Abstract
Small molecules that bind to tubulin exert powerful effects on cell division and apoptosis (programmed cell death). Cell-based high-throughput screening combined with chemo/bioinformatic and biochemical analyses recently revealed a novel compound MI-181 as a potent mitotic inhibitor with heightened activity towards melanomas. MI-181 causes tubulin depolymerization, activates the spindle assembly checkpoint arresting cells in mitosis, and induces apoptotic cell death. C2 is an unrelated compound previously shown to have lethal effects on microtubules in tumorigenic cell lines. We report 2.60 Å and 3.75 Å resolution structures of MI-181 and C2, respectively, bound to a ternary complex of αβ-tubulin, the tubulin-binding protein stathmin, and tubulin tyrosine ligase. In the first of these structures, our crystallographic results reveal a unique binding mode for MI-181 extending unusually deep into the well-studied colchicine-binding site on β-tubulin. In the second structure the C2 compound occupies the colchicine-binding site on β-tubulin with two chemical moieties recapitulating contacts made by colchicine, in combination with another system of atomic contacts. These insights reveal the source of the observed effects of MI-181 and C2 on microtubules, mitosis, and cultured cancer cell lines. The structural details of the interaction between tubulin and the described compounds may guide the development of improved derivative compounds as therapeutic candidates or molecular probes to study cancer cell division.
Keywords: tubulin, microtubule, mitosis inhibitor, cancer, cytoskeleton, crystal structure
Introduction
The development of tubulin-targeting compounds that affect the rapid cell division cycle of cancerous cells is a treatment approach that remains clinically relevant with several successful examples. However, many chemotherapeutic agents in use have problems related to their complex syntheses, toxicities, and resistances observed in clinical cancer settings.1,2 High-throughput screening (HTS) studies have explored many compounds in an effort to identify new, efficacious candidates against cancer cells.3–10 On the basis of the key role microtubules play in cell division, focused screening approaches have examined the effects of diverse compounds on microtubules directly and on the mitotic (M-phase of the cell cycle) events they coordinate. We recently identified a specific M-phase inhibitor, MI-181 (CID 17543402) [Fig. 1(A)], which targets tubulin.11 Extensive characterization revealed inhibition of tubulin polymerization, spindle assembly checkpoint (SAC) activation, mitotic arrest, and induction of apoptosis in cells treated with MI-181. Similarly, prior studies identified C2 (CID 663143) [Fig. 1(A)] as a compound that disrupts microtubule organization and kills cancer cells.12 Fluorescence studies in cultured cells and tubulin polymerization experiments indicated loss of microtubule organization and delayed polymerization kinetics in vitro. However, the compound was postulated to target a protein associated with microtubules and not tubulin directly.
Figure 1.
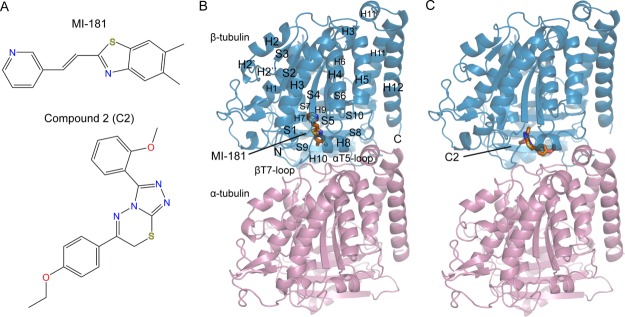
MI-181 and C2 target β-tubulin near the interface between tubulin subunits. A: The chemical structures of MI-181 and C2. B: A single αβ-tubulin heterodimer is shown (α, pink; β, blue) with the two other protein components of the ternary T2R-TTL crystallization assembly removed for clarity. Secondary structural elements, amino-, and carboxy-termini are labeled for β-tubulin. MI-181 (orange) binds deep within the β-subunit away from the αT5 loop. C: Same as B, shown in orange is C2 binding more proximal to the α-subunit in contact with the αT5 loop. Use this link to access the interactive version of this figure
The studies that identified MI-181 and C2 did not determine how each compound bound its prospective target. Structural information from X-ray crystallography enhances HTS techniques by providing chemical insight into how compounds exert their effects. Extensive cryo-electron microscopy analysis of microtubules has been complemented by detailed structural studies of tubulin through stages of advancement from electron crystallography to X-ray crystallography.13–15 Complexation with tubulin binding proteins, in particular stathmin and stathmin-like domains, enabled the first X-ray crystallographic studies of tubulin heterodimers bound to small molecules, which bind to the curved form of stathmin-bound αβ-tubulin (T2R).14,16–21 Stathmin-related domains bind two αβ-tubulin heterodimers in a curved conformation that may represent the unpolymerized population of heterodimers recognized by diverse binding partners.22 Structures of tubulin heterodimers derived from polymerized microtubules exhibit a straight conformation that is not observed in the presence of stathmin or other microtubule-associated proteins.13–15,23–27 Later studies of tubulin modification by tubulin tyrosine ligase (TTL) afforded higher-resolution structures of ternary complexes of tubulin bound to stathmin and TTL (T2R-TTL), and subsequently drug compounds of interest.15,28–30
Here we present the structure of the T2R-TTL ternary tubulin complex bound to MI-181 determined to 2.60 Å resolution. We elucidate the connection between MI-181 binding and its observed effects on mitotic cells by describing the key interactions and the structure of tubulin in the MI-181-bound state. The 3.75 Å resolution crystal structure of T2R-TTL tubulin bound to C2 is also described here. Our work clarifies the binding properties of this compound and reveals the structural basis of its effect on microtubules.
Results and Discussion
Structure determination
The crystal structure of tubulin bound to two ligands described here was obtained using an established system for tubulin crystal growth.15,31 Initial efforts to crystallize tubulin following an approach in which only the stathmin-like domain facilitates crystal growth never recapitulated previously described results.14,32 Briefly, bovine αβ-tubulin, human RB3 or rat STMN4 stathmin-like domain, and chicken TTL reconstitute to form a T2R-TTL complex, which readily crystallizes. The inclusion of TTL enables an alternative crystal packing arrangement resulting in higher resolution diffraction data and has proven effective in studying the binding of diverse microtubule-targeting agents.28–30
A single crystal form grown using T2R-TTL soaked with either MI-181 or C2 gave two crystals from which diffraction data were collected. The crystal soaked with MI-181 exhibited diffraction to at least 2.60 Å and the crystal soaked with C2 diffracted to at least 3.75 Å. The final resolution cutoffs described here were determined optimally from a subset of diffraction images that took into consideration the signal-to-noise ratio for reflection intensities and the random half-data set correlation coefficient, CC1/2.33
The T2R-TTL structures with MI-181 and C2 bound to β-tubulin identify the previously unknown binding sites for each compound [Figs. 1(B) and 1(C)]. Secondary structure elements and sequence numbering of tubulin are based on the initial structural studies of tubulin.13 Both compounds occupy a binding pocket in the intermediate domain of β-tubulin that forms the interface with the α-subunit (residues 206−384). Other structural domains in the β-tubulin subunit include the nucleotide-binding domain (residues 1−205) and a C-terminal helical domain from residue 385 to the C-terminus. Only MI-181 is in proximity to the nucleotide-binding domain, which binds and hydrolyzes GTP within the β-subunit.
Tubulin retains a curved structure in the presence of MI-181 and C2 as observed similarly with colchicine and other molecules that interact with the expansive binding pocket on β-tubulin.16,19,30 Superimposition of the taxol-stabilized straight tubulin heterodimer β-tubulin subunit with the β-tubulin subunit in our heterodimer structures indicates a rotational offset of the α-subunits. An 11° or 9° discrepancy is present depending on whether the respective analysis uses least-squares fitting or mass-weighted axes representing each heterodimer. This is in close structural agreement with the curvature observed in stathmin-bound tubulin with no depolymerizing compounds bound.14,34 The rmsd of β-tubulin over 416 Cα atoms for each compound when superimposed on the apo-T2R-TTL structure is 0.36 Å and 0.34 Å for MI-181 and C2, respectively.15 These results indicate that no large structural rearrangements occur in β-tubulin when bound to MI-181 or C2.
MI-181 and C2 target β-tubulin
The tubulin-MI-181 crystal structure reveals an unanticipated binding mode where MI-181 fills an elongated pocket with contacts to beta strands S5 and S6 of the nucleotide-binding domain of β-tubulin [Fig. 2(A)]. The T5 loop of the α-subunit is not in direct contact with the MI-181 molecule, which contrasts with the interactions that loop makes with many other colchicine-site binding molecules.16,19,30 MI-181 binding does not mimic that of colchicine, whereas it shares similarity with the bound state observed for the molecule TN16.19 A small degree of overlap is observed only between the dimethyl groups of the benzothiazole group of MI-181 and the trimethoxybenzene ring of colchicine [Figs. 3(A) and 3(B)]. The orientation of MI-181 is modeled more confidently than TN16 due to higher resolution diffraction data and a higher degree of asymmetry in the compound chemical structure.
Figure 2.
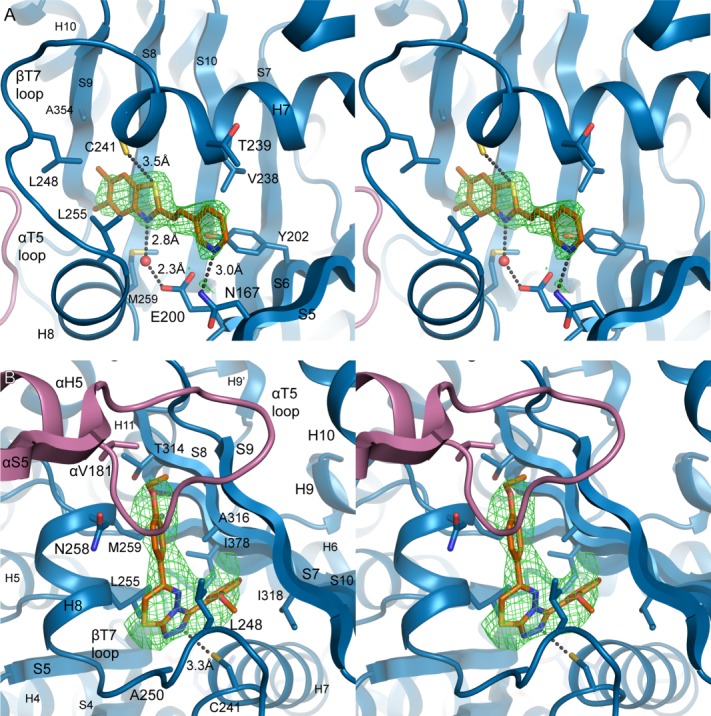
MI-181 and C2 make diverse contacts in the colchicine-binding region of β-tubulin. A: Stereo view of the MI-181 binding site in β-tubulin. Secondary structure and residues interacting with the molecule are labeled. Hydrogen bond distances are indicated and water molecules are shown as red spheres. Overlaid is the σA-weighted Fobs-Fcalc difference electron density, contoured to +3.0σ (green mesh), following molecular replacement and rigid body refinement in the absence of ligands. B: As in A, stereo view of the interactions between C2 and αβ-tubulin. Use this link to access the interactive version of this figure
Figure 3.
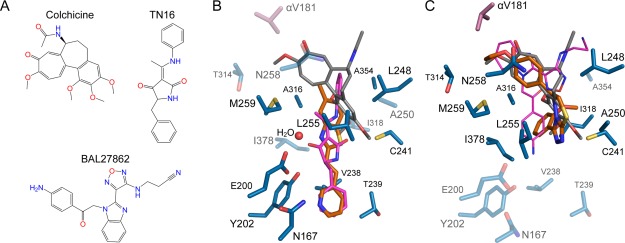
Comparison of MI-181 and C2 binding with colchicine, TN16, and BAL27862 binding on β-tubulin. A: The chemical structures of colchicine, TN16, and BAL27862. B: The MI-181 (orange) binding site with tubulin-bound colchicine (gray) and TN16 (magenta) superimposed using the β-subunit. Residues interacting with MI-181 are shown as sticks and labeled. Residues that interact with C2 are highlighted with transparency. C: As in B, the C2 (orange) binding site superimposed with colchicine (gray) and BAL27862 (magenta). Residues that interact with MI-181 are transparently depicted.
Hydrogen bond acceptors in MI-181 are satisfied by contacts with side chains in β-tubulin as well as one nearby water molecule in the binding site [Fig. 2(A)]. The water molecule is coordinated by the nitrogen of the dimethylbenzothiazole group and the carboxylate of Glu200 on strand 6 with hydrogen bond lengths of 2.8 Å and 2.3 Å, respectively. The nucleotide-binding domain extends an additional residue, Asn167 on strand 5, to hydrogen bond with the pyridine nitrogen (3.0 Å distance between the non-hydrogen atoms). The crystal structure places the thiol group of Cys241 in close proximity to the thiazole sulfur of the bound MI-181 molecule. The geometry and spacing (3.5 Å between heavy atoms) are suggestive of a weak hydrogen bond, with the thiazole sulfur serving as the acceptor. Hydrogen bonding surveys support the occurrence of bona fide hydrogen bonds involving sulfur atoms, but they are relatively uncommon, especially where sulfur acts as the acceptor.35–37 In the present case, for example, where the resolution does not give an indication of the position of the thiol hydrogen, the structural information only suggests a very weak interaction.38 However, Cys241 thiol hydrogen bonding is thought to be a critical bond important in the design of potent microtubule destabilizers.39
Other nonpolar contacts from the surrounding secondary structure line the tubular pocket in which MI-181 binds. These residues include Tyr202 from strand 6, Val238 and Thr239 from helix 7, Leu248 from the T7 loop, Leu255 and Met259 from helix 8, Ala316 from strand 8, and Ala354 from strand 9 [Fig. 2(A)]. The conformation of Leu255 is similar to the conformation observed in the TN16-bound tubulin structure, which differs from that of other colchicine-site ligands.16,19,30 In the bound-state with other compounds, Leu255 of β-tubulin typically occupies the space where the coordinated water molecule is observed near MI-181.
Only one molecule of MI-181 is modeled with full occupancy based on interpretable difference electron density. Inspection of the difference electron density maps during refinement and the introduction of occupancy refinement support the inclusion of a second molecule of MI-181 bound to the second β-tubulin subunit in the asymmetric unit, albeit with less than 100% occupancy. The second molecule is modeled with less than full occupancy and a partially occupied unbound conformation of the T7 loop.
The crystal structure of C2 bound to tubulin elucidated here is similar to structures of colchicine and other compounds bound in the main region of the colchicine-binding site. Two molecules of C2 are bound to the two β-tubulin subunits in the asymmetric unit and were modeled into their respective sites based on difference electron density [Fig. 2(B)]. The diffraction quality of the C2-T2R-TTL crystals warranted cautious placement of the bound molecules that ultimately appears valid at the moderate resolution. The asymmetry of C2, particularly the ethoxyphenyl group and the sulfur-containing thiadiazine ring, enabled unambiguous placement of the molecule. Manual placement was subsequently compared against placement by the LigandFit module of PHENIX and AutoDock Vina.40–42 LigandFit placed the molecule into difference electron density in the same orientation as the final manual choice, and AutoDock Vina failed to dock the molecule in alternative conformations within the binding site.
The three substituent rings of colchicine overlap with the chemical groups of microtubule depolymerizers of diverse chemical structure. C2 overlaps two of the ring structures of colchicine and includes a single group that extends to contact additional secondary structure elements in β-tubulin [Fig. 3(A,C)]. The ethoxyphenyl group of C2 occupies the same binding region as the methoxytropone ring of colchicine, extending towards the α-subunit. Asn258 and Met259 on helix 8, Thr314 on strand 8, and Val181 of the αT5 loop are direct interactions found at the subunit interface. Further into the binding pocket, the triazolothiadiazine group is found roughly in the same position as the trimethoxybenzene ring of colchicine. The pocket of residues surrounding the triazolothiadiazine group includes Cys241 on helix 7 hydrogen-bonding to a triazole nitrogen from 3.3 Å away, Leu248 and Ala250 on the T7 loop, and Leu255 on helix 8.
An additional heptalene ring with an acetamide group in colchicine usually forms more interactions with the protein in this region. However, C2 lacks that moiety and forms new contacts with an extended methoxyphenyl group buried by Ala316 and Ile318 on strand 8, Leu248 on the T7 loop, and Ile378 on strand 10. The amphiphilic aniline group of the microtubule-destabilizing drug BAL27862 was also observed in this region of the binding pocket.30
Biological insights of tubulin-targeting compounds
Two promising compounds identified from HTS with unidentified binding sites have been further elucidated here.11,12 MI-181 interactions with β-tubulin deviate from the footprint of most molecules that bind to the well-studied colchicine-binding site. Of the many crystal structures of tubulin-targeting agents, only the structure of TN16-bound tubulin shares similar binding properties to MI-181.19 TN16 and MI-181 bind in roughly the same spot of the extended pocket near the nucleotide-binding domain of β-tubulin. However, MI-181 is structurally distinct and the benzothiazole scaffold is present in at least eight FDA-approved therapeutics.11 A broad spectrum of melanoma cell lines showed sensitivity to MI-181 with cell-viability IC50 values below 40 nM for most melanomas.11 These properties make MI-181 an attractive candidate for developing new small molecules for the treatment of cancers.
The interactions observed in the crystal structure of C2-bound tubulin directly implicate tubulin as the target of C2. C2 binding to β-tubulin resembles the bound-state of BAL27862, a microtubule depolymerizer of which a prodrug, BAL101553, has entered phase IIa trials in humans.43 The ethoxyphenyl moiety of C2 makes the chemical structure distinct from a panel of caspase activators comprising triazolothiadiazine analogs.44 C2 potently inhibits growth of multiple cell lines derived from fibroblasts and breast tissue.12 The ability to specifically target certain cancerous cell types increases the need to further study the effects of C2 and possible derivatives compatible with the structural details revealed in the crystal structure.
Structural insight into the mechanisms of compounds identified through HTS methods remains important for understanding small molecules with therapeutic potential. Chemical libraries generally contain diverse molecular structures, and this often results in identified hits with unclear targets or binding properties. Structure-activity-relationship (SAR) studies are better guided with the knowledge of what modifications to a molecule may be allowed or forbidden due to the surrounding interactions with its target protein. Other properties of promising candidates may be improved with the development of derivative compounds that show fewer off-target effects, improved solubility, or more desirable absorption, distribution, metabolism, excretion, and toxicity (ADMET) characteristics. The structures of tubulin bound to MI-181 and C2 described here may enable the improvement of these potent, anticancer compounds as additional studies further examine the two compounds.
Materials and Methods
Luria-Bertani medium also known as lysogeny broth (LB) was purchased from EMD Millipore (Gibbstown, NJ).45 Antibiotics, DNase I, and lysozyme were from Sigma Chemical Company (St. Louis, MO). Isopropyl β-D-1-thiogalactopyranoside (IPTG) and dithiothreitol (DTT) were from Gold Biotechnology, Inc. (St. Louis, MO). The compounds MI-181 and C2 (>95% purity) were purchased through MolPort (Riga, Latvia). Protease inhibitor tablets and other chemicals were from Roche and Fisher Scientific, respectively (Indianapolis, IN and Pittsburgh, PA).
The rat STMN4 stathmin-like domain with the mutation Phe20Trp and chicken TTL genes were cloned into pET22b(+) (Novagen) with no additional residues or a C-terminal hexahistidine tag, respectively. The proteins were recombinantly expressed and purified using established methods.15,46 Lyophilized bovine brain tubulin (>99% purity) was purchased from Cytoskeleton, Inc. (Denver, CO), reconstituted to form T2R-TTL complexes, and crystallized with sitting-drop vapor diffusion as described previously.15,31 Crystals were soaked for 24 h in well solution containing 1 mM compound with 10% DMSO. The crystal soaked with MI-181 was cryoprotected in Paratone-N oil and the C2 complex crystal was cryoprotected in well solution with 16% total glycerol then flash-frozen in liquid nitrogen.
Diffraction data were collected at 100K at the Advanced Photon Source (APS) Northeastern Collaborative Access Team (NECAT) beamline 24-ID-C on a DECTRIS PILATUS 6M-F detector. The data collection and refinement statistics are reported in Table1. Data from both crystals were processed using XDS/XSCALE.47 The program Phaser48 was used to solve both structures by molecular replacement (MR) using a high-resolution colchicine-bound structure of T2R-TTL (PDB ID 4O2B) with all nonprotein atoms removed as the search model.30 Both asymmetric units contain one complex of T2R-TTL. Residue numbering for tubulin and stathmin are based on previously established conventions.13,46 MR solutions were initially refined with rigid-body refinement using the phenix.refine module of PHENIX.49 Ligand structures and restraints for MI-181 and C2 were generated with SMILES input for phenix.eLBOW50 using AM1(RM1) geometry optimization, followed by manual restraint of the ethylene linker in MI-181 to the (E)-isomer.51,52 Placement of MI-181 and C2 was carried out manually and subsequently compared to the placement calculated by the LigandFit module of PHENIX.41,42 The placement of C2 was further examined using AutoDock Tools and AutoDock Vina.40 Partial atomic Gasteiger charges and hydrogens were added to models of C2 and the T2R-TTL structure in the absence of C2. The colchicine-binding site was used to center the grid search box with a volume of 40 × 40 × 40 Å using an exhaustiveness parameter of 24. Other ligands in the structures were added early in refinement after inspection of the mFo-DFc difference map in Coot.53
Table 1.
X-ray Data Collection and Refinement Statistics
| T2R-TTL MI-181 | T2R-TTL C2 | |
|---|---|---|
| Data collection | ||
| Space group | P 21 21 21 | P 21 21 21 |
| Unit cell a b c (Å), α = β = γ (°) | 104.83 157.65 181.03, 90 | 105.2 157.69 181.92, 90 |
| Resolution range (Å)a | 90.72−2.60 (2.69−2.60) | 91.07 - 3.75 (3.88 - 3.75) |
| Unique reflections | 92610 (9094) | 31487 (3092) |
| Multiplicity | 6.7 (6.5) | 4.4 (4.6) |
| Completeness (%) | 99.76 (98.99) | 99.25 (99.39) |
| Mean I/σ(I) | 17.89 (1.69) | 4.83 (1.40) |
| Rmergeb | 0.076 (1.076) | 0.371 (1.404) |
| CC1/2c | 0.99 (0.63) | 0.98 (0.49) |
| Res. <I/σ>∼2 (Å)c | 2.65 | 3.90 |
| Wilson B-factor | 62.9 | 81.8 |
| Refinement | ||
| R-work/R-free (%)d | 18.8/23.1 | 23.9 / 27.9 |
| No. of nonhydrogen atoms | 17,335 | 17,035 |
| Macromolecules | 17,000 | 16,823 |
| Ligands | 224 | 196 |
| Water | 111 | 16 |
| Protein residues | 2137 | 2,124 |
| RMSD(bonds) (Å) | 0.002 | 0.002 |
| RMSD(angles) (°) | 0.55 | 0.47 |
| Ramachandran favored (%)e | 97.0 | 96.0 |
| Ramachandran allowed (%) | 3.0 | 4.0 |
| Ramachandran outliers (%) | 0.0 | 0.0 |
| Clashscoree | 2.86 | 3.76 |
| Average B-factor (Å2) | 76.1 | 99.5 |
| Macromolecules | 76.4 | 99.7 |
| Ligands | 67.1 | 83.8 |
| Solvent | 53.9 | 78.4 |
| PDB ID | 4YJ2 | 4YJ3 |
Values in the highest resolution shell are shown in parenthesis.
Rmerge = Σ | I-<I> | / Σ I where I is the integrated intensity of a given reflection.
CC1/2 is the random half-data set correlation coefficient33 and the resolution at which <I/σ>∼2 (Å) is given for interpretation of traditional resolution criteria.
Rfree was calculated using 10% of the data.
Percentages of residues in Ramachandran plot regions and clashscores, or the numbers of unfavorable all-atom steric overlaps ≥ 0.4 Å per 1000 atoms, were determined using MolProbity.58
Both structures were parameterized with individual coordinate and individual (MI-181) or grouped-per-residue (C2) isotropic atomic displacement parameter (ADP) refinement with translation libration screw-motion (TLS) group definitions matching previous T2R-TTL structure group definitions.15,54 Iterative cycles of alternating refinement and model adjustment in Coot were performed using 2mFo-DFc and mFo-DFc difference maps to obtain the final models. Residues primarily in TTL with real-space density correlation coefficients below 0.6 were omitted from the model. The coordinates of the final models and the structure factors have been deposited in the Protein Data Bank with PDB codes 4YJ2 and 4YJ3. Structures were analyzed using Chimera and PyMOL, distance measurements were calculated using β-tubulin from chain B of the structure coordinates, and all figures were prepared in PyMOL.55–57
Acknowledgments
The authors thank Michael Sawaya, Duilio Cascio, Ankur Gholkar, and David Leibly for valuable discussions and help with crystallographic data collection. We also thank M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov and F. Murphy at NECAT beamline 24-ID of the Argonne National Laboratory APS. X-ray diffraction data were collected at the Argonne National Laboratory APS.
Glossary
- ADMET
absorption, distribution, metabolism, excretion, and toxicity
- AMPPCP
adenylylmethylenediphosphonate disodium salt
- GTP
guanosine-5'-triphosphate
- IC50
half-maximal inhibitory concentration
- mM
millimolar
- MR
molecular replacement
- nM
nanomolar
- rmsd
root-mean-square deviation
- SAC
spindle assembly checkpoint
- SAR
structure-activity-relationship
- TTL
tubulin tyrosine ligase
- T2R
a complex of two αβ-tubulin heterodimers bound to one RB3 stathmin-like domain
- T2R-TTL
the T2R complex bound to TTL.
References
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10:194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, Mayer TU, Miyamoto DT, Fathi R, King RW, Mitchison TJ, Schreiber SL. Dissecting cellular processes using small molecules: identification of colchicine-like, taxol-like and other small molecules that perturb mitosis. Chem Biol. 2000;7:275–286. doi: 10.1016/s1074-5521(00)00101-0. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Si Y, Thompsons CM, Smellie A, Ashwell MA, Liu JF, Ye P, Yohannes D, Ng SC. Identification of a small molecule that induces mitotic arrest using a simplified high-content screening assay and data analysis method. J Biomol Screening. 2006;11:21–28. doi: 10.1177/1087057105280726. [DOI] [PubMed] [Google Scholar]
- Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonchick AP, Gerding-Reimers C, Catarinella M, Schurmann M, Preut H, Ziegler S, Rauh D, Waldmann H. Highly enantioselective synthesis and cellular evaluation of spirooxindoles inspired by natural products. Nat Chem. 2010;2:735–740. doi: 10.1038/nchem.730. [DOI] [PubMed] [Google Scholar]
- Chan KS, Koh CG, Li HY. Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis. 2012;3:e411. doi: 10.1038/cddis.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ahn S, Wang J, Lu Y, Dalton JT, Miller DD, Li W. Discovery of novel 2-aryl-4-benzoyl-imidazole (ABI-III) analogues targeting tubulin polymerization as antiproliferative agents. J Med Chem. 2012;55:7285–7289. doi: 10.1021/jm300564b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibbeson BM, Laraia L, Alza E,CJOC, Tan YS, Davies HM, McKenzie G, Venkitaraman AR, Spring DR. Diversity-oriented synthesis as a tool for identifying new modulators of mitosis. Nat Commun. 2014;5:3155. doi: 10.1038/ncomms4155. [DOI] [PubMed] [Google Scholar]
- Laraia L, Stokes J, Emery A, McKenzie GJ, Venkitaraman AR, Spring DR. High content screening of diverse compound libraries identifies potent modulators of tubulin dynamics. ACS Med Chem Lett. 2014;5:598–603. doi: 10.1021/ml5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese S, Lo YC, Huang D, Zangle TA, Gholkar AA, Robert L, Homet B, Ribas A, Summers MK, Teitell MA, Damoiseaux R, Torres JZ. Chemical dissection of the cell cycle: probes for cell biology and anti-cancer drug development. Cell Death Dis. 2014;5:e1462. doi: 10.1038/cddis.2014.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Shimada K, Delva D, Patel M, Ode E, Skouta R, Stockwell BR. Identification of simple compounds with microtubule-binding activity that inhibit cancer cell growth with high potency. ACS Med Chem Lett. 2012;3:35–38. doi: 10.1021/ml200195s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M. The 4 A X-ray structure of a tubulin:stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Zurwerra D, Field JJ, Diaz JF, Altmann KH, Steinmetz MO. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Gigant B, Wang C, Ravelli RB, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- Cormier A, Marchand M, Ravelli RB, Knossow M, Gigant B. Structural insight into the inhibition of tubulin by vinca domain peptide ligands. EMBO Rep. 2008;9:1101–1106. doi: 10.1038/embor.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorleans A, Gigant B, Ravelli RB, Mailliet P, Mikol V, Knossow M. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc Natl Acad Sci USA. 2009;106:13775–13779. doi: 10.1073/pnas.0904223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranaivoson FM, Gigant B, Berritt S, Joullie M, Knossow M. Structural plasticity of tubulin assembly probed by vinca-domain ligands. Acta Cryst D. 2012;68:927–934. doi: 10.1107/S0907444912017143. [DOI] [PubMed] [Google Scholar]
- Maderna A, Doroski M, Subramanyam C, Porte A, Leverett CA, Vetelino BC, Chen Z, Risley H, Parris K, Pandit J, Varghese AH, Shanker S, Song C, Sukuru SC, Farley KA, Wagenaar MM, Shapiro MJ, Musto S, Lam MH, Loganzo F, O'Donnell CJ. Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications. J Med Chem. 2014;57:10527–10543. doi: 10.1021/jm501649k. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Rice LM. The contribution of alphabeta-tubulin curvature to microtubule dynamics. J Cell Biol. 2014;207:323–334. doi: 10.1083/jcb.201407095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecqueur L, Duellberg C, Dreier B, Jiang Q, Wang C, Pluckthun A, Surrey T, Gigant B, Knossow M. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc Natl Acad Sci USA. 2012;109:12011–12016. doi: 10.1073/pnas.1204129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang W, Dreier B, Jiang Q, Pecqueur L, Pluckthun A, Wang C, Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Okada Y, Hirokawa N. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- Asenjo AB, Chatterjee C, Tan D, DePaoli V, Rice WJ, Diaz-Avalos R, Silvestry M, Sosa H. Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep. 2013;3:759–768. doi: 10.1016/j.celrep.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Diaz JF, Marsh M, Cuevas C, Liniger M, Neuhaus C, Andreu JM, Altmann KH, Steinmetz MO. A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proc Natl Acad Sci USA. 2014;111:13817–13821. doi: 10.1073/pnas.1408124111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Northcote PT, Marsh M, Altmann KH, Miller JH, Diaz JF, Steinmetz MO. Structural basis of microtubule stabilization by laulimalide and peloruside A. Angewandte Chemie (International ed in English) 2014;53:1621–1625. doi: 10.1002/anie.201307749. [DOI] [PubMed] [Google Scholar]
- Prota AE, Danel F, Bachmann F, Bargsten K, Buey RM, Pohlmann J, Reinelt S, Lane H, Steinmetz MO. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J Mol Biol. 2014;426:1848–1860. doi: 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Prota AE, Magiera MM, Kuijpers M, Bargsten K, Frey D, Wieser M, Jaussi R, Hoogenraad CC, Kammerer RA, Janke C, Steinmetz MO. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol. 2013;200:259–270. doi: 10.1083/jcb.201211017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier A, Knossow M, Wang C, Gigant B. The binding of vinca domain agents to tubulin: structural and biochemical studies. Methods Cell Biol. 2010;95:373–390. doi: 10.1016/S0091-679X(10)95020-6. [DOI] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Zhou P, Tian F, Lv F, Shang Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009;76:151–163. doi: 10.1002/prot.22327. [DOI] [PubMed] [Google Scholar]
- Iwaoka M, Isozumi N. Hypervalent nonbonded interactions of a divalent sulfur atom. Implications in protein architecture and the functions. Molecules. 2012;17:7266–7283. doi: 10.3390/molecules17067266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaoka M, Takemoto S, Okada M, Tomoda S. Weak nonbonded S --- X (X=O, N, and S) interactions in proteins. Statistical and theoretical studies. B Chem Soc Jpn. 2002;75:1611–1625. [Google Scholar]
- Allen FH, Bird CM, Rowland RS, Raithby PR. Hydrogen-bond acceptor and donor properties of divalent sulfur (Y-S-Z and R-S-H) Acta Cryst B. 1997;53:696–701. [Google Scholar]
- Da C, Telang N, Hall K, Kluball E, Barelli P, Finzel K, Jia X, Gupton JT, Mooberry SL, Kellogg GE. Developing novel C-4 analogues of pyrrole-based antitubulin agents: weak but critical hydrogen bonding in the colchicine site. Med Chem Comm. 2013;4:417–421. doi: 10.1039/C2MD20320K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comp Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Klei H, Adams PD, Moriarty NW, Cohn JD. Automated ligand fitting by core-fragment fitting and extension into density. Acta Cryst D. 2006;62:915–922. doi: 10.1107/S0907444906017161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Moriarty NW, Cohn JD. Ligand identification using electron-density map correlations. Acta Cryst D. 2007;63:101–107. doi: 10.1107/S0907444906046233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molife LR, Imseeh G, Capelan M, El-Khouly F, Cresti N, Smith AD, Averion D, Haris NM, Stimpson SJ, Gumbleton T, Lane HA, Bachmann F, Schmitt-Hoffmann A, Tzankov A, Hannah AL, Anderson S, Bette U, Calvert AH, Plummer R, Kristeleit RS. Phase I/IIa trial of the novel microtubule inhibitor BAL101553 in advanced solid tumors: phase I completed. J Clin Oncol. 2014;32:2562. [Google Scholar]
- Cai S, Drewe J, Zhang HZ, Kasibhatla S, Claassen G, Sirisoma N, Kemnitzer W. 3-Aryl-6-aryl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazines and analogs as activators of caspases and inducers of apoptosis and the use thereof. (2008) Cytovia, Inc [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbaut E, Curmi PA, Ozon S, Lachkar S, Redeker V, Sobel A. Stathmin family proteins display specific molecular and tubulin binding properties. J Biol Chem. 2001;276:16146–16154. doi: 10.1074/jbc.M010637200. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Cryst D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Cryst D. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. AM1: a new general purpose quantum mechanical molecular model. J Am Chem Soc. 1985;107:3902–3909. [Google Scholar]
- Rocha GB, Freire RO, Simas AM, Stewart JJ. RM1: a reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. J Comp Chem. 2006;27:1101–1111. doi: 10.1002/jcc.20425. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System v. 1.5. 2012. (Schrodinger, LLC )
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]


