Significance
In large pelagic fish, as in birds and mammals, significant questions remain concerning the selective pressures that drove the evolution of endothermy. We examined cold tolerance and niche breadth on molecular, organism, and food web levels in three closely related tuna species. We show that energetic benefits of increased cold tolerance in fishes are context dependent: advantageous when quality prey is abundant, but not when such prey is scarce. We broaden our findings to the global level using published literature to show that endothermic bluefin species in all the world’s oceans specialize, to some degree, on high energy prey. Our multifaceted approach demonstrates the concept that the advantage of a “beneficial” adaptation is dependent on environmental and anthropogenic influences.
Keywords: pelagic, tuna, Thunnus, evolution, endothermy
Abstract
Endothermy in vertebrates has been postulated to confer physiological and ecological advantages. In endothermic fish, niche expansion into cooler waters is correlated with specific physiological traits and is hypothesized to lead to greater foraging success and increased fitness. Using the seasonal co-occurrence of three tuna species in the eastern Pacific Ocean as a model system, we used cardiac gene expression data (as a proxy for thermal tolerance to low temperatures), archival tag data, and diet analyses to examine the vertical niche expansion hypothesis for endothermy in situ. Yellowfin, albacore, and Pacific bluefin tuna (PBFT) in the California Current system used more surface, mesopelagic, and deep waters, respectively. Expression of cardiac genes for calcium cycling increased in PBFT and coincided with broader vertical and thermal niche utilization. However, the PBFT diet was less diverse and focused on energy-rich forage fishes but did not show the greatest energy gains. Ecosystem-based management strategies for tunas should thus consider species-specific differences in physiology and foraging specialization.
Endothermy has been described in several lineages of marine teleosts (billfish, tunas, and recently, in the mesopelagic opah Lampris guttatus) and in sharks of the family Lamnidae (1–4). These teleosts and sharks possess convergently evolved mechanisms for conserving heat produced in metabolically active tissues as well as specialized “heater organs.” Heat produced in these tissues is retained via counter current heat exchangers (retia mirabilia) in the vasculature, minimizing heat loss from the gills. In tunas and sharks, the heart receives cooled, deoxygenated blood directly from retia mirabilia; thus cardiac tissues operate at ambient temperatures (5, 6) and must support warmer and energetically demanding body tissues. The cardiac tissues of some tunas are specialized for eurythermal performance, with high tolerance to low temperatures (6–9); however, at lower temperatures, tunas can experience cold-induced bradycardia, limiting cardiac output (6). In bluefin tunas, endothermy is linked to improved cardiac cold tolerance and increased function at lower temperatures (6). These studies indicate that advanced capacity for excitation–contraction (EC) coupling improves cardiac performance and plays an important role in determining the thermal limits of tunas (5, 6, 8).
Increased cold tolerance in vertebrates has been postulated to confer physiological and ecological advantages (1, 10). In fish, one hypothesis is that the evolution of endothermic traits enabled niche expansion both geographically and vertically, increasing access to prey (1, 2). For example, Atlantic bluefin tuna, the most endothermic species of the Thunnus genus, are able to seasonally exploit energetically rich prey (e.g., herring) in subpolar seas (>60°N) (11). Large individuals can make repeated dives to cold waters below the thermocline (as cool as 2–5 °C) to exploit a wider variety of forage species (e.g., herring, squid, mackerel, sand lance) (12). Importantly, tunas have physiological limitations at both high and low temperatures (13, 14), which are linked to constraints on cardiac function (6, 7, 9) and horizontal and vertical movements in the water column, which will in turn impact accessibility of prey.
Forage fishes such as herring, sardines, and anchovy have received attention in the recent scientific literature due to potential overexploitation and the subsequent loss of marine ecosystem services (15, 16). Unlike studies that have documented declines of predatory fish, sea turtles, sharks, marine mammals, and seabirds (17, 18), which are intrinsically valued for their social, ecological, and commercial importance, declines of prey species are usually deemed problematic due to the loss of food resources for apex predators (15, 16). If we really have “fished down marine food webs” (18) to the extent of forage fish overexploitation, it becomes important to understand the strength of trophic linkages between predators and their prey. Determining the relationships between physiology, niche breadth, and foraging success is important if we are to forecast the likely winners and losers following declines in local prey biomass or diversity due to both anthropogenic and environmental influences.
Here, we used expression of specific genes in cardiac tissue as a proxy for enhanced cardiac performance in low temperatures and tested whether this expression is linked to increased vertical niche width, greater access to prey, and increased foraging success. We used three co-occurring species of tunas as a model system. Endothermy in tunas affects numerous biological characteristics including increased body size, enhanced swimming efficiency, complex biomechanics, high migratory capacity, and fecundity (1, 19). Furthermore, foraging strategies are dependent upon geographic location, prey availability, and season. However, by comparing similarly sized, immature Thunnus species with varying cold tolerances that coexist in space and time, many of these confounding variables can be minimized or eliminated, providing an ecological interpretation of how tuna congeners use common physical space. Juveniles of three tuna species, Pacific bluefin tuna Thunnus orientalis (PBFT), yellowfin tuna Thunnus albacares (YFT), and albacore tuna Thunnus alalunga (ALB), overlap seasonally in the California Current Large Marine Ecosystem (CCLME) (13) (SI Appendix, Fig. S1). This co-occurrence provides a rare in situ opportunity to study trophic plasticity and feeding success in closely related species across a gradient of tolerance to low temperatures.
We quantified the expression of specific genes in ventricular tissues of similarly sized juvenile tunas. We chose genes associated with calcium cycling and sarcoplasmic reticulum (SR) stores in cardiac myocytes, which underlie enhanced calcium release, reuptake, and myocyte contractility at low temperatures (9). This study focuses on gene expression of these critical EC-coupling components, which elucidate SR calcium cycling capacity. Whereas phenotypic plasticity can occur, our study examined the three tuna species in a temporal and spatial context in which they were all experiencing a similar physical environment in the relatively stable, late summer CCLME. By archival tagging all species simultaneously and extracting data from a region of spatiotemporal overlap, we compared physical niche (vertical and thermal) of the three species. Finally, we assessed trophic niche breadth by conducting diet analyses in the study region over 3 y. This integrative approach permitted evaluation of comparative cardiac physiological capacities, habitat use, and trophic interactions in the CCLME. We hypothesized that increased expression of cardiac genes conferring cold tolerance of heart function would lead to a broader vertical and thermal niche in PBFT, leading to increased foraging plasticity and success in this species compared with YFT and ALB, which we expected to be vertically, thermally, and trophically more limited in scope.
Results and Discussion
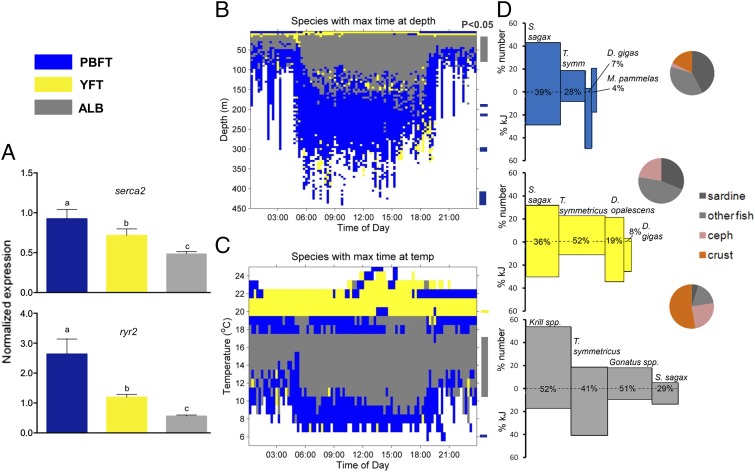
PBFT, YFT, and ALB showed significant variation in mRNA expression levels for two proteins associated with excitation–contraction coupling and calcium cycling across the cardiac SR. Results indicate that PBFT hearts maintain significantly greater expression of SR Ca2+ ATPase (serca2) and ryanodine receptor 2 (ryr2) genes compared to cardiac tissues from YFT (ANOVA followed by Tukey’s test, P = 0.0011, P = 1 × 10−5) and ALB (P = 1 × 10−5, P = 4 × 10−5) (Fig. 1A). These two proteins, cardiac isoforms of SERCA2 and RYR2, are involved in SR Ca2+ storage and release, respectively. They play a key role in maintaining SR function and overall Ca2+ homeostasis in cardiac myocytes and increase the capacity for active vertebrate hearts to maintain excitation–contraction coupling at low temperatures (7, 20, 21). Elevated expression of these proteins, particularly SERCA2, is strongly correlated with higher heart rates and enhanced SR function, which together maintain cardiac performance at low temperatures in endothermic tunas and sharks (7, 8, 20, 22).
Fig. 1.
Comparison of three co-occurring tunas [Pacific bluefin tuna (PBFT), blue; yellowfin tuna (YFT), yellow; albacore tuna (ALB), gray] on the molecular, behavioral, and ecological levels. (A) Expression of mRNA for serca2 and ryr2 genes that are involved in Ca2+ storage and cycling in the sarcoplasmic reticulum and contractility of cardiac myocytes. Letters above bars represent significant differences between species (ANOVA followed by Tukey’s test). (B and C) Temperature and depth data from the time series recorded by electronic tags. Colors represent which one of the three tuna species (blue: PBFT; yellow: YFT; gray: ALB) spent the most proportional time, relative to the other two species, at depth (B) or temperature (C) for a given time of day (x axis). Bars (colored by species) to the right of B and C show depths and temperatures at which one tuna species spent significantly more time than the other two species (one-way ANOVA, P < 0.05). (D) Index of relative importance (IRI) for each of the three tuna species while co-occurring in the study region. Polygon area is derived from percentage of number (y axis 0↑), percentage of total energy (kJ) (y axis 0↓), and frequency of occurrence (x axis) for the four most prevalent prey items of each tuna species. Pie charts show relative importance (by number) of fish, cephalopods, and crustaceans in diet, with the relative proportion of sardine also shown.
The mRNA expression patterns for the cardiac genes examined (PBFT > YFT > ALB) corresponded to a species-specific thermal niche as determined from electronic tag depth and temperature data. We examined 1,518 d of archival tag time series records (SI Appendix, Table S2 shows breakdown by species) to discern if niche separation occurred between these three juvenile Thunnus species in California Current waters. Tagging results here are from 2004–2005, as ALB were primarily tagged during these years; further analysis of the entire dataset (2002–2009) demonstrated that patterns of PBFT and YFT depth and temperature distribution were consistent across the entire tag dataset. Whereas depth utilization data showed overlap between tuna species in vertical and thermal habitat, especially at shallow depths and higher water temperatures (SI Appendix, Fig. S3), each tuna species clearly predominated at different ranges of depth and temperature (Fig. 1 B and C and SI Appendix, Table S3 for results of statistical analyses). YFT primarily occupied shallow, warmer waters of the mixed layer (<25 m, water temperatures of 19–24 °C) while making brief occasional dives into deeper, colder waters below the thermocline (Fig. 1B and SI Appendix, Fig. S3). Compared with the other two species, ALB spent relatively more time at intermediate, thermocline-associated depths (20–90 m) and temperatures (11–17 °C) and spent the least time at deepest depths and lowest temperatures (Fig. 1 B and C and SI Appendix, Fig. S3 and Table S3). Overall, PBFT used deeper and colder waters, spending the most time at greater depths (190–450 m; Fig. 1B) and using waters of lower ambient temperatures (5–8 °C; Fig. 1C and SI Appendix, Table S3) than the other two tuna species (SI Appendix, Table S3).
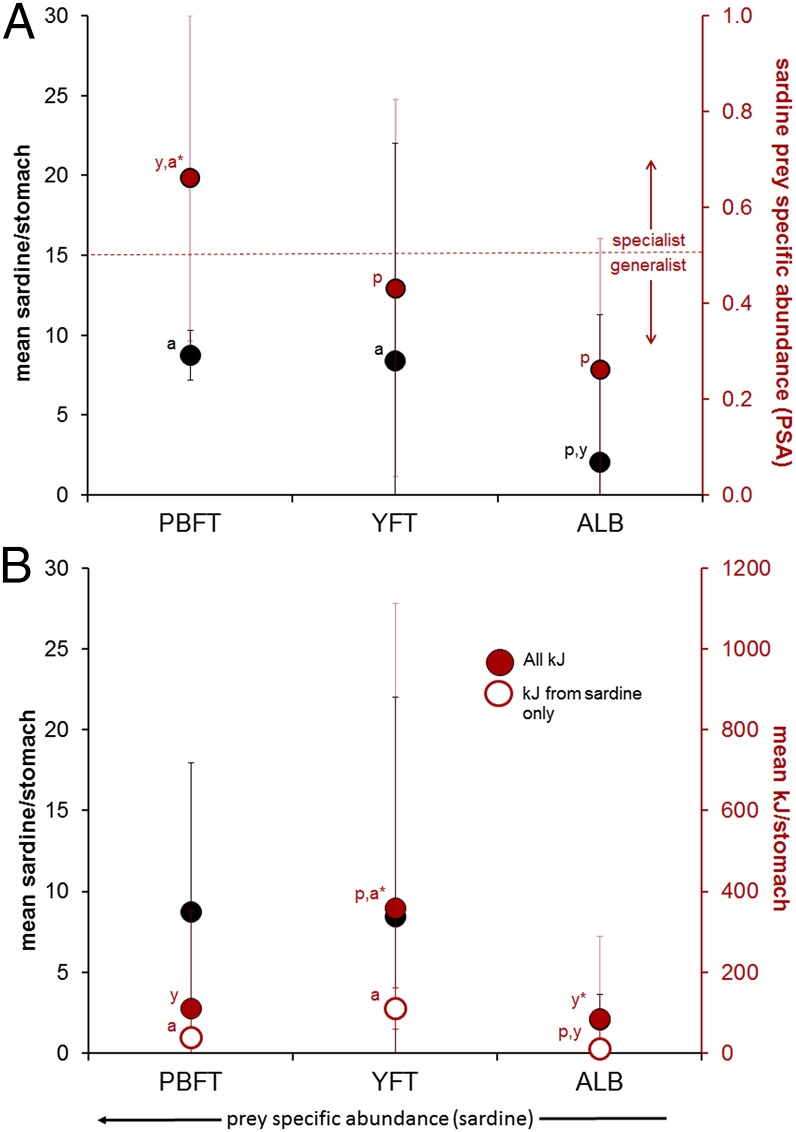
Despite having the widest vertical and thermal niche of the three Thunnus species in the CCLME, PBFT diet in the California Current system at the times observed was the most narrow (Shannon–Weiner Information Measure or H = 1.19). YFT and ALB diets had more even contributions by multiple prey species (YFT H = 1.30, ALB H = 1.25; Fig. 1D and SI Appendix, Tables S4 and S5). Albacore diet showed high contributions from crustaceans, fishes, and cephalopods, and YFT diet was most diverse (Fig. 1D and SI Appendix Tables S5 and S6) consisting of the most evenly distributed assemblage of forage fish, cephalopod, and crustacean species. From 2008 to 2010, PBFT diet showed evidence for specialization (prey-specific abundance or PSA > 0.5) (23) on a specific prey item, sardine (Fig. 2A). There was no evidence for diet specialization in YFT or ALB. The high PSA for sardines indicates that PBFT selectively fed on sardines when they were encountered in the CCLME and exploited other prey species to a lesser degree than did ALB and especially YFT, which fed successfully on sardines but also concurrently on other prey species (Fig. 2A). Estimates of prey consumption (kJ stomach−1) were highly variable for PBFT (range: 1–1,091 kJ; mean ± SD: 110 ± 241 kJ), YFT (1–1,756 kJ; 358 ± 755 kJ) and ALB (1–1,451 kJ; 85 ± 204 kJ) (Fig. 2B and SI Appendix, Fig. S4). Although archival tag data indicated that PBFT occupied the widest vertical and thermal niche (Fig. 1) and showed evidence for specialization on an energy-rich prey item (Fig. 2A), PBFT did not show the highest estimated kJ stomach−1. YFT consumed the most kJ stomach−1 (Kruskal–Wallis test, P = 0.007), and energy intake by PBFT and ALB was not statistically different (Kruskal–Wallis test, P = 0.53, Fig. 2B). Thus, relative sardine specialization by PBFT did not lead to higher absolute values of sardine consumption, nor did it lead to higher energetic consumption overall in PBFT (Fig. 2). Estimates of kJ stomach−1 were similar to results using the same methods in similarly sized tunas (yellowfin and bigeye tuna) in other regions (24); however, visceral endothermy in PBFT has been associated with more rapid digestion rates than other tunas (25, 26). Higher digestion rates could cause PBFT stomach contents to evacuate more rapidly and thus represent shorter time frames of recent diet. However, YFT kJ stomach−1 estimates were more than threefold higher than those of PBFT (Fig. 2); it is therefore unlikely that different gastric evacuation rates would completely explain the differences in kJ stomach−1 estimates for PBFT and YFT. Percentage of empty stomachs was similar across species, although PBFT had slightly more (15%) than YFT (14%) or ALB (11%).
Fig. 2.
Consumption of sardines and overall feeding success by co-occurring Pacific bluefin (PBFT), yellowfin (YFT), and albacore (ALB) tuna. (A) Mean values of prey-specific abundance and sardines found in each stomach of PBFT, YFT, and ALB. Prey-specific abundance (PSA) value > 0.5 suggests specialization (23). Letters above each point indicate statistically significant difference from indicated species (p = PBFT, y = YFT, a = ALB). *P < 0.001 (Kruskal–Wallis test). (B) Although PBFT had the highest PSA for sardine and were putative specialists, they did not consume more sardine-derived kilojoules per individual than YFT, and YFT consumed significantly more kilojoules overall than both ALB and PBFT. Note the relatively high variability in kJ stomach−1 for YFT.
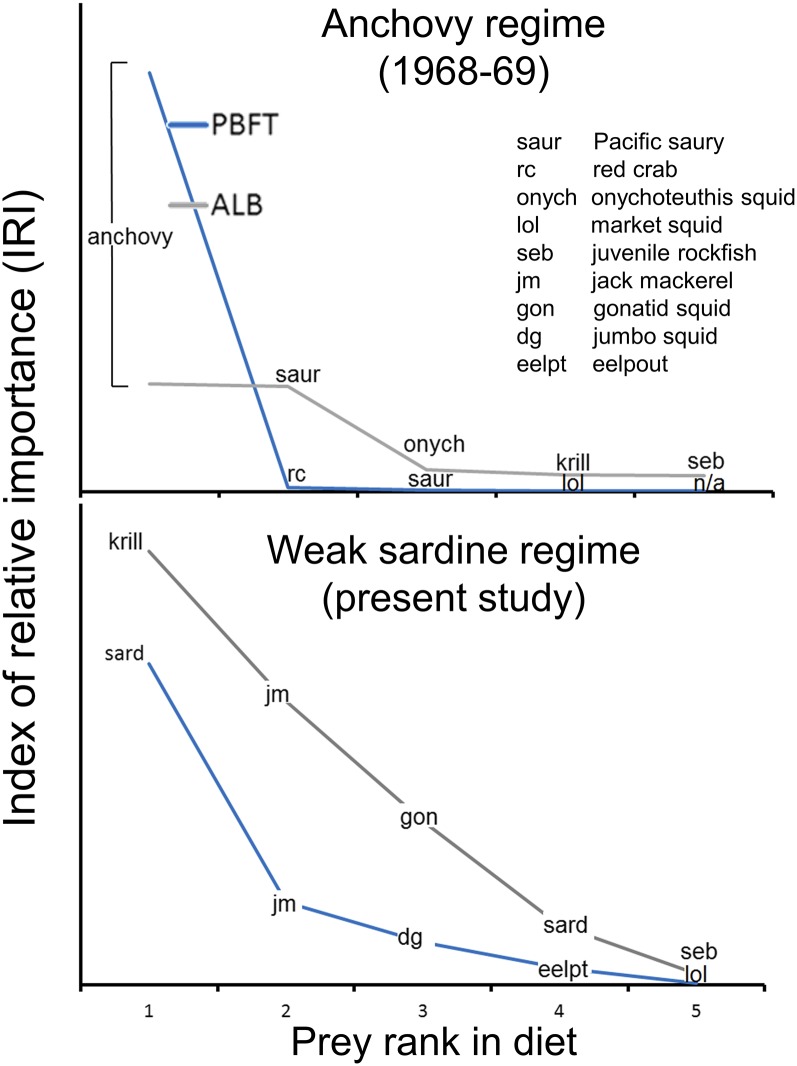
Dietary data for all three species were collected during a declining sardine regime (27). Anchovies were absent from predator diets and sardines were not as abundant as during sardine regimes in the CCLME, suggesting that our study took place during a transitional phase of the anchovy/sardine cycle (28). A previous study exists on the diets of PBFT and ALB in the CCLME (29) to which we can compare our feeding data. During the strong anchovy regime in 1968–1969, anchovy showed the highest index of relative importance (IRI) in PBFT diet, and both PBFT and ALB fed predominately on anchovy (29) (Fig. 3). However, throughout these years (1968–1969) other prey were still of relative importance to ALB diet but negligible in PBFT diet (Fig. 3). In the years of our study (2008–2010), PBFT maintained a similar pattern to that of the prior 1968–1969 anchovy regime: high importance of one energetically rich prey species and minimal importance of other prey items (Fig. 3 and SI Appendix, Table S4). In this study (2008–2010) ALB switched to a different primary prey (krill), with jack mackerel (Trachurus symmetricus) and gonatid squids (Gonatus spp.) contributing more to ALB diets than sardine (Fig. 3 and SI Appendix, Table S5). Thus, ALB seem to more readily switch foraging strategies in the face of cyclical changes in the anchovy or sardine cycle in the eastern Pacific Ocean (EPO) (Fig. 3), although diet data here, previous studies, and author observations suggest that all three species feed to some degree on readily available prey during relative scarcity of sardine and/or anchovy (30). For example, in this study (2008–2010) all three tuna species fed on juvenile jack mackerel (PBFT: 6.9 IRI; YFT: 23.4 IRI; ALB: 16.7; SI Appendix, Tables S4–S6); this prey species can be inferred to have been abundant in the CCLME during the years of this study, but was of much less relative importance to PBFT than to YFT or ALB (Figs. 1D and 3).
Fig. 3.
Index of relative importance for Pacific bluefin tuna (PBFT, blue line) and albacore tuna (ALB, grey line) during a strong anchovy regime (1968–1969) and a weak sardine regime (2008–2010). PBFT primarily consumed energetically rich fishes (anchovy or sardine) during both periods. ALB showed greater dietary breadth and foraging plasticity during the weak sardine regime (present study), subsidizing their diet with other species more than PBFT. ALB changed primary prey from anchovy during the anchovy regime to krill during the weak sardine regime.
Fig. 4.
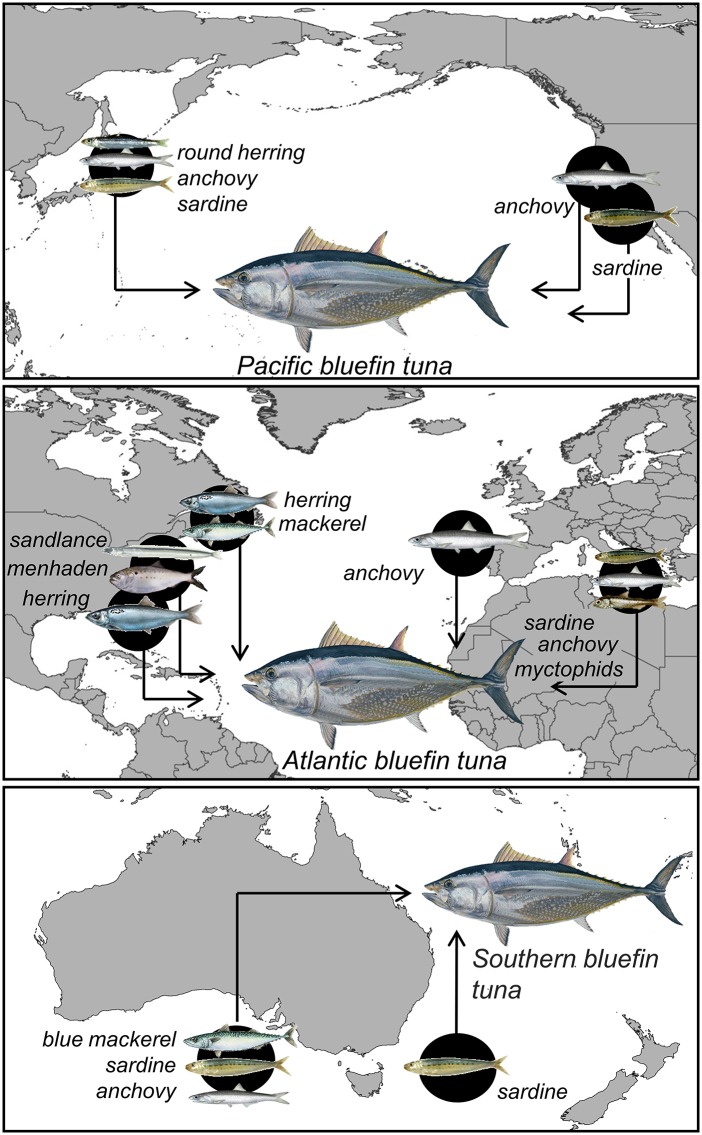
Prey preferences of Pacific, Atlantic, and Southern bluefin tuna in various ocean regions. Feeding studies have indicated that Pacific, Atlantic, and Southern bluefin diets tend to be dominated by one or two prey items, usually energetically rich, schooling fishes, in many feeding areas. Changes in residency patterns and/or declines in physical conditions of bluefin tuna have been shown in some of these areas when target prey is scarce. See SI Appendix, Table S7 for references and energy density of preferred prey.
Our results do not represent a comprehensive comparison of behavior and diet of PBFT, YFT, and ALB throughout their entire ranges or ontogeny. Larger PBFT migrate to both cold temperate foraging grounds and subtropical spawning grounds (11); large YFT in the CCLME remain largely residential to EPO waters (14), and large albacore migrate to subtropical waters in the Pacific Ocean (31). The relative benefits of cardiac performance, concordant water column utilization, and foraging capacity have not been comparatively assessed in these adult tunas and are outside the scope of this study. Importantly, the sardine in tuna diets were young of the year (YOY), and the vertical distribution of YOY sardine in the pelagic CCLME is not well characterized. In CCLME waters, sardine and other energy-rich forage fish (e.g., jack mackerel, Pacific mackerel, Pacific herring) may seek daytime refuge in subthermocline waters (32–36). The deeper-diving behavior of PBFT in this study could be due to the pursuit of diel-vertically migrating forage species (e.g., forage fish or squid) at depth, or it could be demonstrative of searching behavior rather than successful foraging. At times or in areas where primary prey (sardines or anchovy) are scarce, PBFT may be foraging on deeper-dwelling fish, cephalopods, and/or crustaceans (Fig. 1D). Whereas new bioenergetics models indicate that PBFT feeding success is high in the CCLME (37), PBFT utilization of deeper waters in this study did not translate to increased diet diversity or higher dietary energetic input in the region and time period studied in the CCLME (Figs. 1 and 2).
The relative trophic specialization of PBFT suggests that juvenile PBFT do not extensively alter foraging behavior to the extent of other Thunnus species, even when it may be to their benefit to do so (Figs. 2 and 3). Other diet studies of PBFT and closely related Atlantic bluefin (Thunnus thynnus) and Southern bluefin (Thunnus maccoyii) point to similar patterns across multiple oceanic regions and ontogeny (subadults and adults). Bluefin species globally prefer a few species of schooling, energetically rich fishes as their primary prey (anchovy and sardine in Japan and in the CCLME, herring, menhaden, mackerel and/or sandlance in the western Atlantic, anchovy and/or sardine in the eastern Atlantic and Mediterranean, and sardines, anchovy, and mackerel in the South Pacific; Fig. 3 and SI Appendix, Table S7); this is in contrast to YFT, which in the Pacific, Atlantic, and Indian oceans feed on a large number of different species of fish, cephalopods, and crustaceans (38–43), many of which have relatively low energy density (e.g., flying fish, halfbeaks, pelagic red crabs; SI Appendix, Table S7). Studies of ALB diet across various ocean regions (Pacific Ocean, North Atlantic Ocean, and Mediterranean Sea) reveal that ALB diet is diverse but often dominated by cephalopod and crustacean species (30, 41, 44, 45). Fish that commonly occur in ALB diets in those studies are largely mesopelagic (e.g., lightfish and barracudinas; SI Appendix, Table S7) (46, 47), in accordance with ALB vertical habitat described here. Occasionally ALB forage extensively on anchovy when this prey is highly abundant (29, 48, 49).
Importantly, in our study and the diet studies above, all bluefin species demonstrated trophic plasticity and fed on a variety of prey items across regions. We recognize that bluefin tunas can feed successfully in the absence of the most energetically rich prey; using stable isotope analysis, Madigan et al. estimated a mixed diet in PBFT, which likely feed at certain times throughout the year on DSL-associated organisms (e.g., euphausiids, squid), particularly during years of low sardine and/or anchovy abundance (30). That study also showed interannual plasticity of YFT diet, whereas ALB and PBFT diets were mixed, but consistent, between years (30). However, it is likely that seasonal abundance of energetically rich prey represent crucial feeding periods, especially for juvenile bluefin tunas, in which growth is maximized by optimal feeding conditions at preferred sea surface temperatures (13). For example, herring abundance can affect both Atlantic bluefin tuna condition (50) and distribution (51). Depletion of optimal prey species can thus negatively affect individual bluefin tuna condition, population growth, and/or alter migratory patterns (50–52).
All bluefin tuna species face uncertain futures due to exploitation and environmental change. Declines in biomass have been demonstrated in Atlantic and Southern bluefin tunas (53, 54), although there is also recent evidence for potential recovery (55, 56). In the most recent stock assessments, PBFT were declared to be overfished with estimated spawning biomass population declines of 94% of prefished levels (57). Our data suggest that overexploitation of energetically rich prey can have unequal effects on species of ostensibly similar trophic ecology, as the lack of juvenile PBFT diet diversity indicates that they may rely more heavily on sardine and/or anchovy abundance in the CCLME. During periods of localized primary prey scarcity, increased cardiac tolerance to the cold may facilitate juvenile PBFT horizontal niche expansion to locate preferred prey; as PBFT body size and thermal inertia increase, the benefits of cold tolerance may increase by facilitating foraging at higher latitudes on energetically rich prey by giant, mature bluefin tuna. Removal of forage fish by commercial fisheries have impacts on marine predators, and such effects can be difficult to measure (15, 16, 50). PBFT are in decline, and eastern Pacific sardine may soon collapse (58). The status of primary forage species (sardine and anchovy in the CCLME) should become a component of ecosystem-based management for overfished species.
Materials and Methods
Electronic Tagging.
The Animal Care and Use program at Stanford University provided Administrative Panel on Laboratory Animal Care (APLAC) oversight for tuna tagging and collection. Archival tag data were used from 87 electronic tags for a total of 1,518 d. Sample size n and days for each species are as follows: PBFT (n = 43 individuals, 725 d), YFT (n = 31 individuals, 583 d), ALB (n = 13 individuals, 210 d) (size of all species ranged from 66 to 83 cm at time of tagging). Archival tag (Lotek 2310 LTD tags) data for PBFT, YFT, and ALB tuna were collected between 2002 and 2009 as part of the Tagging of Pacific Predators (TOPP) project (13). Geolocation estimates were generated using methodology described in Block et al. (13) and used to identify periods of spatial and temporal overlap in the CCLME between these three tuna species. We defined overlap as periods when all three tuna species occurred within a 0.5 × 0.5 degree area during a 7-d period, and used the maximum and minimum possible latitude and longitude of fish within that cell based on estimated geolocation errors (±1.9° latitude, ±0.8° longitude) (13) (SI Appendix, Fig. S1). Only data from months for which stomach content data were collected (July–October) were included (SI Appendix, Table S2). Depth data from fish within this overlap area and period were analyzed for tag-measured differences in ambient temperature and depth utilization. To describe each species’ vertical and thermal distribution during periods of overlap, we calculated proportion of time spent at depth (TAD) and temperature (TAT) by time of day in 15-min intervals for each individual. Mean TAD, TAT, and comparative figures showing which species spent the most time at particular depths/temperatures were generated. Temperature-depth profiles were constructed for each species by calculating mean (±SD) temperature at depth for each day for each species; temperature-depth profiles for all days of data for each species were compared, which validate that all three tuna species were in areas with similar water column thermal structure (SI Appendix, Fig. S2). To assess statistically significant interspecific differences for TAD and TAT, we calculated the TAT and TAD for depth bins of 10 m and temperature bins of 1 °C and 5 °C (SI Appendix, Table S3) for PBFT (n = 43), YFT (n = 31), and ALB (n = 13). Proportions of TAD and TAT for each species were arcsine transformed and compared using one-way ANOVA for each depth and temperature bin. Tukey’s honest significant difference criterion was used for the multiple comparison test. Significant differences are reported for α = 0.05 and α = 0.1 (SI Appendix, Table S3). See SI Appendix, section 1.1 for full description of cross-species comparison of archival tag data.
Cardiac Gene Expression.
PBFT (n = 7), YFT (n = 5), and ALB (n = 8) individuals were collected in June 2008 by hook-and-line fishing in the CCLME, simultaneously with diet data collection reported below. Fish were placed in a seawater-filled vinyl sling, lifted aboard the vessel, and immediately killed. Cardiac tissues were sampled within 15 min and preserved in RNAlater (Applied Biosystems) as per the manufacturer recommendation. RNA was extracted from the tissues using TRIzol reagent (Invitrogen) in accordance with the manufacturer’s protocol. RNA quantity and quality was assessed using spectrophotometric and gel-based methods. mRNA was transcribed to cDNA in preparation for cloning and qPCR. Where necessary, degenerate primers were designed and used in PCR with the appropriate cDNA to clone and sequence genes of interest. Relative transcript abundance was assessed using qPCR techniques. qPCR assays were performed for each gene and species using standard dilution curves with a minimum of four points, dissociation curves, and no-template and no-reverse-transcriptase controls. Relative expression values for each gene were calculated using a standard curve method. To compare gene expression values across species, reference gene beta-actin in each species was calculated and used to normalize transcript abundance of serca2 and ryr2 in the corresponding species. Beta-actin normalized gene expression data followed a Gaussian distribution and was analyzed based on ordinary one-way ANOVA followed by Tukey's test to correct for multiple comparisons. Multiplicity adjusted P values are reported. See SI Appendix, section 1.2 for a full description of the sampling and qPCR assays.
Diet Analysis.
Stomachs were collected from PBFT (n = 82), YFT (n = 75), and ALB (n = 86) tunas from the fishing vessel R/V Shogun during the months of July–October in the years 2008–2010. Stomachs were collected from each species only during periods when all three species were being caught in the same general area (July–October), according to recreational catch data. Only one stomach sample for each tuna species was analyzed per sampling site to eliminate pseudoreplication of a single day’s feeding conditions. Cumulative prey curves indicate that sample sizes were adequate to describe tuna diets over the spatiotemporal scale of the study (SI Appendix, Fig. S3). Stomach contents were rinsed into sieves; large, freshly consumed sardines (used as bait on fishing vessels) were easily identified and removed. Prey species were identified by hard parts. Length, mass, and energy value (kJ⋅g−1) of prey were calculated using published algorithms (SI Appendix, section 1.3 for full description of prey length, mass, and energetic reconstruction). A modified IRI (29), using percentage of prey energetic content in place of percentage of volume (SI Appendix, section 1.3), was calculated for the top four items in each predator diet. Shannon–Weiner information measures (H) and PSA were calculated for the top four prey items in each tuna species to assess specialization on certain prey. Mean kJ stomach−1 and sardine PSA values were compared across tuna species using the nonparametric Kruskal–Wallis test, with significance at α = 0.05. Prey of high importance to Pacific, Atlantic, and Southern bluefin in other oceanic regions were assessed from the literature (SI Appendix, Table S7).
Supplementary Material
Acknowledgments
We thank the captains and crew of the F/V Shogun for assistance with sample collection and for electronic tagging support; O. Snodgrass for providing training in diet analysis; M. Hines, J. Adelson, and S. Crawford for assisting with diet analysis; C. Farwell, A. Norton, L. Rodriguez, R. Schallert, M. Price, N. Arnoldi, and E. Estess for assisting with sample collection and bluefin, yellowfin, and albacore tagging; Stanford student A. Mah for assistance with qPCR assay development; the Mexican government for permitting the collection and tagging of tunas and sampling in their waters; and the Monterey Bay Aquarium Foundation for support of the research cruises aboard the Shogun. The animal care and use program at Stanford University provided APLAC oversight to B.A.B. and D.J.M. for tuna collections. Electronic tagging was supported by the Tagging of Pacific Pelagics program, via grants to B.A.B. from the Office of Naval Research and from the Gordon and Betty Moore, David and Lucile Packard, and Alfred P. Sloan foundations.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500524112/-/DCSupplemental.
References
- 1.Block BA, Finnerty JR, Stewart AF, Kidd J. Evolution of endothermy in fish: Mapping physiological traits on a molecular phylogeny. Science. 1993;260(5105):210–214. doi: 10.1126/science.8469974. [DOI] [PubMed] [Google Scholar]
- 2.Graham JB, Dickson KA. Tuna comparative physiology. J Exp Biol. 2004;207(Pt 23):4015–4024. doi: 10.1242/jeb.01267. [DOI] [PubMed] [Google Scholar]
- 3.Carey FG. A brain heater in the swordfish. Science. 1982;216(4552):1327–1329. doi: 10.1126/science.7079766. [DOI] [PubMed] [Google Scholar]
- 4.Wegner NC, Snodgrass OE, Dewar H, Hyde JR. Whole-body endothermy in a mesopelagic fish, the opah, Lampris guttatus. Science. 2015;348(6236):786–789. doi: 10.1126/science.aaa8902. [DOI] [PubMed] [Google Scholar]
- 5.Block BA, Stevens ED, editors. Tuna: Physiology, Ecology, and Evolution. Vol 19 Academic; San Diego: 2001. [Google Scholar]
- 6.Blank JM, et al. In situ cardiac performance of Pacific bluefin tuna hearts in response to acute temperature change. J Exp Biol. 2004;207(Pt 5):881–890. doi: 10.1242/jeb.00820. [DOI] [PubMed] [Google Scholar]
- 7.Shiels HA, Maio AD, Thompson S, Block BA. Warm fish with cold hearts: Thermal plasticity of excitation-contraction coupling in bluefin tuna. P R Soc B. 2011;278(1702):18–27. doi: 10.1098/rspb.2010.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayasundara N, Gardner LD, Block BA. Effects of temperature acclimation on Pacific bluefin tuna (Thunnus orientalis) cardiac transcriptome. Am J Physiol Regul Integr Comp Physiol. 2013;305(9):R1010–R1020. doi: 10.1152/ajpregu.00254.2013. [DOI] [PubMed] [Google Scholar]
- 9.Galli GLJ, Lipnick MS, Shiels HA, Block BA. Temperature effects on Ca2+ cycling in scombrid cardiomyocytes: A phylogenetic comparison. J Exp Biol. 2011;214(Pt 7):1068–1076. doi: 10.1242/jeb.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett AF, Ruben JA. Endothermy and activity in vertebrates. Science. 1979;206(4419):649–654. doi: 10.1126/science.493968. [DOI] [PubMed] [Google Scholar]
- 11.Block BA, et al. Electronic tagging and population structure of Atlantic bluefin tuna. Nature. 2005;434(7037):1121–1127. doi: 10.1038/nature03463. [DOI] [PubMed] [Google Scholar]
- 12.Lawson GL, Castleton MR, Block BA. Movements and diving behavior of Atlantic bluefin tuna Thunnus thynnus in relation to water column structure in the northwestern Atlantic. Mar Ecol Prog Ser. 2010;400:245–265. [Google Scholar]
- 13.Block BA, et al. Tracking apex marine predator movements in a dynamic ocean. Nature. 2011;475(7354):86–90. doi: 10.1038/nature10082. [DOI] [PubMed] [Google Scholar]
- 14.Schaefer KM, Fuller DW, Block BA. Movements, behavior, and habitat utilization of yellowfin tuna (Thunnus albacares) in the Pacific Ocean off Baja California, Mexico, determined from archival tag data analyses, including unscented Kalman filtering. Fish Res. 2011;112(1-2):22–37. [Google Scholar]
- 15.Cury PM, et al. Global seabird response to forage fish depletion—one-third for the birds. Science. 2011;334(6063):1703–1706. doi: 10.1126/science.1212928. [DOI] [PubMed] [Google Scholar]
- 16.Smith ADM, et al. Impacts of fishing low-trophic level species on marine ecosystems. Science. 2011;333(6046):1147–1150. doi: 10.1126/science.1209395. [DOI] [PubMed] [Google Scholar]
- 17.Myers RA, Worm B. Rapid worldwide depletion of predatory fish communities. Nature. 2003;423(6937):280–283. doi: 10.1038/nature01610. [DOI] [PubMed] [Google Scholar]
- 18.Pauly D, Christensen V, Dalsgaard J, Froese R, Torres F., Jr Fishing down marine food webs. Science. 1998;279(5352):860–863. doi: 10.1126/science.279.5352.860. [DOI] [PubMed] [Google Scholar]
- 19.Dickson KA, Graham JB. Evolution and consequences of endothermy in fishes. Physiol Biochem Zool. 2004;77(6):998–1018. doi: 10.1086/423743. [DOI] [PubMed] [Google Scholar]
- 20.Landeira-Fernandez AM, Morrissette JM, Blank JM, Block BA. Temperature dependence of the Ca2+-ATPase (SERCA2) in the ventricles of tuna and mackerel. Am J Physiol Regul Integr Comp Physiol. 2004;286(2):R398–R404. doi: 10.1152/ajpregu.00392.2003. [DOI] [PubMed] [Google Scholar]
- 21.Landeira-Fernandez AM, Castilho PC, Block BA. Thermal dependence of cardiac SR Ca2+-ATPase from fish and mammals. J Therm Biol. 2012;37(3):217–223. [Google Scholar]
- 22.Weng KC, et al. Satellite tagging and cardiac physiology reveal niche expansion in salmon sharks. Science. 2005;310(5745):104–106. doi: 10.1126/science.1114616. [DOI] [PubMed] [Google Scholar]
- 23.Amundsen PA, Gabler HM, Staldvik FJ. A new approach to graphical analysis of feeding strategy from stomach contents data—modification of the Costello (1990) method. J Fish Biol. 1996;48(4):607–614. [Google Scholar]
- 24.Ménard F, Stéquert B, Rubin A, Herrera M, Marchal É. Food consumption of tuna in the Equatorial Atlantic ocean: FAD-associated versus unassociated schools. Aquat Living Resour. 2000;13(04):233–240. [Google Scholar]
- 25.Clark TD, Farwell CJ, Rodriguez LE, Brandt WT, Block BA. Heart rate responses to temperature in free-swimming Pacific bluefin tuna (Thunnus orientalis) J Exp Biol. 2013;216(Pt 17):3208–3214. doi: 10.1242/jeb.086546. [DOI] [PubMed] [Google Scholar]
- 26.Whitlock RE, et al. Quantifying energy intake in Pacific bluefin tuna (Thunnus orientalis) using the heat increment of feeding. J Exp Biol. 2013;216(Pt 21):4109–4123. doi: 10.1242/jeb.084335. [DOI] [PubMed] [Google Scholar]
- 27.Zwolinski JP, et al. Distributions and abundances of Pacific sardine (Sardinops sagax) and other pelagic fishes in the California Current Ecosystem during spring 2006, 2008, and 2010, estimated from acoustic–trawl surveys. Fish Bull. 2012;110(1):110–122. [Google Scholar]
- 28.Chavez FP, Ryan J, Lluch-Cota SE, Niquen C M. From anchovies to sardines and back: Multidecadal change in the Pacific Ocean. Science. 2003;299(5604):217–221. doi: 10.1126/science.1075880. [DOI] [PubMed] [Google Scholar]
- 29.Pinkas L, Oliphant MS, Iverson ILK. Food habits of albacore, bluefin tuna, and bonito in California waters. Fish Bull Cal Dept Fish Game. 1971;152:1–105. [Google Scholar]
- 30.Madigan DJ, et al. Stable isotope analysis challenges wasp-waist food web assumptions in an upwelling pelagic ecosystem. Sci Rep. 2012;2(654):654. doi: 10.1038/srep00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Childers J, Snyder S, Kohin S. Migration and behavior of juvenile North Pacific albacore (Thunnus alalunga) Fish Oceanogr. 2011;20(3):157–173. [Google Scholar]
- 32.Sepulveda C, Kohin S, Chan C, Vetter R, Graham J. Movement patterns, depth preferences, and stomach temperatures of free-swimming juvenile mako sharks, Isurus oxyrinchus, in the Southern California Bight. Mar Biol. 2004;145(1):191–199. [Google Scholar]
- 33.Demer DA, et al. Advanced Survey Technologies Program. Southwest Fisheries Science Center; La Jolla, CA: 2011. Acoustic-trawl surveys of Pacific sardine (Sardinops sagax) and other pelagic fishes in the California Current ecosystem: Part 1, Methods and an example application. [Google Scholar]
- 34.Lassuy DR, Moran D. 1989. Species profiles: Life histories and environmental requirements of coastal fishes and invertebrates (Pacific Northwest). Pacific herring. in US Fish Wildl Serv Biol Rep (US Army Corps of Engineers, Washington, DC), p 18.
- 35.Mais KF. Pelagic fish surveys in the California Current. Fish Bull. 1974;152:1–79. [Google Scholar]
- 36.Robinson CJ, Arenas FV, Gomez GJ. Diel vertical and offshore-inshore movements of anchovies off the central Baja California coast. J Fish Biol. 1995;47(5):877–892. [Google Scholar]
- 37.Whitlock RE, et al. Direct quantification of energy intake in an apex marine predator suggests physiology is a key driver of migrations. Sci Adv. 2015 doi: 10.1126/sciadv.1400270. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham BS, Grubbs D, Holland K, Popp BN. A rapid ontogenetic shift in the diet of juvenile yellowfin tuna from Hawaii. Mar Biol. 2007;150(4):647–658. [Google Scholar]
- 39.Potier M, et al. Forage fauna in the diet of three large pelagic fishes (lancetfish, swordfish and yellowfin tuna) in the western equatorial Indian Ocean. Fish Res. 2007;83(1):60–72. [Google Scholar]
- 40.Rudershausen PJ, et al. Feeding Ecology of Blue Marlins, Dolphinfish, Yellowfin Tuna, and Wahoos from the North Atlantic Ocean and comparisons with other oceans. Trans Am Fish Soc. 2010;139(5):1335–1359. [Google Scholar]
- 41.Young JW, et al. Feeding ecology and niche segregation in oceanic top predators off eastern Australia. Mar Biol. 2010;157(11):2347–2368. [Google Scholar]
- 42.Cole JS. 1980. Synopsis of biological data on the yellowfin tuna, Thunnus albacares (Bonnaterre, 1788), in the Pacific Ocean. in Synopses of biological data on eight species of scombrids (Inter-Am. Trop. Tuna Comm., La Jolla, CA), pp 71–150.
- 43.Olson RJ, et al. Decadal diet shift in yellowfin tuna Thunnus albacares suggests broad-scale food web changes in the eastern tropical Pacific Ocean. Mar Ecol Prog Ser. 2014;497:157–178. [Google Scholar]
- 44.Goñi N, Logan J, Arrizabalaga H, Jarry M, Lutcavage M. Variability of albacore (Thunnus alalunga) diet in the Northeast Atlantic and Mediterranean Sea. Mar Biol. 2011;158(5):1057–1073. [Google Scholar]
- 45.Laurs RM, Lynn RJ. Biology, Oceanography, and Fisheries of the North Pacific Transition Zone and Subarctic Frontal Zone. In: Wetherall JA, editor. NOAA Technical Report. National Marine Fisheries Service, Honolulu, HI; 1991. p. 97. [Google Scholar]
- 46.Pusineri C, et al. Food and feeding ecology of juvenile albacore, Thunnus alalunga, off the Bay of Biscay: A case study. ICES J Mar Sci. 2005;62(1):116–122. [Google Scholar]
- 47.Consoli P, et al. Feeding habits of the albacore tuna Thunnus alalunga (Perciformes, Scombridae) from central Mediterranean Sea. Mar Biol. 2008;155(1):113–120. [Google Scholar]
- 48.Watanabe H, Kubodera T, Masuda S, Kawahara S. Feeding habits of albacore Thunnus alalunga in the transition region of the central North Pacific. Fish Sci. 2004;70(4):573–579. [Google Scholar]
- 49.Bernard HJ, Hedgepeth JB, Reilly SB. Stomach contents of albacore, skipjack, and bonito caught off southern California during summer 1983. CalCOFI Report. 1985;(26):175–183. [Google Scholar]
- 50.Golet WJ, Cooper AB, Campbell R, Lutcavage ME. Decline in condition of northern bluefin tuna (Thunnus thynnus) in the Gulf of Maine. Fish Bull. 2007;105(3):390–395. [Google Scholar]
- 51.Schick RS, Lutcavage ME. Inclusion of prey data improves prediction of bluefin tuna (Thunnus thynnus) distribution. Fish Oceanogr. 2009;18(1):77–81. [Google Scholar]
- 52.Fromentin J-M. Lessons from the past: Investigating historical data from bluefin tuna fisheries. Fish Fish. 2009;10(2):197–216. [Google Scholar]
- 53.Collette B, et al. 2011. Thunnus thynnus. The IUCN Red List of Threatened Species. Version 2014.3. Available at www.iucnredlist.org. Accessed May 30, 2015.
- 54.Collette B, et al. 2011. Thunnus maccoyii. The IUCN Red List of Threatened Species. Version 2014.3. Available at www.iucnredlist.org. Accessed May 30, 2015.
- 55.Taylor NG, McAllister MK, Lawson GL, Carruthers T, Block BA. Atlantic bluefin tuna: A novel multistock spatial model for assessing population biomass. PLoS ONE. 2011;6(12):e27693. doi: 10.1371/journal.pone.0027693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fromentin J-M, Bonhommeau S, Arrizabalaga H, Kell LT. The spectre of uncertainty in management of exploited fish stocks: The illustrative case of Atlantic bluefin tuna. Mar Policy. 2014;47(0):8–14. [Google Scholar]
- 57.ISC . Stock Assessment of Bluefin Tuna in the Pacific Ocean in 2014. International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean, La Jolla, CA; 2014. [Google Scholar]
- 58.Zwolinski JP, Demer DA. A cold oceanographic regime with high exploitation rates in the Northeast Pacific forecasts a collapse of the sardine stock. Proc Natl Acad Sci USA. 2012;109(11):4175–4180. doi: 10.1073/pnas.1113806109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.