Significance
The natural product leinamycin (LNM), upon reductive activation by cellular thiols, exerts its antitumor activity by an episulfonium ion-mediated DNA alkylation. Manipulation of the LNM biosynthesis in Streptomyces atroolivaceus S-140 yielded a recombinant strain that produced an LNM biosynthetic intermediate, leinamycin E1 (LNM E1). Complementary to the reductive activation of LNM by cellular thiols, LNM E1 can be oxidatively activated by cellular reactive oxygen species (ROS) to generate a similar episulfonium ion intermediate, thereby alkylating DNA and leading to eventual cell death. The feasibility of exploiting LNM E1 as an anticancer prodrug activated by ROS was demonstrated in two prostate cancer cell lines, LNCaP and DU-145. The structure of LNM E1 also reveals critical new insights into LNM biosynthesis.
Keywords: cancer targeting, drug discovery, natural product, pathway engineering, sulfur metabolism
Abstract
Leinamycin (LNM) is a potent antitumor antibiotic produced by Streptomyces atroolivaceus S-140, featuring an unusual 1,3-dioxo-1,2-dithiolane moiety that is spiro-fused to a thiazole-containing 18-membered lactam ring. Upon reductive activation in the presence of cellular thiols, LNM exerts its antitumor activity by an episulfonium ion-mediated DNA alkylation. Previously, we have cloned the lnm gene cluster from S. atroolivaceus S-140 and characterized the biosynthetic machinery responsible for the 18-membered lactam backbone and the alkyl branch at C3 of LNM. We now report the isolation and characterization of leinamycin E1 (LNM E1) from S. atroolivacues SB3033, a ΔlnmE mutant strain of S. atroolivaceus S-140. Complementary to the reductive activation of LNM by cellular thiols, LNM E1 can be oxidatively activated by cellular reactive oxygen species (ROS) to generate a similar episulfonium ion intermediate, thereby alkylating DNA and leading to eventual cell death. The feasibility of exploiting LNM E1 as an anticancer prodrug activated by ROS was demonstrated in two prostate cancer cell lines, LNCaP and DU-145. Because many cancer cells are under higher cellular oxidative stress with increased levels of ROS than normal cells, these findings support the idea of exploiting ROS as a means to target cancer cells and highlight LNM E1 as a novel lead for the development of anticancer prodrugs activated by ROS. The structure of LNM E1 also reveals critical new insights into LNM biosynthesis, setting the stage to investigate sulfur incorporation, as well as the tailoring steps that convert the nascent hybrid peptide–polyketide biosynthetic intermediate into LNM.
Leinamycin (LNM) (Fig. 1A, 1) is an antitumor antibiotic produced by Streptomyces atroolivaceus S-140 (1). It features an unusual 1,3-dioxo-1,2-dithiolane moiety that is spiro-fused to a thiazole-containing 18-membered lactam ring, a molecular architecture not found to date in any other natural product (Fig. 1A). LNM shows potent antitumor activity in vitro and in vivo and is active against tumors that are resistant to clinically important anticancer drugs, such as cisplatin, doxorubicin, mitomycin, and cyclophosphamide; therefore 1 has been pursued as a promising anticancer drug lead (2–4).
Fig. 1.
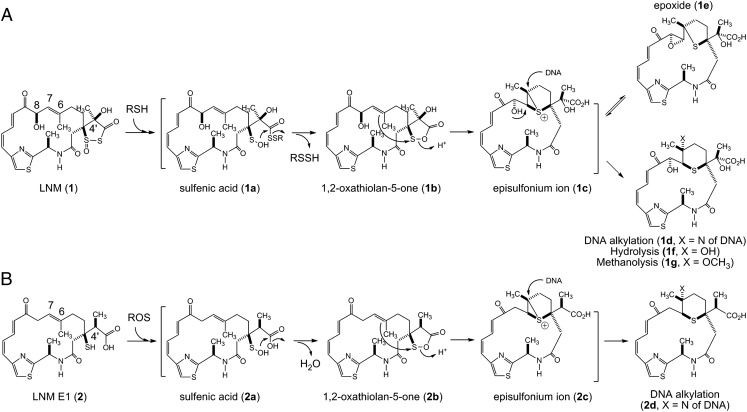
LNM (1) and LNM E1 (2) as novel anticancer drug leads via episulfonium ion-mediated DNA alkylation upon complementary activation: (A) reductive activation of 1 by cellular thiols and (B) oxidative activation of 2 by cellular ROS, affording a pair of episulfonium ions that efficiently alkylate the N7 position of deoxyguanosine bases of DNA, thereby causing DNA cleavage and eventual cell death.
The mode of action of 1 has been extensively investigated. The 1,3-dioxo-1,2-dithiolane moiety of 1 is essential for its antitumor activity. Upon reductive activation in the presence of thiol agents, 1 exerts its antitumor activity by an episulfonium ion-mediated DNA alkylation, a mode of action that is unprecedented among all DNA-damaging natural products (Fig. 1A). Thus, under a reductive cellular environment, the 1,3-dioxo-1,2-dithiolane moiety of 1 is first attacked by a thiol to produce a sulfenic acid intermediate (1a) that can cyclize, by ejecting a persulfide, to form a 1,2-oxathiolan-5-one intermediate (1b). Subsequent rearrangement of 1b, through an intramolecular attack of the 1,2-oxathiolan-5-one moiety by the C-6/C-7 alkene, affords an episulfonium ion intermediate (1c), which can efficiently alkylate the N7 position of deoxyguanosine bases of DNA (1d), ultimately causing DNA cleavage and cell death. The episulfonium ion 1c exists in equilibrium with an epoxide 1e through the intramolecular backside attack by the C-8-hydroxyl group (Fig. 1A) (4–11). Although 1c formation does not require DNA, because the hydrolysis product (1f) forms rapidly in the absence of DNA (5), noncovalent DNA binding by the Z,E-5-(thiazol-4-yl)-penta-2,4-dienone moiety of 1 significantly enhances its DNA alkylation activity (9). In addition, the persulfides generated in the formation of 1b also contribute to the observed DNA damaging activity of 1, but via an independent mechanism mediated by reactive oxygen species (ROS) (4, 12–14).
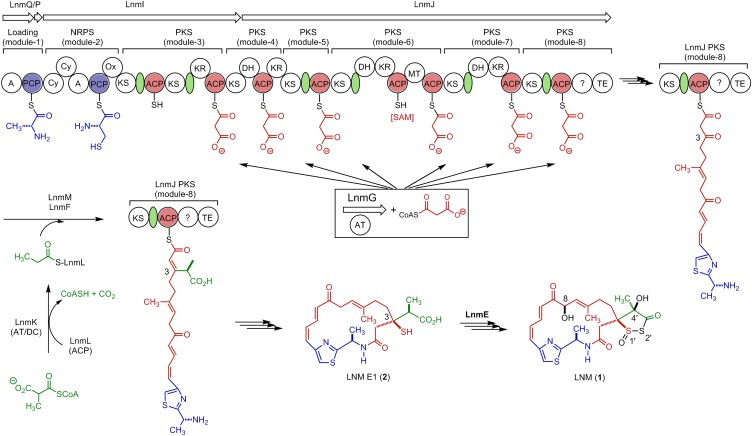
We have previously cloned and sequenced the lnm biosynthetic gene cluster from S. atroolivaceus S-140, which consists of 27 ORFs (orfs) (15–17). In vivo and in vitro characterizations of the 1 biosynthetic machinery have since established that (i) the thiazole-containing 18-membered lactam backbone of 1 is synthesized by a hybrid nonribosomal peptide synthetase (NRPS)-acyltransferase (AT)–less type I polyketide synthase (PKS), consisting of LnmQ (adenylation protein), LnmP [peptidyl carrier protein (PCP)], LnmI (a hybrid NRPS-AT–less type I PKS), LnmJ (AT-less type I PKS), and LnmG (AT) (17–19) and (ii) the alkyl branch at C-3 of 1 is installed by a novel pathway for β-alkylation in polyketide biosynthesis, featuring LnmK (acyltransferase/decarboxylase), LnmL [acyl carrier protein (ACP)], LnmM [hydroxymethylglutaryl-CoA synthase (HCS)], and LnmF [enoyl-CoA hydratase (ECH)] (Fig. 2) (20–22). However, the mechanism of sulfur incorporation to form the 1,3-dioxo-1,2-dithiolane moiety, which is essential for the DNA damage activity of 1, as well as the tailoring steps that convert the nascent hybrid peptide–polyketide intermediate into 1, remains elusive.
Fig. 2.
Proposed biosynthetic pathway for LNM (1) featuring (i) the LnmQPIJ hybrid NRPS-AT–less type I PKS with the discrete LnmG AT loading the malonyl CoA extender units to all six PKS modules, (ii) the LnmKLMF enzymes catalyzing the β-alkyl branch at C3, and (iii) LNM E1 (2) as a key intermediate for 1 biosynthesis, setting the stage to investigate the mechanism of sulfur incorporation and the tailoring steps that convert 2 to 1, requiring minimally hydroxylations at C-8 and C-4′, oxidation at S-1′, S-insertion at 2′, and 1,3-dioxo-1,2-dithiolane ring formation. Color coding indicates the moieties installed by NRPS (blue), PKS (red), β-alkyl branch (green), and other tailoring enzymes (black). SAM, S-adenosylmethionine. The green oval denotes an AT docking domain. Domain abbreviations are: A, adenylation; AT, acyltransferase; Cy, cyclization; DH, dehydratase; KR, ketoreductase; KS, ketosynthase; MT, methyltransferase, Ox, oxidation; PCP, peptidyl carrier protein; TE, thioesterase; ?, domain of unknown function.
To shed light into these steps, we systematically inactivated all genes within the lnm cluster whose functions or roles in 1 biosynthesis cannot be readily predicted on the basis of bioinformatics analysis alone (16, 17). Each of the mutant strains was then fermented, with the S. atroolivaceus S-140 WT as a control, to isolate new metabolites accumulated to account for their roles in 1 biosynthesis. Here, we report the isolation and characterization of leinamycin E1 (LNM E1, 2) from SB3033, a ΔlnmE mutant strain of S. atroolivaceus S-140. Significantly, the structure of 2 reveals critical new insights into 1 biosynthesis, setting the stage to investigate sulfur incorporation, as well as the tailoring steps that convert the nascent hybrid peptide–polyketide biosynthetic intermediate into 1 (Fig. 2). Most strikingly, 2 can be readily oxidized into a sulfenic acid intermediate (2a), which, in a mechanistic analogy to 1a and via a similar 1,2-oxathiolan-5-one intermediate (2b) to 1b, can undergo further rearrangement to form an episulfonium ion intermediate (2c). Thus, complementary to the reductive activation of 1 by cellular thiols to generate an episulfonium ion intermediate, 2 can be oxidatively activated by cellular ROS to generate a similar episulfonium ion intermediate, thereby alkylating DNA and leading to eventual cell death (Fig. 1B). The feasibility of exploiting 2 as an anticancer prodrug activated by ROS was demonstrated in two prostate cancer cell lines, LNCaP and DU-145, that are known to exist under higher oxidative stress than normal tissues. Because many cancer cells are under higher cellular oxidative stress with increased levels of ROS than normal cells (23, 24), our results suggest a means of exploiting ROS to target cancer cells and highlight 2 as a novel lead for the development of anticancer prodrugs activated by ROS.
Results and Discussion
Inactivation of lnmE Abolishing 1 Production and Accumulating 2 as the Major Metabolite in SB3033.
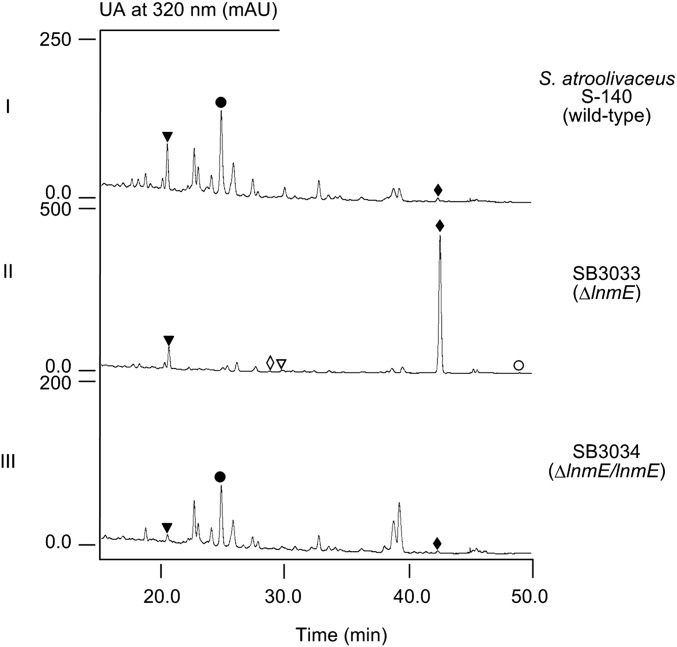
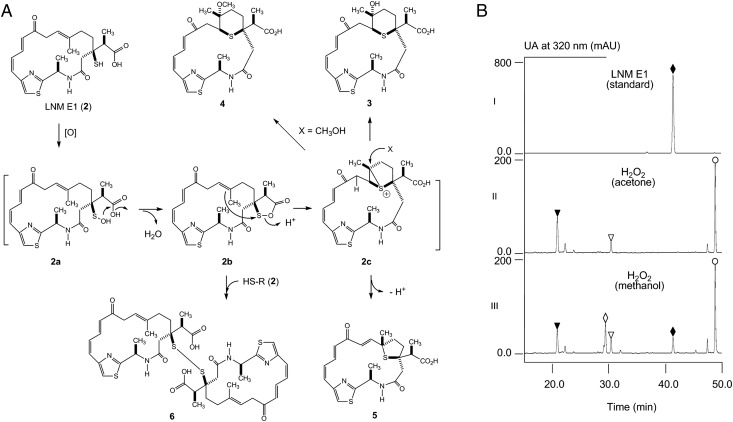
The lnm biosynthetic gene cluster consists of 27 orfs, and 9 of them (i.e., lnmQPIJGKLMF) have been characterized for their roles in 1 biosynthesis (15–22). The functions of the remaining 18 orfs in 1 biosynthesis cannot be readily predicted by bioinformatics analysis alone (16, 17). The lnmE gene, 1 of the 18 orfs, encodes a protein of 307 amino acids, showing no significant homology to any functionally characterized protein. To determine the role of lnmE in 1 production, an lnmE in-frame deletion mutant strain SB3033 was constructed (SI Appendix), the genotype of which was confirmed by Southern analysis (SI Appendix, Fig. S1). SB3033 was fermented according to previous conditions (15–21) using the S. atroolivaceus S-140 WT as a control (SI Appendix). HPLC analysis showed that SB3033 abolished 1 production but accumulated one major new metabolite, named LNM E1 (2), with the same UV absorption spectrum as 1 (Fig. 3, traces I and II). LNM E1 was purified by a combination of chromatographic methods, and its structure was determined by spectroscopic methods (SI Appendix).
Fig. 3.
HPLC analysis of the crude products isolated from fermentation cultures of S. atroolivaceus S-140 WT (I), SB3033 (i.e., ΔlnmE) (II), and SB3034 (i.e., ΔlnmE/lnmE) (III). The metabolites with similar UV absorption are 1 (●), 2 (♦), 3 (▼), 4 (◊), 5 (∇), and 6 (○).
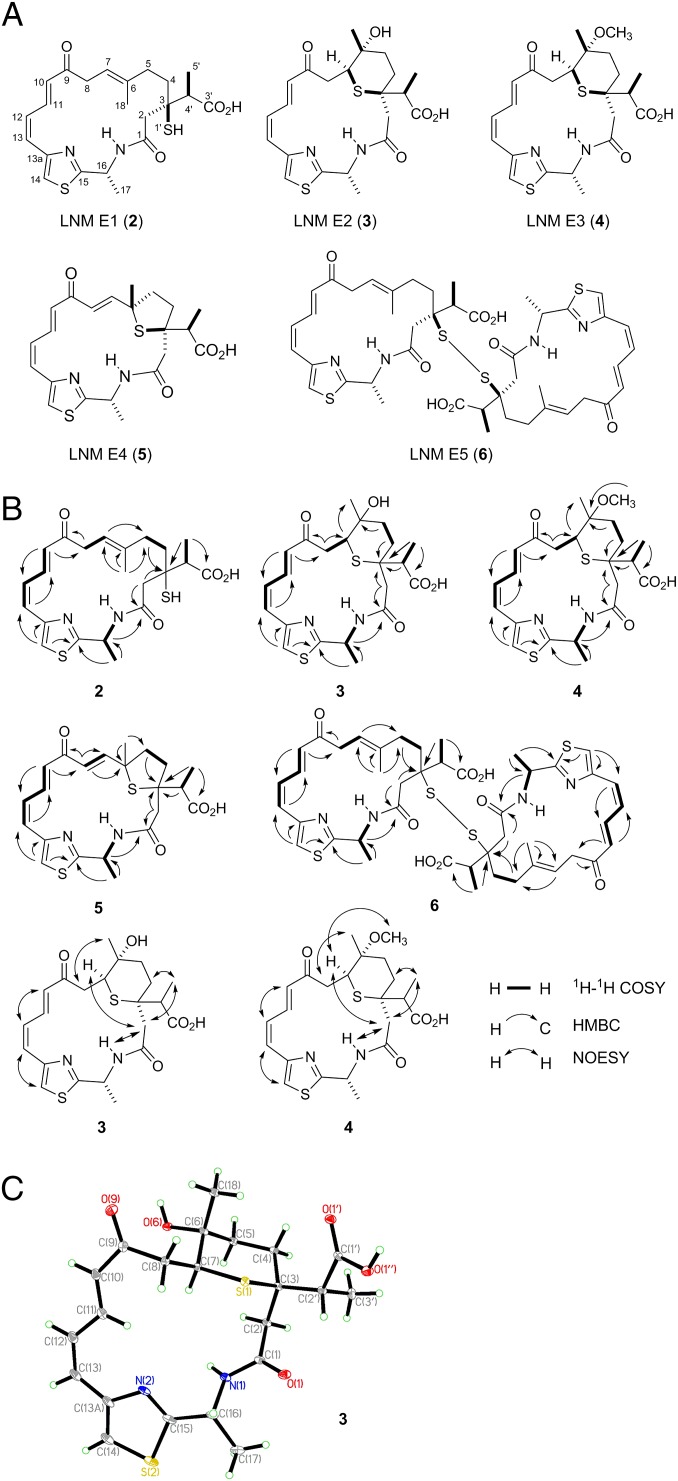
LNM E1 was isolated as a yellow solid and had the molecular formula of C22H28O4N2S2 as determined by analysis of electrospray ionization-mass spectrometry (ESI-MS) and NMR spectroscopic data, which was verified by high resolution ESI-MS (HR-ESI-MS) (found m/z 449.1562, calculated [M + H]+ ion at m/z 449.1563) (SI Appendix, Fig. S2), requiring 10 degrees of unsaturation. Its UV absorption and NMR spectroscopic data (SI Appendix, Table S2 and Figs. S3–S7) were analogous to those of 1. The main differences between 2 and 1 are two hydroxylated carbons and the 1,3-dioxo-1,2-dithiolane moiety. Two hydroxylated carbons (C-8 and C-4′) and a thioester carbonyl carbon (C-3′, δC around 205) of 1 were absent in the NMR spectra of 2 whereas an sp3 methylene (C-8, δH 3.34 and 3.26; δC 40.6) and an sp3 methine (C-4′, δH 2.84; δC 46.3) and a carbonyl carbon (C-3′, δC 175.1) were present in 2. This information led to the conclusion that C-8 and C-4′ were not oxygenated and C-3′ was a free carboxylic acid. Also, the chemical shift of quaternary carbon C-3 (δC 50.2) of 2 was shifted upfield ∼25 ppm in comparison with 1, which indicated that the C-3 was substituted with a thiol moiety. Detailed 2D NMR [COSY, heteronuclear multiple-bond correlation spectroscopy (HMBC), and heteronuclear multiple-quantum correlation spectroscopy (HMQC)] experiments confirmed above assignments (Fig. 4). The absolute configuration of C-4′ was established as S based on X-ray crystallographic analysis of 3.
Fig. 4.
Metabolites isolated from the ΔlnmE mutant strain S. atroolivaceus SB3033: (A) structures of LNM E1 (2) and its minor cometabolites LNM E2 (3), LNM E3 (4), LNM E4 (5), and LNM E5 (6), (B) key 1H-1H COSY, 1H-13C HMBC, and 1H-1H NOESY correlations supporting their structural elucidation (SI Appendix, Figs. S3–S27), and (C) X-ray structure of 3 (SI Appendix), establishing the absolute stereochemistry of 3, as well as 2, 4, 5, and 6 on the basis of their shared biosynthetic origin.
To complement the ΔlnmE mutation in SB3033, pBS3108, a construct in which the expression of lnmE is under the control of the constitutive ErmE* promoter (25), was introduced into SB3033 by conjugation to afford SB3034 (SI Appendix). SB3034 was fermented under the conditions known for 1 production (15–22) with the S. atroolivaceus WT as a control. HPLC analysis showed that SB3034 restored 1 production to a level comparable with that of the WT (Fig. 3, traces I and III). Collectively, these data unambiguously established the intermediacy of 2 in 1 biosynthesis, suggesting that one of the two sulfur atoms is most likely incorporated after the β-alkylation steps but before the thioesterase (TE)-mediated 18-membered lactam ring formation. On the basis of these findings, we now propose 2 as the nascent product of the LNM hybrid NRPS-AT–less type I PKS, setting the stage to investigate the mechanism of sulfur incorporation, as well as the tailoring steps that convert 2 to 1, requiring minimally hydroxylations at C-8 and C-4′, oxidation at S-1′, S-insertion at 2′, and 1,3-dioxo-1,2-dithiolane ring formation (Fig. 2).
Oxidative Activation of 2 Resulting in an Episulfonium Ion Intermediate Similar to That Generated in Reductive Activation of 1.
The potent antitumor activity of 1 has been attributed to its ability, upon reductive activation in the presence of thiols, to form an episulfonium ion intermediate, which can efficiently alkylate DNA, ultimately causing DNA cleavage and cell death. In the absence of DNA, the episulfonium ion intermediate can also react with H2O or CH3OH to rapidly form 1f or 1g, respectively, which has served as the evidence supporting the involvement of the episulfonium ion intermediate (Fig. 1A) (4–11).
A close examination of the HPLC profile of SB3033 revealed at least four additional minor metabolites, 3–6, with the same UV absorption as 1 and 2, whose relative abundance, however, seemed to be dependent on the solvents and time used to prepare the crude products (Fig. 3, trace II). With the exception of 4, which was produced in trace quantity but can be readily detected by liquid chromatography-mass spectrometry (LC-MS) analysis, 3, 5, and 6, were individually purified, together with 2, from the large-scale fermentation of SB3033 (SI Appendix).
The structures of 3, 5, and 6 were established by the HR-ESI-MS (SI Appendix, Fig. S2) and comprehensive NMR spectroscopic analyses (Fig. 4 and SI Appendix, Table S2 and Figs. S8–S13 and S20–S27). The relative stereochemistry of C-3, C-6, and C-7 in 3 was established using information from the NOESY spectrum (Fig. 4B) and comparison of its spectroscopic data to those of 1g (Fig. 1A). The absolute stereochemistry of C-3 and C-16 in 3 as in 1 was assigned based on a shared biosynthetic origin. The strong NOESY correlations of H-7 with H-2a and of H3-18 with H-8b defined the relative stereochemistry of C-6, C-7, and C-3 in 3 (Fig. 4B). The last question to be settled was the absolute configuration of the branch carbon, C-4′, in 2–6, which could not be determined from the NMR data alone due to the free rotation of the carbon–carbon bond between C-4′ and C-3. Thus, a single crystal of 3 was successfully obtained from a mixture of methanol/water (10:0.2), and X-ray crystallographic analysis established its absolute configuration at C-4′ as S (SI Appendix), thereby the same S configuration at C-4′ for 2, 4, 5, and 6 on the basis of their shared biosynthetic origin (Fig. 4).
The structures of 3 and 5, which are similar to that of 1f and 1e, respectively, indicate that they most likely come from the rearrangement through a similar episulfonium ion intermediate as 1c (Fig. 1). We also found that 6 increased over time in solution presumably due to air oxidation of 2. These two pieces of evidence prompted us to propose that 2, the major product from SB3033, may proceed through oxidative activation to form a similar sulfenic acid (2a), which, in a mechanistic analogy to 1a, could undergo further rearrangement to afford an episulfonium ion 2c, leading to the eventual formation of 3, 4, and 5 (Figs. 1 and 5A).
Fig. 5.
Formation of 3–6 via an oxidative activation and episulfonium ion-mediated rearrangement of 2: (A) A proposed mechanism to account for the formation of 3–6 from 2 and (B) HPLC analysis of the reaction mixtures, confirming rapid conversion of 2 (♦) to 3 (▼), 4 (◊), 5 (∇), and 6 (○) upon exposure to H2O2 in acetone (II) or methanol (III). [O], oxidation as exemplified by H2O2.
To test this hypothesis, we incubated 2 with hydrogen peroxide (H2O2), mimicking oxidative activation (SI Appendix). When the reaction is carried out in acetone, compounds 3, 5, and 6 were indeed produced (Fig. 5B, trace II), whose identities were confirmed by HR-ESI-MS and HPLC analysis in comparison with authentic standards. When the reaction was carried out in methanol, a new compound was produced, in addition to 3, 5 and 6 (Fig. 5B, trace III). This new compound was isolated, and its structure as 4, the methoxyl-adduct of 2c (Fig. 5A), was similarly established on the basis of HR-ESI-MS (SI Appendix, Fig. S2) and NMR analyses (Fig. 4 and SI Appendix, Table S2 and Figs. S14–S19).
LNM E1 (2) Showing Potent Cytotoxicity Against Prostate Cancer Cell Lines LNCaP and DU-145 with Increased Levels of ROS.
The fact that 2, upon oxidative activation, can rapidly form an episulfonium ion intermediate (2c) suggests that 2 could be exploited as an anticancer prodrug activated by ROS. We reasoned that 2 will remain inactive in normal cells, thereby exhibiting minimal systemic toxicity, and could be activated in situ if cells contain high levels of ROS; the resultant 2c may then alkylate DNA, in a mechanistic analogy to 1c, resulting in DNA cleavage and cell death but only targeting cancer cells under high oxidative stress (Fig. 1).
It is now well known that many cancer cells possess higher cellular oxidative stress with increased levels of ROS than normal cells (23, 24). We have also demonstrated that ROS levels in androgen-sensitive LNCaP prostate cancer cells or androgen-insensitive DU-145 prostate cancer cells could be further modulated by selected androgen (26, 27) or polyamine analogs (28, 29), respectively, as well as polyamine oxidase inhibitors (30). We have reported that ROS levels in intact LNCaP and DU-145 cells can be readily estimated by the fluorescent dye 2′,7′-dichloroflurescin diacetate (DCFH) oxidation assay following a published procedure (26, 31). Thus, we first determined the relative levels of ROS in DU-145 and LNCaP cells, with and without the treatments of selected agents, with the normal human prostate epithelial cells (HPECs) as controls (SI Appendix). As summarized in SI Appendix, Table S3, (i) the ROS levels in both LNCaP and DU-145 cell lines are relatively higher than in normal HPEC, (ii) LNCaP cells have relatively higher ROS levels that DU-145 cells, (iii) the ROS levels in DU-145 cells are moderately enhanced after the treatment of 0.1 μM BE-3-3-3 (a polyamine analog), and the observed enhancement is completely abolished by the cotreatment with 25 μM MDL 72527 (a polyamine oxidase inhibitor), and (iv) the ROS levels in LNCaP cells are significantly enhanced after the treatment of either 1 nM R1881 (an androgen analog) or 0.1 μM BE-3-3-3, and the observed enhancements are markedly reduced by the cotreatment with 15 μM vitamin E (an antioxidant) or 25 μM MDL 72527, respectively.
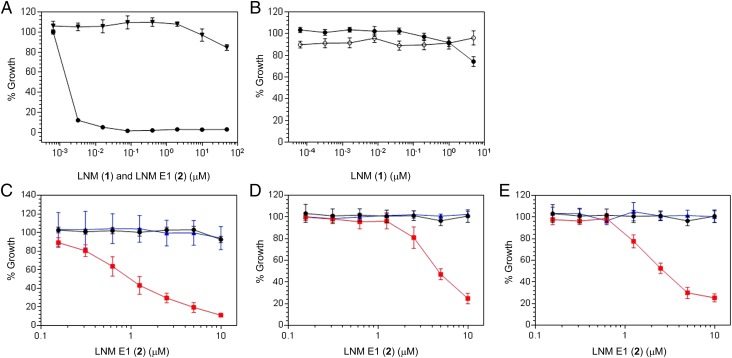
Encouraged by the sharp differences of ROS levels between normal (HPEC) and cancer (DU-145 and LNCaP) cells and our ability to modulate ROS levels using selected agents, we next proceeded to test whether 2 could exert cytotoxicity against cancer cells with elevated levels of ROS (SI Appendix). DNA levels were measured as an indicator of cell proliferation, and 1 was used as a control (Fig. 6). (i) For normal HPEC, 1 was a potent growth inhibitor (IC50 ∼2 nM) whereas 2 showed little growth inhibitory activity even at μM concentration (Fig. 6A). These findings would be expected with the reductive cellular environment of normal HPEC (SI Appendix, Table S3). (ii) In contrast, 1 was essentially inactive against DU-145 and LNCaP cells. These findings are in good agreement with the fact that these two cell lines are under high oxidative stress (Fig. 6B and SI Appendix, Table S3). (iii) However, 2 was also inactive against DU-145 and LNCaP cells, suggesting that the basal levels of ROS in these two cell lines are not high enough to activate 2 (Fig. 6 C–E and SI Appendix, Table S3). (iv) At the elevated ROS levels upon treatments of DU-145 cells with BE-3-3-3 or LNCaP cells with either R1881 or BE-3-3-3, 2 indeed potently inhibited the growth of both DU-145 (IC50 ∼1 μM) and LNCaP cells (IC50 ∼4 μM and ∼2 μM, respectively, with R1881 and BE-3-3-3). Significantly, the observed growth inhibitory activities of 2 against both cell lines were completely abrogated by the cotreatments of DU-145 with BE-3-3-3 and MDL 72527 or LNCaP with BE-3-3-3 and MDL 72527 or R1881 and vitamin E, respectively (Fig. 6 C–E). Both MDL 72527, a polyamine oxidase inhibitor, and vitamin E, an antioxidant, are known to reduce ROS levels in DU-145 or LNCaP cells (SI Appendix, Table S3) (26, 27, 30). These data therefore unambiguously correlated the cytotoxicity of 2 with ROS-mediated activation in cells and supported the development of 2 as an anticancer prodrug exploiting the elevated levels of ROS in many cancer cells. Anticancer prodrugs activated by ROS in selected cancer cell lines are known but are few (32–36). The unprecedented mode of action of 2, via oxidative activation and episulfonium ion-mediated DNA alkylation, is very exciting (Fig. 1), providing an outstanding opportunity to explore the novel scaffold of 2 and related natural products for anticancer drug discovery.
Fig. 6.
Growth inhibitory activity for LNM E1 (2) against normal (HPEC) and cancer (DU-145 and LNCaP) cells treated with selected agents modulating ROS levels and compared with LNM (1) as a control. DNA levels were measured as an indicator of cell growth, and all data were normalized to growth effect with vehicle as controls. (A) Effects of 1 (●) and 2 (▼) on normal HPEC. (B) Effects of 1 on DU-145 (●) or LNCaP (○) cells. (C) Effects of 2 on DU-145 cells (●) or DU-145 cells treated with BE-3-3-3 (red filled square) or BE-3–3-3 and MDL 72527 (blue triangle). (D) Effects of 2 on LNCaP cells (●) or LNCaP cells treated with R1881 (red filled square) or R1881 and vitamin E (blue triangle). (E) Effects of 2 on LNCaP cells (●) or LNCaP cells treated with BE-3-3-3 (red filled square) or BE-3-3-3 and MDL 72527 (blue triangle).
Methods
Materials, methods, and detailed experimental procedures are provided in SI Appendix. SI Appendix, Table S1 contains plasmids and strains used in this study, SI Appendix, Table S2 compiles all of the 1H and 13C NMR data for 2–6, and SI Appendix, Table S3 summarizes ROS levels in human prostate cells under the conditions studied in DCF fluorescent units estimated by the DCFH oxidation assay. SI Appendix, Fig. S1 depicts the construction and confirmation of the ΔlnmE mutant strains SB3032 and SB3033. SI Appendix, Fig. S2 summarizes all of the HR-ESI-MS spectra of 2–6. SI Appendix, Figs. S3–S7, Figs. S8–S13, Figs. S14–S19, Figs. S20–S24, and Figs. S25–S27 are 1H and 13C NMR spectra of 2–6, respectively, supporting their structural determination.
Supplementary Material
Acknowledgments
We thank Kyowa Hakko Kogyo Co. Ltd. for the WT S. atroolivaceus S-140 strain, the NMR Core facility at The Scripps Research Institute for obtaining 1H and 13C NMR data, and Dr. Xiao-Nian Li (Kunming Institute of Botany) for collecting the X-ray data of 3. This work was supported in part by NIH Grant CA106150.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Cambridge Structural Database, Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CSD reference no. 1036770).
See Commentary on page 8164.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506761112/-/DCSupplemental.
References
- 1.Hara M, et al. Leinamycin, a new antitumor antibiotic from Streptomyces: Producing organism, fermentation and isolation. J Antibiot (Tokyo) 1989;42(12):1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 2.Hara M, Saitoh Y, Nakano H. DNA strand scission by the novel antitumor antibiotic leinamycin. Biochemistry. 1990;29(24):5676–5681. doi: 10.1021/bi00476a005. [DOI] [PubMed] [Google Scholar]
- 3.Ashizawa T, et al. Antitumor activity of KF22678, a novel thioester derivative of leinamycin. Anticancer Drugs. 1999;10(9):829–836. doi: 10.1097/00001813-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Gates KS. Mechanisms of DNA damage by leinamycin. Chem Res Toxicol. 2000;13(10):953–956. doi: 10.1021/tx000089m. [DOI] [PubMed] [Google Scholar]
- 5.Asai A, et al. Thiol-mediated DNA alkylation by the novel antitumor antibiotic leinamycin. J Am Chem Soc. 1996;118(28):6802–6803. [Google Scholar]
- 6.Breydo L, Zang H, Mitra K, Gates KS. Thiol-independent DNA alkylation by leinamycin. J Am Chem Soc. 2001;123(9):2060–2061. doi: 10.1021/ja003309r. [DOI] [PubMed] [Google Scholar]
- 7.Nooner T, Dutta S, Gates KS. Chemical properties of the leinamycin-guanine adduct in DNA. Chem Res Toxicol. 2004;17(7):942–949. doi: 10.1021/tx049964k. [DOI] [PubMed] [Google Scholar]
- 8.Viswesh V, Gates K, Sun D. Characterization of DNA damage induced by a natural product antitumor antibiotic leinamycin in human cancer cells. Chem Res Toxicol. 2010;23(1):99–107. doi: 10.1021/tx900301r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fekry MI, et al. Noncovalent DNA binding drives DNA alkylation by leinamycin: Evidence that the Z,E-5-(thiazol-4-yl)-penta-2,4-dienone moiety of the natural product serves as an atypical DNA intercalator. J Am Chem Soc. 2011;133(44):17641–17651. doi: 10.1021/ja2046149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viswesh V, Hays AM, Gates K, Sun D. DNA cleavage induced by antitumor antibiotic leinamycin and its biological consequences. Bioorg Med Chem. 2012;20(14):4413–4421. doi: 10.1016/j.bmc.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivaramakrishnan S, Breydo L, Sun D, Gates KS. The macrocycle of leinamycin imparts hydrolytic stability to the thiol-sensing 1,2-dithiolan-3-one 1-oxide unit of the natural product. Bioorg Med Chem Lett. 2012;22(11):3791–3794. doi: 10.1016/j.bmcl.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra K, Kim W, Daniels JS, Gates KS. Oxidative DNA cleavage by the antitumor antibiotic leinamycin and simple 1,2-dithiolan-3-one 1-oxides: Evidence for thio-dependent conversion of molecular oxygen to DNA-cleaving oxygen radicals mediated by polysulfides. J Am Chem Soc. 1997;119(48):11691–11692. [Google Scholar]
- 13.Chatterji T, et al. Small molecules that mimic the thiol-triggered alkylating properties seen in the natural product leinamycin. J Am Chem Soc. 2003;125(17):4996–4997. doi: 10.1021/ja029169y. [DOI] [PubMed] [Google Scholar]
- 14.Keerthi K, Rajapakse A, Sun D, Gates KS. Synthesis and characterization of a small analogue of the anticancer natural product leinamycin. Bioorg Med Chem. 2013;21(1):235–241. doi: 10.1016/j.bmc.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng YQ, Tang GL, Shen B. Identification and localization of the gene cluster encoding biosynthesis of the antitumor macrolactam leinamycin in Streptomyces atroolivaceus S-140. J Bacteriol. 2002;184(24):7013–7024. doi: 10.1128/JB.184.24.7013-7024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci USA. 2003;100(6):3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang GL, Cheng YQ, Shen B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem Biol. 2004;11(1):33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Tang GL, Cheng YQ, Shen B. Polyketide chain skipping mechanism in the biosynthesis of the hybrid nonribosomal peptide-polyketide antitumor antibiotic leinamycin in Streptomyces atroolivaceus S-140. J Nat Prod. 2006;69(3):387–393. doi: 10.1021/np050467t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang GL, Cheng YQ, Shen B. Chain initiation in the leinamycin-producing hybrid nonribosomal peptide/polyketide synthetase from Streptomyces atroolivaceus S-140: Discrete, monofunctional adenylation enzyme and peptidyl carrier protein that directly load D-alanine. J Biol Chem. 2007;282(28):20273–20282. doi: 10.1074/jbc.M702814200. [DOI] [PubMed] [Google Scholar]
- 20.Liu T, Huang Y, Shen B. Bifunctional acyltransferase/decarboxylase LnmK as the missing link for beta-alkylation in polyketide biosynthesis. J Am Chem Soc. 2009;131(20):6900–6901. doi: 10.1021/ja9012134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, et al. Characterization of the lnmKLM genes unveiling key intermediates for β-alkylation in leinamycin biosynthesis. Org Lett. 2011;13(3):498–501. doi: 10.1021/ol102838y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohman JR, Bingman CA, Phillips GN, Jr, Shen B. Structure of the bifunctional acyltransferase/decarboxylase LnmK from the leinamycin biosynthetic pathway revealing novel activity for a double-hot-dog fold. Biochemistry. 2013;52(5):902–911. doi: 10.1021/bi301652y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerutti PA. Prooxidant states and tumor promotion. Science. 1985;227(4685):375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 24.Pizzimenti S, Toaldo C, Pettazzoni P, Dianzani MU, Barrera G. The “two-faced” effects of reactive oxygen species and the lipid peroxidation product 4-hydroxynonenal in the hallmarks of cancer. Cancers (Basel) 2010;2(2):338–363. doi: 10.3390/cancers2020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser T, Bibb MJ, Butter MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 26.Ripple MO, Henry WF, Rago RP, Wilding G. Prooxidant-antioxidant shift induced by androgen treatment of human prostate carcinoma cells. J Natl Cancer Inst. 1997;89(1):40–48. doi: 10.1093/jnci/89.1.40. [DOI] [PubMed] [Google Scholar]
- 27.Ripple MO, Henry WF, Schwarze SR, Wilding G, Weindruch R. Effect of antioxidants on androgen-induced AP-1 and NF-kappaB DNA-binding activity in prostate carcinoma cells. J Natl Cancer Inst. 1999;91(14):1227–1232. doi: 10.1093/jnci/91.14.1227. [DOI] [PubMed] [Google Scholar]
- 28.Kee K, et al. Metabolic and antiproliferative consequences of activated polyamine catabolism in LNCaP prostate carcinoma cells. J Biol Chem. 2004;279(26):27050–27058. doi: 10.1074/jbc.M403323200. [DOI] [PubMed] [Google Scholar]
- 29.Kee K, et al. Activated polyamine catabolism depletes acetyl-CoA pools and suppresses prostate tumor growth in TRAMP mice. J Biol Chem. 2004;279(38):40076–40083. doi: 10.1074/jbc.M406002200. [DOI] [PubMed] [Google Scholar]
- 30.Casero RA, Jr, et al. The role of polyamine catabolism in anti-tumour drug response. Biochem Soc Trans. 2003;31(2):361–365. doi: 10.1042/bst0310361. [DOI] [PubMed] [Google Scholar]
- 31.Basu HS, et al. A small molecule polyamine oxidase inhibitor blocks androgen-induced oxidative stress and delays prostate cancer progression in the transgenic adenocarcinoma of the mouse prostate model. Cancer Res. 2009;69(19):7689–7695. doi: 10.1158/0008-5472.CAN-08-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng X, Gandhi V. ROS-activated anticancer prodrugs: A new strategy for tumor-specific damage. Ther Deliv. 2012;3(7):823–833. doi: 10.4155/tde.12.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Major Jourden JL, Cohen SM. Hydrogen peroxide activated matrix metalloproteinase inhibitors: A prodrug approach. Angew Chem Int Ed Engl. 2010;49(38):6795–6797. doi: 10.1002/anie.201003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang Y, Balakrishnan K, Gandhi V, Peng X. Hydrogen peroxide inducible DNA cross-linking agents: Targeted anticancer prodrugs. J Am Chem Soc. 2011;133(48):19278–19281. doi: 10.1021/ja2073824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao S, Wang Y, Peng X. ROS-inducible DNA cross-linking agent as a new anticancer prodrug building block. Chemistry. 2012;18(13):3850–3854. doi: 10.1002/chem.201200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagen H, et al. Aminoferrocene-based prodrugs activated by reactive oxygen species. J Med Chem. 2012;55(2):924–934. doi: 10.1021/jm2014937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.