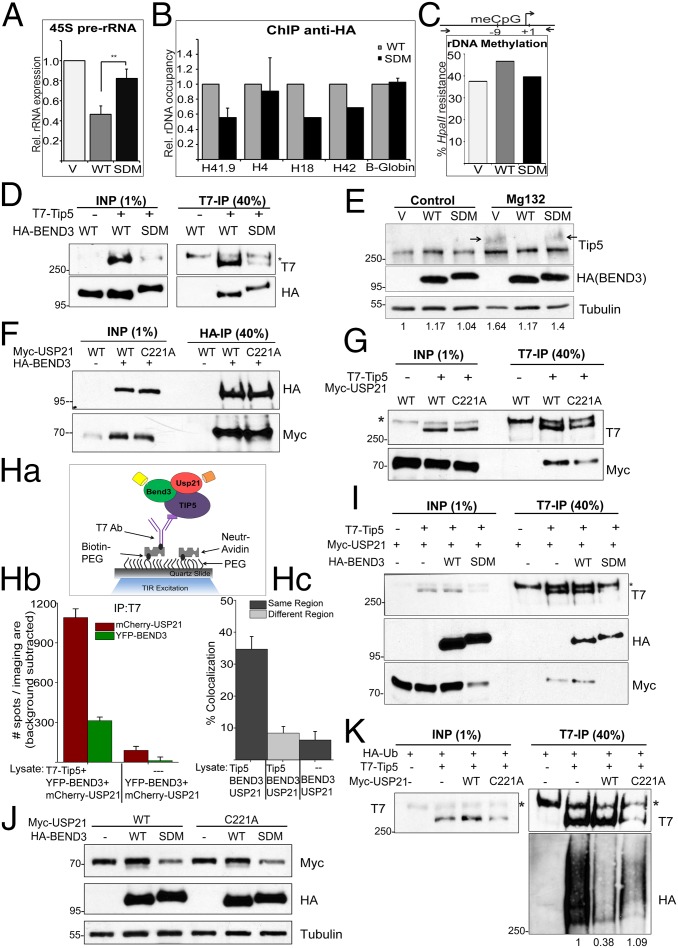
BEND3 is SUMOylated at K20 and K512 sites, and these two sites are crucial for its role in mediating transcription repression at a reporter locus (11). We generated a SUMO double mutant (SDM), BEND3.K20R;K512R and addressed if SUMOylation of BEND3 is required for rDNA silencing in vivo. Overexpression of BEND3.SDM somewhat relieved the rDNA repression and the levels of 45S pre-rRNA reverted close to the levels observed in cells that were transfected with the empty vector (Fig. 4A). ChIP in cells expressing HA-BEND3.WT or HA.BEND3.SDM revealed that there was a reduction in the association of SDM mutant to the rDNA locus, especially to the promoter regions (Fig. 4B). Consistent with a relief of rDNA repression in BEND3.SDM-expressing cells, we also found reduced rDNA methylation at rDNA promoters in these cells compared with cells expressing BEND3.WT (Fig. 4C). These results indicate that loss of rDNA silencing in BEND3.SDM-expressing cells could be due to the inability of BEND3.SDM to associate with the rDNA locus.
Fig. 4.
BEND3 stabilizes Tip5 via USP21 deubiquitinase. (A) Relative levels of 45S pre-rRNA transcript in cells transfected with pCGN (V) or HA-BEND3-WT (WT) or HA-BEND3 sumo double mutant (SDM) as assayed by qRT-PCR analysis. Error bars represent SD; n = 3. **P value < 0.01. (B) ChIP anti-HA in cells transfected with HA-BEND3.WT (WT) or HA-BEND3.SDM (SDM). Data are represented as percentage input normalized to WT. (C) rDNA promoter methylation levels assayed by methylation-sensitive restriction analysis in cells transfected with pCGN (V) or BEND3.WT or BEND3.SDM. (Data are representative of two independent experiments.) (D) Immunoprecipitation of T7-Tip5 with HA-BEND3.WT or HA-BEND3.SDM using T7 Ab. (E) Levels of Tip5 in cells expressing pCGN (V) or HA-BEND3 or HA-BEND3.SDM in control and upon MG132 treatment. Arrow denotes accumulation of ubiquitinated forms of Tip5. Relative intensity of Tip5 (as quantified by Image J) is shown at the bottom. Values are normalized to V in control cells. (F) BEND3 associates with USP21 deubiquitinase. Immunoprecipitation of HA-BEND3 and Myc-USP21 or USP21.C221A mutant using HA antibody. (G) Tip5 associates with USP21. Immunoprecipitation of T7-Tip5 and Myc-USP21 using T7 antibody. (Ha–Hc) Determination of Tip5 complexes containing both BEND3 and USP21 by SiMPull and colocalization analyses. (Ha) Schematic of YFP and mCherry molecules pulled down from U2OS cell lysates expressing T7-Tip5, YFP-BEND3, and mCherry-USP21 using biotinylated T7 Ab. Cell lysate expressing YFP-BEND3 and mCherry-USP21 incubated with biotinylated T7 Ab served as the control. (Hb) Average number of YFP and mCherry fluorescent molecules per imaging area (5,000 µm2). (Hc) Note 35 ± 4% overlap. (I) Immunoprecipitation from cells expressing T7-Tip5, Myc-USP21, and HA-BEND3 or HA-BEND3.SDM using T7 Ab. (J) SUMOylated BEND3 stabilizes USP21. Total levels of Myc-USP21 or Myc-USP21.C221A in cells expressing HA-BEND3 or HA-BEND3.SDM. (K) Ubiquitination assay in cells expressing HA-Ub, T7-Tip5, and Myc-USP21 or Myc-USP21.C221A. The Tip5 ubiquitination levels shown at the bottom of the gel were quantitated using ImageJ and normalized to the respective Input and IP levels.
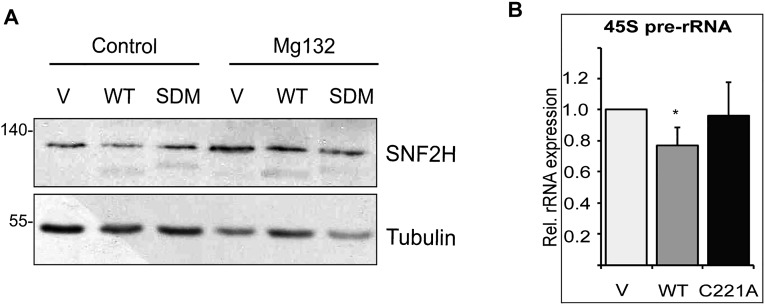
Next, we first asked if BEND3.SDM associates with the NoRC complex. IP of BEND3.WT or BEND3.SDM with Tip5 showed that both the WT and mutant BEND3 could efficiently associate with Tip5 (Fig. 4D). Interestingly, total levels of exogenous T7-Tip5 were consistently and dramatically reduced/destabilized in the presence of HA.BEND3.SDM, but not in the presence of HA.BEND3.WT (Fig. 4D). Similarly, we observed that cells expressing BEND3.WT showed marginally and reproducibly elevated levels of endogenous Tip5 protein (Fig. 4E). Based on these observations, we hypothesized that the SUMOylated BEND3 associates with Tip5 and in turn stabilizes Tip5, perhaps by preventing Tip5 degradation. To test this, we treated cells expressing vector control, BEND3.WT, or BEND3.SDM with MG132 to prevent proteasomal degradation of ubiquitinated proteins. We observed stabilization of Tip5 in MG132-treated vector as well as BEND3.SDM-expressing cells, compared with the untreated cells, suggesting that Tip5 is ubiquitinated (Fig. 4E; note the arrow). However, MG132-treated cells expressing BEND3.WT did not stabilize Tip5. Furthermore, there was no evidence of higher-molecular-weight forms of Tip5 (arrow) in these samples, suggesting that BEND3.WT by interacting with Tip5 prevents Tip5 ubiquitination. However, BEND3 expression did not alter the levels of Snf2h in cells (Fig. S4A).
Fig. S4.
(A) Relative levels of SNF2H in cells expressing HA-BEND3 or HA-BEND3.SDM. (B) 45S prerRNA levels in cells expressing Vector or USP21 or USP21.C221A.
To better understand how SUMOylated-BEND3 stabilizes Tip5, whether BEND3 prevents ubiquitination of Tip5, and how the association of BEND3 to Tip5 facilitates the rapid deubiquitination of Tip5, we examined the STRING association network for BEND3 (string-db.org/newstring_cgi/show_input_page.pl?UserId=_lyRGtIZs8Pz&sessionId=u8tmVQ4USCAv). We found that BEND3 is known to associate with the deubiquitinase USP21, a known H2A deubiquitinase (22). IP results reveal that both BEND3 and Tip5 interact with the WT as well as the catalytic null mutant (C221A) of USP21, suggesting that this interaction is independent of USP21 catalytic activity (Fig. 4 F and G).
Next, we investigated whether BEND3, Tip5, and USP21 exist in a single complex by performing Single Molecule Pull down (SiMPull) (23, 24) using biotin-conjugated anti-T7 antibody with lysates from cells triply transfected with YFP-BEND3, mCherry-USP21, and T7-Tip5. SimPull results revealed that 35 ± 4% of YFP-BEND3 molecules were found to colocalize with mCherry-USP21 molecules, indicating that in cells a subset of Tip5, BEND3, and USP21 exists in a single complex (Fig. 4H).
Interestingly, expression of BEND3.SDM caused destabilization of both WT-USP21 and USP21.C221A mutant (Fig. 4 I and J). Furthermore, as mentioned above, expression of BEND3.SDM caused the levels of USP21 to drop significantly with a concomitant decrease in the levels of Tip5 (Fig. 4 I and J). To test directly whether USP21 deubiquitinates Tip5 and thereby stabilizes Tip5, we assayed the extent of Tip5 ubiquitination by performing T7 immunoprecipitation in cells expressing HA-Ub and T7-Tip5 with or without Myc.USP21 (WT or C221A mutant). As shown in Fig. 4K, in the absence of USP21, T7-Tip5 was found to be highly ubiquitinated. However, in cells expressing Myc-USP21.WT there was a clear reduction in the levels of Tip5 ubiquitination with concomitant stabilization of Tip5 levels (Fig. 4K). Interestingly, in the presence of the catalytic mutant USP21.C221A, the ubiquitination levels of T7-Tip5 were restored, suggesting that USP21 specifically deubiquitinates and thereby stabilizes Tip5 (Fig. 4K). Consistent with this, we observed that rRNA levels decreased in cells expressing USP21 but not USP21.C221A (Fig. S4B). Based on these results, we propose that SUMOylated BEND3 stabilizes USP21 in this complex, which is then responsible for deubiquitinating Tip5, preventing Tip5 degradation (Fig. 5). In the absence of SUMOylated BEND3, USP21 levels plummet, and Tip5 is now poly-ubiquitinated and degraded, causing relief of transcription repression. We propose that BEND3 associates with and stabilizes the NoRC complex and that these activities together are required to establish rDNA silencing.
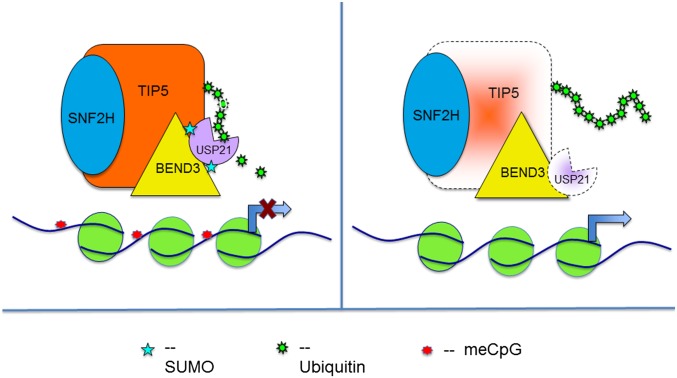
Fig. 5.
Cartoon demonstrating the role of SUMOylated BEND3 in stabilizing USP21 and hence Tip5 levels.
Discussion
Chromatin architecture dictates gene expression, and the accurate regulation of transcription is crucial for genomic stability (25). Heterochromatin represents a major repressive chromatin domain in the cell nucleus and typically consists of repetitive elements (26, 27). In human cells, heterochromatic regions are located predominantly at centromeres, telomeres, and at rDNA loci. We previously demonstrated that BEND3 associated with heterochromatic regions in mouse cells and repressed transcription at an in vivo reporter locus (11). However, the role of BEND3 in vivo and its potential repressive nature in the endogenous context remained to be addressed.
We demonstrate that a fraction of BEND3 localizes to nucleoli, decorates the rDNA cluster, and binds to the rDNA promoter. Furthermore, BEND3-overexpressed cells showed reduced H3K4Me3 and acetylH4, with a concomitant increase in H4K20me3 and H3K27me3 at rDNA loci, and was consistent with the down-regulation of rDNA transcription. Depletion of BEND3 showed elevated levels of pre-rRNA and increased H3K4me3, suggesting that BEND3 functions as an rDNA transcription repressor. Our results are consistent with the recent observations that BEND3 allows polycomb recruitment and H3K27me3 to specific chromatin to generate repressive chromatin, especially in the absence of H3K9me3 and DNA methylation (15).
Transcription of rRNA gene loci by RNA polymerase I is modulated by several players. rDNA loci switch between an active and a silenced state, and more than 50% of the nucleolar organizing regions contain active rDNA clusters. However, both the active and inactive rRNA genes are thought to associate with one another to form higher-order chromatin within the nucleoli (6). NoRC, a chromatin-remodeling complex comprising of the ATPase SNF2h and TIP5, is required for establishing repressive chromatin structure at rDNA promoters (9). Elegant work conducted almost a decade ago by Grummt and colleagues demonstrated that NoRC establishes heterochromatic histone modifications and de novo DNA methylation at rDNA loci and that this leads to chromatin compaction and transcription silencing of the rDNA repeats (9). Whether NoRC silences active rRNA genes, maintains the silent state of inactive genes, or is required for both activities remains to be determined (2, 6, 28). Recent work argues that NoRC is required for the inheritance or maintenance of silent epigenetic status at rDNA gene clusters (21).
We observed that BEND3 associates with NoRC and that, in the absence of BEND3, Tip5 association to the rDNA promoter is severely compromised. Similarly, NoRC coordinates the binding of BEND3 to the rDNA promoter. These data suggest that BEND3 and NoRC cooperate to establish rDNA silencing. Our results demonstrate that BEND3 stabilizes the Tip5 component of NoRC by modulating ubiquitination of Tip5. Furthermore, SUMOylation of BEND3 is critical for preventing this modification on Tip5. We suggest that SUMOylation of BEND3 is important for stabilizing NoRC and that this in turn is critical for rDNA silencing (Fig. 5).
Protein ubiquitination is a reversible posttranslational modification (PTM) that plays a critical role in many cellular processes. A class of proteases known as deubiquitinases (DUBs) catalyzes the removal of ubiquitin from target proteins (29–31). Of these, ubiquitin-specific proteases (USP) form the largest group that is characterized by the presence of a USP domain (32, 33). USP21 was initially identified as a histone H2A deubiquitinase (34), but has been shown to regulate the deubiquitination of several other substrates including RIG-1, RIP1, and GATA-3 (35–37). USP21 is thought to modulate target protein stability by modulating E3 deubiquitinase activity, thereby affecting the transfer of ubiquitin from the E3 ligase to its target protein (36). We have identified USP21 as a specific DUB of the Tip5/NoRC complex. USP21 interacts with BEND3, and the levels of USP21 are strongly affected by the presence of BEND3. A dramatic destabilization of USP21 was observed in the presence of BEND3-SDM, suggesting that the USP21 level is modulated by the SUMOylation status of BEND3. USP21 can interact with and deubiquitinate Tip5, thereby stabilizing the total levels of Tip5. We propose that SUMOylated BEND3 stabilizes the Tip5/NoRC complex by stabilizing USP21 deubiquitinase. At present, it is not clear how SUMOylated BEND3 stabilizes USP21. Although it is not uncommon for DUBs to undergo PTMs that affect their stability as well as activity, there is a lack of precedence in the case of USP21 to be modulated through any such modifications (38).
We also observed that overexpression of BEND3 causes hyper-heterochromatinization and chromatin compaction. The fact that BEND3 associates with most heterochromatic regions, and recalling the similarities between the genomic organization at centromeres, telomeres, and rRNA genes, we suggest that BEND3 may play important roles in assembling higher-order chromatin structure. The general role of NoRC in the establishment of higher-order chromatin structure by acting as a scaffold to coordinate modification of histones has recently been proposed (39). This has been supported by the role of Tip5 in maintaining the functional integrity of telomeres and centromeres (39). Many other mechanisms have been implicated in the targeting of NoRC to specific heterochromatic sites. The recruitment of NoRC to the rDNA promoter is mediated by the transcription factor TTF1, whereas Trf2 and CENP-A are known to regulate NoRC association to telomeres and centromeres, respectively (7, 28, 39). Furthermore, nucleolar retention of Tip5 has been shown to require pRNA, and this depends on PARP1, a protein recently shown to be critical for the inheritance of silent chromatin (21, 40). That enhanced expression of BEND3 changes the chromatin environment at the rDNA is also evident by an increased rDNA methylation state, suggesting that BEND3 is a potent repressor of transcription. Furthermore, loss of BEND3 results in hyperactivation of transcription at the rDNA loci. This is similar to what has been observed for JHDM1B, a demethylase implicated in transcriptional repression of rRNA genes (41). Taken together, we propose that BEND3 and NoRC play a highly concerted role in the maintenance of heterochromatin architecture at several genomic regions and that the rDNA locus provides an excellent insight into this phenomenon.
Materials and Methods
A detailed description of all of the plasmids, antibodies, primers, and experimental procedures can be found in SI Materials and Methods. Experimental procedures include immunoprecipitation, ChIP, immunofluorescence, FISH, DNA methylation assay, SimPull, EMSA, and psoralen cross-linking assay.
SI Materials and Methods
Plasmids and Antibodies.
YFP-BEND3 plasmids have been described previously (11). Flag-HA-Tip5 was a gift from Ingrid Grummt, German Cancer Research Center, Heidelberg. Tip5 was recloned into pCGT. Flag-SNF2H was kindly provided by Patrick Varga-Weisz, Babraham Institute, Babraham, Cambridgeshire, United Kingdom. Myc-USP21 plasmids were a gift from Jianhua Yang, Baylor College of Medicine, Houston. The rDNA-containing plasmid for DNA FISH was a gift from Brian McStay, National University of Ireland, Galway, Ireland.
Co-Immunoprecipitation.
Cells were cotransfected with YFP-BEND3 and HA/T7-tagged Tip5 or Flag-Snf2h and harvested after 24 h. Cells were lysed in buffer containing 50 mM Tris, pH 7.4, 10% (vol/vol) glycerol, 0.1 Triton-X, 250 mM NaCl, 1 mM CaCl2, and protease inhibitors for 15 min at 4 °C followed by micrococcal nuclease treatment for 30 min at room temperature (RT). The lysate was precleared using Gammabind Sepharose beads for 30 min followed by incubation with appropriate antibody overnight at 4 °C. Antibody-bound protein complex was pulled down using Gammabind Sepharose beads for 1.5 h followed by three washes using IP buffer before denaturation in Laemmli buffer and Western blot analysis.
Chromatin Immunoprecipitation.
Cells were fixed in 1% formadehyde for 10 min at RT and quenched with 0.125 M glycine for 5 min. Cells were harvested and lysed in buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.0. Chromatin was sonicated using Diagenode Bioruptor for 20–30 cycles of 30 s ON and 30 s OFF to obtain 200–500 bp. Sheared chromatin was precleared using protein G Dynabeads (Life Technologies) for 2 h before incubation with appropriate antibody overnight at 4 °C. Antibody-bound chromatin was pulled down using preblocked Dynabeads for 1.5 h. Beads were washed once each with low salt buffer (0.1% SDS; 1% Triton; 2 mM EDTA; 20 mM Tris, pH 8, +150 mM NaCl), once with high salt buffer (500 mM NaCl), once with LiCl wash buffer (10 mM Tris, pH 8.0; 1% Na-deoxycholate; 1% NP-40, 250 mM LiCl; 1 mM EDTA), and twice with TE (10 mM Tris, pH 8.0, 1 mM EDTA). Chromatin was eluted in elution buffer (1% SDS, 0.1 M sodium bicarbonate) at 65 °C twice, 10 min each. Cross-link was reversed with 0.2 M NaCl overnight at 65 °C. The eluted material was Rnase A-treated followed by Proteinase K treatment at 42 °C for 2 h. DNA was purified using a QIAquick PCR purification kit (Qiagen), and qPCR was performed using SYBR Green PCR Master mix and analyzed on a 7300 PCR System (Applied Biosystems). ChIP-qPCR results were calculated as percentage of IP/input signal (% input).
Immunofluorescence.
U2OS cells were grown on coverslips for 16–24 h before fixing in 2% (vol/vol) paraformaldehyde. Cells were permeabilized with 0.5% Triton-X for 7 min on ice followed by blocking with 1% PBS-normal goat serum for 30 min at RT. Cells were incubated with specific primary antibodies for 1 h at RT followed by an appropriate fluorophore-conjugated secondary antibody for 30 min. Cells were then stained with DAPI and mounted in either p-phenylenediamine or Vectashield (Vector Laboratories Inc.). Images were acquired using a Delta Vision optical-sectioning deconvolution instrument (Applied Precision) on an Olympus microscope. For FUrd labeling, cells were pulsed 48 h posttransfection with 2 mM FUrd for 10 min followed by immunostaining with BrdU antibody.
FISH.
RNA FISH was performed as described earlier (42). Briefly, cells were fixed in 4% (vol/vol) formaldehyde before permeabilization in 0.5% TritonX-100, 5 mM vanadyl ribonucleoside complex on ice for 10 min. Cells were then rinsed with PBS 3× 10 min and once in 2× SSC before hybridization at 37 °C for 12–16 h with a nick-translated probe.
For DNA FISH, cells were fixed in 4% (vol/vol) formaldehyde, and DNA was denatured using heat denaturation [4 min at 72 °C in 70% (vol/vol) formamide, 2× SSC] and hybridized using Spectrum red-labeled rDNA probe. The probe was generated using a nick translation kit (Abbott).
DNA Methylation Assay.
Genomic DNA (1 μg) was treated with or without HpaII at 37° C overnight. DNA was then purified, and qPCR was performed using previously reported (43) primers that span a HpaII site within the rDNA promoter (Table S1). Undigested control was used to normalize the qPCR results.
ChIP primers for rDNA locus and qRT primers for rRNA transcript are as previously reported (44, 45)
SiMPull.
Quartz microscopic slides were used to make flow chambers for the SiMPull experiments. The slides were passivated with methoxy-PEG doped with 1% biotin-PEG (Lysan Bio, Inc.) (24). Biotinylated-T7 antibody (Novagen) was immobilized on a PEG-passivated surface at ∼20 nM concentration for 20 min after coating the flow chambers with 0.2 mg/mL NeutrAvidin (Thermo) for 5 min.
For investigating the Tip5-BEND3-USP21 multimeric complex, SiMPull was carried out using cells that were either triply transfected with T7-Tip5, YFP-BEND3, and mCherry-USP21 or doubly transfected with YFP-BEND3 and mCherry-USP21. Cells were collected 24 h posttransfection and were lysed in the MNase digestion buffer: 50 mM Tris⋅HCl, pH 7.4, 200 mM NaCl, 0.2% Triton, 10% (vol/vol) glycerol, 1 mM CaCl2. Four units of MNase was then added for every 108 cells, and the lysates were incubated at RT for 20 min. The reaction was then stopped by adding EDTA (final concentration of 5 mM). The lysates were spun at 12,500 × g for 10 min, and the supernatant was used for subsequent SiMPull analyses. The lysates were appropriately diluted in T200 buffer (20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl) and incubated in the SiMPull chamber for 20 min, followed by two washes of T200 buffer (20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl) to remove unbound lysate.
Single-molecule fluorescence data were acquired by a prism-type total internal reflection fluorescence (TIRF) microscope and analyzed using scripts written in Matlab.
SiMPull Data Analysis.
Single-molecule data were acquired as the average number of YFP or mCherry fluorescent spots per imaging area (5,000 µm2). The subtracted background fluorescence that results from nonspecific binding of lysate was calculated by adding the triple lysate on the surface immobilized with control anti-HA antibody. The background fluorescence of YFP was determined to be 93 ± 21 and, for mCherry, 209 ± 21 per imaging area. Percentage colocalization between the co-immunoprecipitated YFP-BEND3 and mCherry-USP21 was calculated as the number of coaligned molecules of one fluorescent molecule (mCherry-USP21) with respect to the fluorescent molecules found in lower density on the surface (YFP-BEND3). This was necessary because YFP-BEND3 and mCherry-USP21 were not pulled down to the same extent by T7-Tip5 due to their independent interactions with T7-Tip5. A cutoff of 2 pixels was set for checking colocalization as the value corresponds to a diffraction limited spot (∼300 nm) for our TIRF setup. Error bars represent SD of the mean values obtained from three independent experiments.
EMSA.
Probes were generated by PCR from U2OS genomic DNA in the presence of [α-32P]dCTP. The same primer sets used in qPCR were used. Probes were purified using a gel purification column (Qiagen) after PCR, and 40- to 80-ng probes were used in each EMSA-binding reaction. EMSA-binding reactions were assembled in a 25-μl reaction volume. Protein extracts were incubated with 1 µg BSA, 500 ng poly(dI-dC) (Sigma) as nonspecific competitor and a 32P-labeled probe in binding buffer [20 mM Hepes, pH 7.9, 150 mM KCl, 1 mM EDTA, 0.5 mM DTT, and 8% (vol/vol) glycerol] for 30 min at RT. The binding reactions were then loaded onto a 5% (vol/vol) nondenaturing polyacrylamide (19:1) gel that had been prerun for 1 h at 240 V in 0.5× Tris/borate/EDTA at 4 °C. The samples were electrophoresed at 240 V for 1 h at 4 °C. The gel was then dried at 80 °C for 1 h and exposed to a Phosphor Imager screen or X-ray films.
Psoralen Cross-Linking Assay.
Cells were subjected to psoralen cross-linking 48 h posttransfection. Psoralen cross-linking and Southern were performed as described earlier with some modifications (46, 47). Briefly, cells were either untreated (control) or treated with 1/20th volume of Trioxsalen (200 μg/mL) for 20 min on ice and then irradiated with 366 nm UV for 10 min at a distance of 5 cm from lamps in Stratalinker 2400. Irradiation was repeated three more times with a fresh Trioxsalen addition. Genomic DNA was isolated and digested with BamHI overnight. Ten micrograms of DNA was run in 1% agarose gels in Tris/acetate/EDTA at 2 V⋅cm−1 for 18–20 h. Gel was subsequently stained with EtBr and de-cross-linked in Stratalinker2400 (254 nm UV) for a total of 4,000 mJ⋅cm−2. DNA was transferred and hybridized using a 2.2-kb probe that detects a BamHI fragment from the coding region of rRNA genes.
Supplementary Material
Acknowledgments
We thank members of the S.G.P. laboratory for discussions and suggestions; Drs. A. Belmont, B. Freeman, I. Grummt, B. McStay, T. Moss, R. Santoro, Jianhua Yang, and P. Varga-Weisz for providing reagents and suggestions; and Drs. D. Rivier and Paula Bubulya for critical reading of the paper. This work was supported by the National Science Foundation (NSF) Center for the Physics of Living Cells (PHY1147498) and NIH (AG042331) awards (to T.H.); American Cancer Society (RSG11-174-01RMC) and NIH (1RO1GM088252) awards (to K.V.P.); and Campus Research Board University of Illinois at Urbana–Champaign, NSF career (1243372), and NIH (1RO1GM099669) awards (to S.G.P.). T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424705112/-/DCSupplemental.
References
- 1.Grummt I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17(14):1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 2.Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum Mol Genet. 2007;16(Spec No 1):R21–R27. doi: 10.1093/hmg/ddm020. [DOI] [PubMed] [Google Scholar]
- 3.Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol. 2003;4(8):641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence RJ, Pikaard CS. Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle. 2004;3(7):880–883. [PubMed] [Google Scholar]
- 5.McStay B. Nucleolar dominance: A model for rRNA gene silencing. Genes Dev. 2006;20(10):1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 6.McStay B, Grummt I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 7.Strohner R, et al. NoRC: A novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20(17):4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strohner R, et al. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol. 2004;24(4):1791–1798. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32(3):393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21(17):4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathyan KM, Shen Z, Tripathi V, Prasanth KV, Prasanth SG. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci. 2011;124(Pt 18):3149–3163. doi: 10.1242/jcs.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abhiman S, Iyer LM, Aravind L. BEN: A novel domain in chromatin factors and DNA viral proteins. Bioinformatics. 2008;24(4):458–461. doi: 10.1093/bioinformatics/btn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korutla L, Degnan R, Wang P, Mackler SA. NAC1, a cocaine-regulated POZ/BTB protein interacts with CoREST. J Neurochem. 2007;101(3):611–618. doi: 10.1111/j.1471-4159.2006.04387.x. [DOI] [PubMed] [Google Scholar]
- 14.Sreenath K, et al. Nuclear matrix protein SMAR1 represses HIV-1 LTR mediated transcription through chromatin remodeling. Virology. 2010;400(1):76–85. doi: 10.1016/j.virol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 15.Saksouk N, et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell. 2014;56(4):580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Dai Q, et al. BEND6 is a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Development. 2013;140(9):1892–1902. doi: 10.1242/dev.087502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai Q, et al. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 2013;27(6):602–614. doi: 10.1101/gad.213314.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8(3):719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 19.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 20.Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9(8):774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell. 2012;45(6):790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 22.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giri S, et al. The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. eLife. 2015;4:4. doi: 10.7554/eLife.06496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473(7348):484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108(4):475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 26.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447(7143):399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 28.Guetg C, et al. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29(13):2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 30.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009;109(4):1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Y, Scheel H, Hofmann K, Komander D. Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst. 2009;5(12):1797–1808. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- 33.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa T, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22(1):37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan Y, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211(2):313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, et al. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. J Biol Chem. 2013;288(13):9373–9382. doi: 10.1074/jbc.M112.374744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu G, et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285(2):969–978. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessler BM, Edelmann MJ. PTMs in conversation: Activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem Biophys. 2011;60(1-2):21–38. doi: 10.1007/s12013-011-9176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postepska-Igielska A, Grummt I. NoRC silences rRNA genes, telomeres, and centromeres. Cell Cycle. 2014;13(4):493–494. doi: 10.4161/cc.27783. [DOI] [PubMed] [Google Scholar]
- 40.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22(3):351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 41.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450(7167):309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 42.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietrzak M, Rempala G, Nelson PT, Zheng JJ, Hetman M. Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS ONE. 2011;6(7):e22585. doi: 10.1371/journal.pone.0022585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan BC, et al. Epigeneitc silencing of ribosomal RNA genes by Mybbp1a. J Biomed Sci. 2012;19:57. doi: 10.1186/1423-0127-19-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011;39(12):4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol Cell. 2009;35(4):414–425. doi: 10.1016/j.molcel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Cong R, et al. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012;40(19):9441–9454. doi: 10.1093/nar/gks720. [DOI] [PMC free article] [PubMed] [Google Scholar]