Significance
This research uncovers the mechanisms of an ancient arms race between butterflies and plants, seen today in countless gardens as caterpillars of cabbage butterflies that devour cabbage crop varieties. Nearly 90 million years ago, the ancestors of Brassica (mustards, cabbage) and related plants developed a chemical defense called glucosinolates. While very toxic to most insects, humans experience glucosinolates as the sharp taste in wasabi, horseradish and mustard. Here we report that this triggered a chemical arms race that escalated in complexity over time. By investigating the evolutionary histories of these plants and insects, we found that major increases in chemical defense complexity were followed by butterflies evolving countertactics to allow them to continue to attack and feed on the plants.
Keywords: coevolution, phylogenomics, evolutionary novelty, chemical defenses, diversification
Abstract
Coevolutionary interactions are thought to have spurred the evolution of key innovations and driven the diversification of much of life on Earth. However, the genetic and evolutionary basis of the innovations that facilitate such interactions remains poorly understood. We examined the coevolutionary interactions between plants (Brassicales) and butterflies (Pieridae), and uncovered evidence for an escalating evolutionary arms-race. Although gradual changes in trait complexity appear to have been facilitated by allelic turnover, key innovations are associated with gene and genome duplications. Furthermore, we show that the origins of both chemical defenses and of molecular counter adaptations were associated with shifts in diversification rates during the arms-race. These findings provide an important connection between the origins of biodiversity, coevolution, and the role of gene and genome duplications as a substrate for novel traits.
Over half a century ago, Ehrlich and Raven (1) coined the term ‘coevolution’ and proposed that coevolutionary interactions between species with close ecological relationships generated much of the eukaryotic biodiversity on Earth. One of their primary examples of coevolution was the chemically mediated interactions between butterflies of the subfamily Pierinae (Pieridae, Lepidoptera) and their angiosperm host-plants in the order Brassicales. Members of the plant order Brassicales are united by their production of secondary metabolites called glucosinolates (i.e., mustard oils). Upon tissue damage, glucosinolates are modified into toxins long studied for their defensive properties and flavor (e.g., mustard and horseradish) (2). In the Arabidopsis thaliana (thale cress) genome, at least 52 genes are involved in glucosinolate biosynthesis (3, 4) and some exhibit strong evidence of adaptive evolution that is attributed to herbivore mediated selection (5, 6). Pierinae caterpillars detoxify the glucosinolates of their Brassicales host-plants by redirecting these otherwise toxic breakdown products to inert metabolites using a gene that encodes a nitrile-specifier protein (7). The key innovation of the Brassicales, defensive glucosinolates, evolved roughly 90 million years ago (Ma); within 10 million years, Pierinae responded with their own key innovation, the nitrile-specifier protein, and colonized the Brassicales. Subsequently, Pierinae net diversification rates increased compared with that of their sister clade Coliadinae, whose members did not colonize Brassicales (8).
Although these studies provide “perhaps the most convincing example” that the evolution of a key innovation resulted in an increased net diversification rate (9), much remains unknown about the origins and subsequent evolutionary dynamics of the key innovations that have had macroevolutionary consequences. To address this gap in the literature, here we further investigate these key innovations in the aforementioned plant and butterfly lineages by (i) assessing if these innovations increased in complexity over time and are associated with shifts in net diversification rates in both coevolutionary partners, (ii) identifying genomic mechanisms that facilitated the appearance and escalation of innovations that mediated the observed coevolutionary dynamics, and (iii) testing whether any observed increases in net diversification rates are associated with adaptive evolution.
Brassicales species synthesize more than 120 different glucosinolate compounds (10, 11). To investigate their evolution and potential escalation, we placed this diversity into a robust phylogenetic framework (Fig. 1). Inferring these relationships requires significantly more data than generated in previous studies, where phylogenetic estimates lack sufficient statistical support to adequately describe the last 90 million years of Brassicales evolution (SI Appendix, Text S1) (12). To resolve the phylogenetic backbone of Brassicales, we generated transcriptomes and identified orthologs from taxa across 14 families (average de novo assembled transcripts per species, 42,657; average length, 794 bp), and combined these with orthologs from nine available genomes (SI Appendix, Text S1). A time-calibrated phylogenomic tree was then generated and family level glucosinolate diversity and species richness were mapped onto it to identify the evolutionary points of origin for novel glucosinolate groups and shifts in net diversification rates (i.e., speciation minus extinction rates) (Fig. 1). We also localized the two ancient whole genome duplications (i.e., WGD or polyploid events) of Brassicales, named At-β and At-α (Fig. 1) (13), using further analyses of age distribution of gene duplicates, assessment of their gene trees, and a comparison of genomes (SI Appendix, Text S2). The exact phylogenetic placement of both the At-β and At-α has been a mystery due to a lack of a robust phylogenetic framework and a dearth of genomic resources from the vast majority of families in Brassicales. In addition, we performed evolutionary analyses of the core biosynthetic pathways across Brassicales families to investigate the origin of pathways that encode novel glucosinolate chemical classes (Fig. 2 and SI Appendix, Text S3). Similarly, we assembled a dataset from eight gene regions (6,716 bp) and 96 specimens to estimate the divergence times and diversification dynamics of the major lineages of Pieridae butterflies on a phylogeny, which has recently been well resolved at the genus level (14) (SI Appendix, Text S4). To investigate the evolutionary dynamics of the nitrile-specifier protein gene (i.e., counter adaptation against glucosinolates), we surveyed nine phylogenetically important species using transcriptome and whole genome sequencing (Fig. 2 and SI Appendix, Text S5) and tested the activity of identified nitrile-specifier proteins for two pivotal genera (Anthocharis and Pieris) using glucosinolate-myrosinase assays on gut extract and expressed proteins.
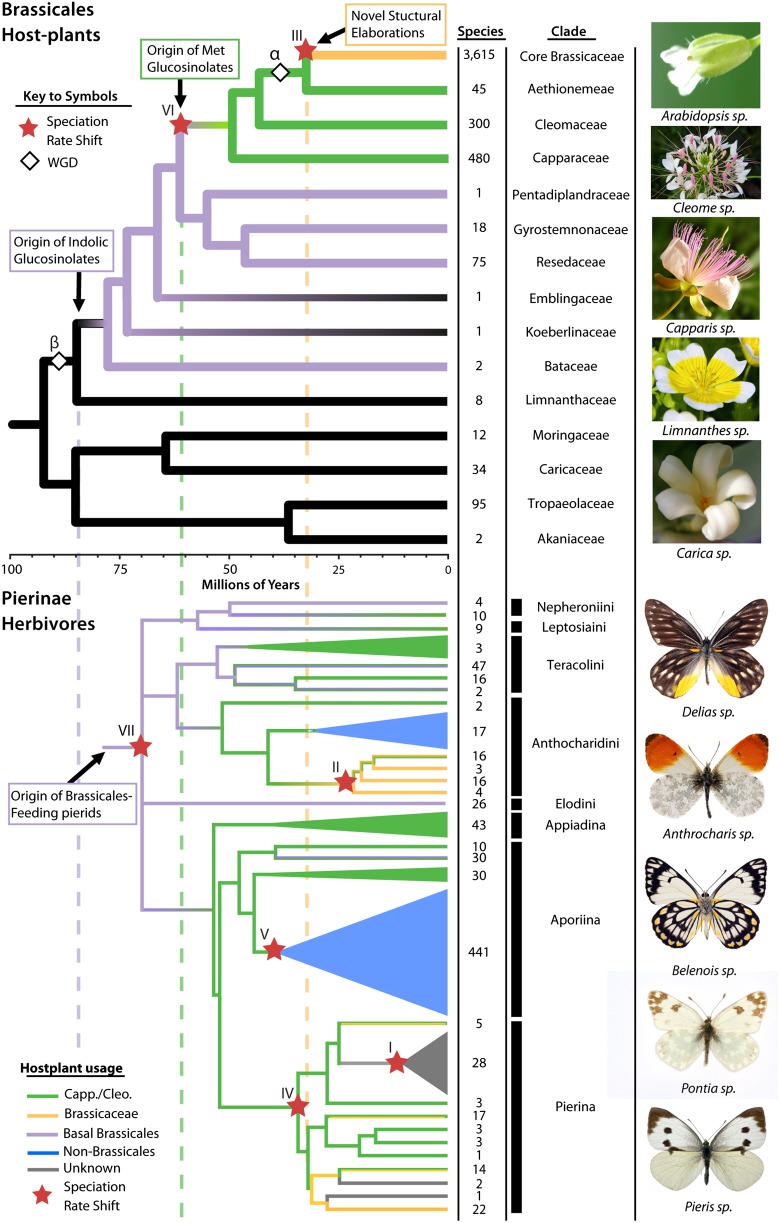
Fig. 1.
Phylogeny and diversity. A chronogram of both Brassicales families (Upper) and Pierinae butterfly genera (Lower), showing species numbers and identification of clades in the adjacent table. A common temporal scale is provided between the two chronograms. The branches in the Brassicales phylogeny are colored to indicate the origin of indolic glucosinolates (purple), methionine derived glucosinolates (green), and novel structural elaborations to glucosinolates unique to the core Brassicaceae lineage (orange). Vertical dashed lines also indicate the origin of these novel chemical groups. Primary host–plant associations of various Pierinae lineages are similarly colored: orange (Brassicaceae), green (Capparaceae or Cleomaceae), orange-green (mixture of previous), purple (more basal Brassicales that synthesize indolic glucosinolates), blue (non-Brassicales feeding) and gray (unknown). The phylogenetic positions for the At-α and At-β WGDs are depicted with white diamond symbols, and significant net diversification rate shifts with red star symbols.
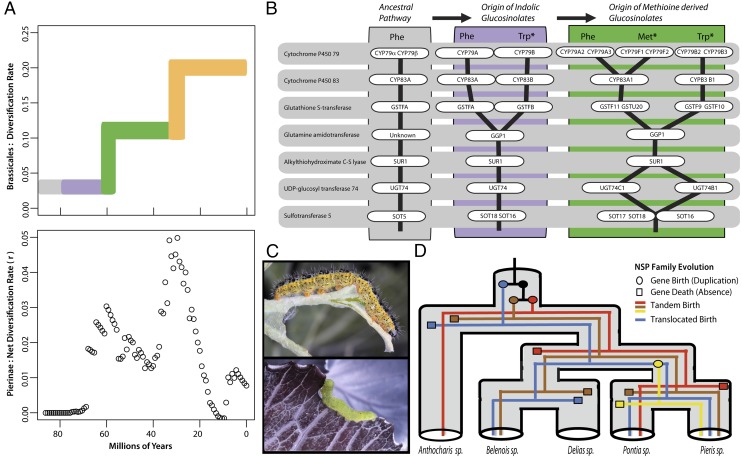
Fig. 2.
Evolution of glucosinolate biosynthethic pathways and detoxification nitrile-specifier protein (NSP) family. (A) Shifts in diversification rates during Brassicales evolution are highly punctuated (Upper). Lines are colored identical to lineages in Fig. 1. (Lower) How Pierinae butterflies have diversified during the same period. Time estimates are shown at the bottom. (B) An illustration of the evolution of core glucosinolate pathways across Brassicales; with substrates tryptophan (Trp), phenylalanine (Phe), and methionine (Met) shown at the top, enzymes depicted as white ovals, and each pathway as black vertical lines. (C) Photographs show Pieris brassicae (Upper) and Pieris rapae (Lower) caterpillar feeding on Brassica oleracea (European cabbage). (D) Evolution of the NSP gene family is shown across select Pierinae genera, indicating the birth and death dynamics of four paralogous clades.
Our results identify three important steps of escalation in the evolution of Brassicales that are reflected in ancestral Pierinae feeding patterns (Fig. 1). Brassicales, which appeared 92 Ma (median crown age; 95% HPD 50–133 Ma), initially were only able to synthesize glucosinolates from phenylalanine and branched chain amino acids (Fig. 2B; ancestral pathway). After the At-β WGD event 77.5 Ma (95% HPD 42–112 Ma), indolic glucosinolates appeared, which are synthesized from the amino acid tryptophan. Use of this novel substrate, as well as the previous ones, is restricted to the descendant families of the At-β event. These families show a near uniform retention of gene duplicates across the beginning of the core pathway, whose biosynthetic steps involve the conversion of either tryptophan or one of the more ancestral amino acid substrates into an intermediate aldoxime compound. No such retention of duplicates is seen in other metabolic pathways (SI Appendix, Text S3). Rather than simply functioning as redundant copies, the retained gene duplicates of the glucosinolate core pathway, including the cytochrome P450 79 (CYP79) step, have distinct substrates and products that are independently regulated by their own transcription factors (i.e., they have nonoverlapping functions) (SI Appendix, Text S3) (15). Here, we infer that the ability to synthesize glucosinolates from these aldoxime products is the ancestral state for Brassicales. This conclusion is supported by a recent analysis of the CYP79 family in a distant relative (Poplar; Populus trichocarpa), which revealed that this gene family in other plant orders is involved in the biosynthesis of herbivore-induced volatile aldoximes from phenylalanine and branched chain amino acids (16). Our results further indicate that the escalation of glucosinolate diversity, using new amino acid substrates to produce novel classes of compounds used as defenses in the arms-race against the butterflies, occurred via both single gene and whole genome duplications, involving the retention and neofunctionalization of both regulatory and core biosynthesis genes.
Nearly all Brassicales-feeding Pierinae butterflies consume families of Brassicales that synthesize indolic glucosinolates, and these compounds are well documented as important cues for host recognition and oviposition. Our mean estimate of the colonization of Brassicales by Pierinae butterflies is 68 Ma (Pierinae crown age; 95% HPD 60–75 Ma), concordant with the appearance of glucosinolate detoxification and an increase in their diversification rate (8). Together these insights provide a more detailed understanding of the colonization dynamics of Brassicales than previously described (8) by associating the diversification of Pierinae not with the timing of the origin of Brassicales per se, but rather with the origin of indolic glucosinolates. Indolic glucosinolates elicited an evolutionary response in Pierinae that highlights a dynamic identified by Erhlich and Raven (1), where a formerly toxic compound becomes an attractant after colonization.
A second major phase of escalation occurred when the ancestors of the plant lineages Capparaceae and Cleomaceae evolved a new suite of glucosinolate compounds derived from another novel substrate (methionine) (Fig. 1). This escalation in glucosinolate diversity occurred again via gene duplications followed by the neofunctionalization of both regulatory and core biosynthesis genes (Fig. 2 and SI Appendix, Text S1) and coincides with a significant increase in net diversification rate near the origin of these plant families (P value < 0.01) (Fig. 1 and SI Appendix, Text S1). Although the Pierinae that colonized the Brassicales necessarily fed on plants containing indolic glycosinolates (Fig. 1), today there are significantly fewer species feeding on the indolic-only lineages compared to the Capparaceae and Cleomaceae (30 vs. 133 species, respectively, P value < 0.0001).
The final escalation event we discovered appeared 32 Ma (95% HPD 17–46 Ma) with the evolution of Brassicaceae (mustard family), which have the greatest diversity of glucosinolates within Brassicales. We find this glucosinolate diversity to be derived from genes originating via tandem duplications and retention following the second major WGD of Brassicales, the At-α event (17, 18). This chemical defense escalation coincides with a dramatic increase in Brassicaceae diversification (P value < 0.01; Fig. 2; SI Appendix, Text S1). Shortly after Brassicaceae appeared, two independent lineages of Pierinae herbivores colonized them: one from the Anthocharidini and one from the Pierina butterfly lineages (Fig. 1 and SI Appendix, Text S4). Both butterfly colonizations coincide with increased diversification rates (P value < 0.01) (SI Appendix, Text S5). Thus, we have uncovered a dynamic escalation process that appears to have affected the macroevolution of Brassicales plants and their Pierinae herbivores. This process was facilitated by gene and genome duplications in Brassicales. Finally, comparison of macroevolutionary patterns of both groups shows a striking temporal concordance consistent with the expected outcome of diffuse coevolutionary interactions (Fig. 2).
Having established that Brassicaceae harbor the greatest glucosinolate diversity among plants and that this diversity arose via gene and genome duplications, we investigated whether butterfly detoxification mechanisms reflected a corresponding escalation in trait complexity. All of the Brassicales-feeding Pierinae could be using the same detoxification gene, evolving via allelic turnover, or there could be different gene copies having independent functional roles (e.g., specific to different types of glucosinolates). The latter condition would be evidence of an escalation at the molecular level via gene-family dynamics.
Our results show that rather than every butterfly using the same nitrile-specifier gene for glucosinolate detoxification, species have lineage-specific copies derived from a combination of at least four paralogous clades that differ in the glucosinolates they detoxify (Fig. 2). Of the two independent colonizations of Brassicaceae by Pierinae, the Anthocharidini lineage exemplar species (Anthocharis cardamines) has two nitrile-specifier genes derived from a single clade; they differ by 16 amino acids and their nitrile-forming activity against different glucosinolates is different (Benzyl: ancestral compound and 4-methylsulfuinylbutyl: novel methionine derived compound; SI Appendix, Text S5). In the Pierina exemplar species (Pieris rapae), there are two nitrile-specifier genes, but they come from different gene family clades. Although nitrile-forming activity is found in the paralogous clade not found in A. cardamines, similar nitrile-forming activity could not be detected in the gene from the paralogous clade most closely related to the A. cardamines clade (SI Appendix, Text S5). Thus, not only do different nitrile-specifier gene lineages exhibit functional differences against glucosinolates, but the two independent colonizations of the Brassicales were facilitated by independent nitrile-specifier gene lineages. We further assessed the relationship between nitrile-specifier genes and host-plant glucosinolate content by investigating how these detoxification genes evolved after butterfly lineages colonized plants without a glucosinolate defense system (Fig. 1). Aporini stopped using Brassicales 37 Ma (95% HPD 34–42 Ma) and in that lineage’s exemplar, Delias nigrina, neither nitrile-specifier genes were found in its genome nor any detectable enzymatic activity in its larval midgut (SI Appendix, Text S5). Thus, the two independent lineages that colonized Brassicaceae used independent copies of the nitrile-specifier protein, and feeding upon glucosinolate-containing plants appears necessary to maintain members of the nitrile-specifier gene family within a butterfly genome.
Our genomic data also provides a means of testing whether these patterns of escalation are associated with neutral or adaptive evolution. In Pierinae, we asked whether evolutionary dynamics of the nitrile-specifier protein departed from neutrality in the lineages leading to the two independent functional loci in Anthocharidini and Pierina. Using a maximum likelihood analysis of codon evolution (site-branch analyses), we found evidence of greater than expected levels of amino acid substitutions (i.e., diversifying selection) in both lineages (P value < 0.0001; SI Appendix, Text S5), consistent with adaptive evolution. In Brassicales, we asked whether species diversification in Brassicaceae was driven due to a neutral, random process (i.e., reciprocal gene loss) (19) or an adaptive process (i.e., facilitated by the origin of a novel trait) following the At-α event (Fig. 1 and SI Appendix, Text S6). Reciprocal gene loss is a mechanism of speciation, which has led to the emergence of new yeast species following whole genome duplication (19), that involves the passive loss of alternate copies of duplicated genes among different populations leading to their reproductive isolation (20). Because the At-α event predates the split between the Arabidopsis (4) and Aethionema (21) lineages, by quantifying the genome-wide duplication status of these earliest diverging Brassicaceae we can test whether reciprocal gene loss is associated with the increased diversification rates seen in the Arabidopsis lineage, which has 3615 species compared with 45 species in the Aethionema lineage. Of At-α duplicate genes, both lineages share the duplication status for 74% (53% reverted to singletons, 21% retained as duplicates), whereas the Aethionema and Arabidopsis lineages had 18% and 8% of these genes uniquely lost, respectively. Thus, lineage specific loss of duplicates was inversely correlated with shifts in diversification rates, and a putative genome-wide level of reciprocal gene loss was estimated at less than 0.4%. We conclude that reciprocal gene loss following the At-α event was therefore not a major contributor to diversification, as the vast majority of genes quickly reverted to single copy status after the At-α event and only a subset of genes, such as those in the glucosinolate pathway were retained as duplicates. Consistent with the hypothesis that retention of duplicates after WGD is driven by selective benefits (22), previous analyses indicates a high metabolic cost of glucosinolate production (23), a result incompatible with the retention of glucosinolate duplicate being neutral.
Half a century ago, Ehrlich and Raven (1) hypothesized that plant–insect interactions driven by diffuse coevolution over long periods of evolutionary time could be a major source of terrestrial biodiversity. Here, our analyses find strong support for this hypothesis in the repeated escalation of key innovations and bursts of diversification in Brassicales plants and Pierinae butterflies over the past 80 Myr. Importantly, gene and genome duplication events, via birth–death dynamics, have had a central role in these macroevolutionary events on each side of the plant–insect interaction. Although gene birth–death dynamics are known to be important in immune response and xenobiotic interactions (24, 25), our observation of similar dynamics in the nitrile-specifier gene family of butterflies provides a much needed data point from a novel gene family. These observed gene birth–death dynamics suggest that allelic change within genes may not be sufficient for long-term success during direct, as well as diffuse, coevolutionary interactions, extending Ohno’s seminal idea on the role of gene duplications in macroevolution (26), where evolutionary novelty more often occurs by gene duplication rather than by allelic variation of existing genes. The birth–death dynamics of certain gene families likely also drives the coevolutionary interactions described in other well studied host plant and herbivore systems (27), including between Apiaceae (Parsley family) and Papilionidae butterflies (28) or Oecophoridae moths (29). Interestingly, the cytochome P450 gene family appears to also play a crucial role in these systems as in the Brassicales. For example, the biosynthesis of furanocoumarins in Apiaceae but also the detoxification of these compounds by both Papilionidae and Oecophoridae involve cytochrome P450s (27). Rather than identifying patterns that are unique to these clades, we take these findings to suggest that such gene and genome duplication dynamics may be a general feature of the coevolutionary interactions that generated much of the diversity on Earth.
Materials and Methods
Transcriptomes were sequenced, assembled de novo across Brassicales, and combined with available plant genomes to: (i) Resolve phylogenetic relationships among families using 1155 single copy genes and divergence dates estimated with fossil calibrations (SI Appendix, Text S1); (ii) detect and phylogenetically localize two ancient whole genome duplications (SI Appendix, Text S2); and (iii) investigate the origin and evolution of glucosinolate pathways (SI Appendix, Text S3). We identified shifts in speciation rates following the origin of these traits (SI Appendix, Text S1), where chemical complexity appears to have escalated over time, and that Brassicales-feeding Pieridae butterflies have evolved counteradaptations from duplicate genes and coradiated with plant hosts. Parallel phylogenetic and temporal estimates of Pieridae used 7 nuclear genes and 1 mitochondrial gene, with divergence dates estimated with fossil calibrations (SI Appendix, Text S4). Pierinae species diversity and host-plant use were based on existing databases (SI Appendix, Text S4). The evolutionary dynamics of the nitrile-specifier protein were reconstructed using whole genome and transcriptome sequencing coupled with sequence analysis of codon evolutionary dynamics, with functional performance assayed on heterologously expressed proteins using established methods (SI Appendix, Text S5). Lastly, comparative genomic analyses were conducted to investigate whether observed net diversification rate shifts could be associated with neutral gene loss events following the most recent whole genome duplication at the base of Brassicaceae (SI Appendix, Text S6).
Supplementary Material
Acknowledgments
We thank Tao Shi for his assistance with phylogenetic analyses, Laura Williams for providing tissue of Koeberlina, and Goran Hellekant for providing Pentadiplandra fruits. This work was supported by National Science Foundation Grants PGRP 1202793 (to P.P.E.) and DEB 1146603 (to J.C.P.), Knut and Alice Wallenberg Foundation 2012.0058 (to C.W.W.), and the Academy of Finland 131155-Swedish Research Council 2012-3715 (to C.W.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information BioProject database (accession no. PRJNA283303).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503926112/-/DCSupplemental.
References
- 1.Ehrlich PR, Raven PH. Butterflies and plants: A study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 2.Gadamer J. Ueber das sinigrin. Ber Dtsch Chem Ges. 1897;30:2322–2327. [Google Scholar]
- 3.Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 4.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 5.Benderoth M, et al. Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci USA. 2006;103(24):9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad KVSK, et al. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science. 2012;337(6098):1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittstock U, et al. Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc Natl Acad Sci USA. 2004;101(14):4859–4864. doi: 10.1073/pnas.0308007101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheat CW, et al. The genetic basis of a plant-insect coevolutionary key innovation. Proc Natl Acad Sci USA. 2007;104(51):20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Futuyma DJ, Agrawal AA. Macroevolution and the biological diversity of plants and herbivores. Proc Natl Acad Sci USA. 2009;106(43):18054–18061. doi: 10.1073/pnas.0904106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56(1):5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 11.Mithen R, Bennett R, Marquez J. Glucosinolate biochemical diversity and innovation in the Brassicales. Phytochemistry. 2010;71(17-18):2074–2086. doi: 10.1016/j.phytochem.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107(43):18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 14.Wahlberg N, Rota J, Braby MF, Pierce NE, Wheat CW. Revised systematics and higher classification of pierid butterflies (Lepidoptera: Pieridae) based on molecular data. Zool Scr. 2014;46(6):641–650. [Google Scholar]
- 15.Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci. 2010;15(5):283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Irmisch S, et al. Two herbivore-induced cytochrome P450 enzymes CYP79D6 and CYP79D7 catalyze the formation of volatile aldoximes involved in poplar defense. Plant Cell. 2013;25(11):4737–4754. doi: 10.1105/tpc.113.118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofberger JA, Lyons E, Edger PP, Chris Pires J, Eric Schranz M. Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol Evol. 2013;5(11):2155–2173. doi: 10.1093/gbe/evt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliebenstein DJ. A role of gene duplication and natural variation of gene expression in the evolution of metabolism. PloS ONE. 2008;3(3):e1838. doi: 10.1371/journal.pone.0001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440(7082):341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- 20.Werth CR, Windham MD. A model for divergent, allopatric speciation of polyploid pteridophytes resulting from silencing of duplicate-gene expression. Am Nat. 1991;137(4):515–526. [Google Scholar]
- 21.Haudry A, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- 22.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16(7):805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 23.Bekaert M, Edger PP, Hudson CM, Pires JC, Conant GC. Metabolic and evolutionary costs of herbivory defense: Systems biology of glucosinolate synthesis. New Phytol. 2012;196(2):596–605. doi: 10.1111/j.1469-8137.2012.04302.x. [DOI] [PubMed] [Google Scholar]
- 24.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6(6):551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 25.Ingelman-Sundberg M. Polymorphism of cytochrome P450 and xenobiotic toxicity. Toxicology. 2002;181-182:447–452. doi: 10.1016/s0300-483x(02)00492-4. [DOI] [PubMed] [Google Scholar]
- 26.Ohno S. Evolution by Gene Duplication. 1st Ed Spinger; New York: 1970. [Google Scholar]
- 27.Berenbaum MR, Zangerl AR. Facing the future of plant-insect interaction research: Le retour à la “raison d’être”. Plant Physiol. 2008;146(3):804–811. doi: 10.1104/pp.107.113472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berenbaum MR. Chemical mediation of coevolution: Phylogenetic evidence for Apiaceae and associates. Ann Mo Bot Gard. 2011;88:45–59. [Google Scholar]
- 29.Li W, Zangerl AR, Schuler MA, Berenbaum MR. Characterization and evolution of furanocoumarin-inducible cytochrome P450s in the parsnip webworm, Depressaria pastinacella. Insect Mol Biol. 2004;13(6):603–613. doi: 10.1111/j.0962-1075.2004.00518.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.