Abstract
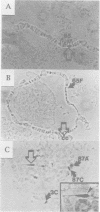
Heterochromatic position-effect variegation (PEV) describes the mosaic phenotype of a euchromatic gene placed next to heterochromatin. Heterochromatin-mediated silencing has been studied extensively in Drosophila, but the lack of a ubiquitous reporter gene detectable at any stage has prevented a direct developmental characterization of this phenomenon. Current models attribute variegation to the establishment of a heritable silent state in a subset of the cells and invoke differences in the timing of silencing to explain differences in the patch size of various mosaic patterns. In order to follow the course of heterochromatic silencing directly, we have generated Drosophila lines variegating for a lacZ reporter that can be induced in virtually all cells at any developmental stage. Our data indicate that silencing begins in embryogenesis and persists in both somatic and germline lineages. A heterogeneity in the extent of silencing is also revealed; silencing is suppressed in differentiated tissues but remains widespread in larval imaginal discs containing precursor cells for adult structures. Using eye development as an example, we propose that the mosaic phenotype is determined during differentiation by a variegated relaxation in heterochromatic silencing. Though unpredicted by prevailing models, this mechanism is evident in other analogous systems.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker W. K. A clonal system of differential gene activity in Drosophila. Dev Biol. 1967 Jul;16(1):1–17. doi: 10.1016/0012-1606(67)90014-0. [DOI] [PubMed] [Google Scholar]
- Baker W. K. Position-effect variegation. Adv Genet. 1968;14:133–169. [PubMed] [Google Scholar]
- Belyaeva E. S., Zhimulev I. F. Cytogenetic and molecular aspects of position effect variegation in Drosophila. III. Continuous and discontinuous compaction of chromosomal material as a result of position effect variegation. Chromosoma. 1991 Aug;100(7):453–466. doi: 10.1007/BF00364556. [DOI] [PubMed] [Google Scholar]
- Bonner J. J., Parks C., Parker-Thornburg J., Mortin M. A., Pelham H. R. The use of promoter fusions in Drosophila genetics: isolation of mutations affecting the heat shock response. Cell. 1984 Jul;37(3):979–991. doi: 10.1016/0092-8674(84)90432-x. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M. Position effect variegation in the mouse. Genet Res. 1974 Jun;23(3):291–306. doi: 10.1017/s0016672300014932. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Rastelli L., Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994 Jun 1;13(11):2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer D. R., Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994 Jul 1;77(7):993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Dura J. M. Stage dependent synthesis of heat shock induced proteins in early embryos of Drosophila melanogaster. Mol Gen Genet. 1981;184(3):381–385. doi: 10.1007/BF00352509. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Hartnett T. A heat shock-activated cDNA rescues the recessive lethality of mutations in the heterochromatin-associated protein HP1 of Drosophila melanogaster. Mol Gen Genet. 1993 Sep;240(3):333–338. doi: 10.1007/BF00280383. [DOI] [PubMed] [Google Scholar]
- Ephrussi B, Herold J L. Studies of Eye Pigments of Drosophila. I. Methods of Extraction and Quantitative Estimation of the Pigment Components. Genetics. 1944 Mar;29(2):148–175. doi: 10.1093/genetics/29.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvarque M. O., Dura J. M. polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 1993 Aug;7(8):1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- Fjose A., Polito L. C., Weber U., Gehring W. J. Developmental expression of the white locus of Drosophila melanogaster. EMBO J. 1984 Sep;3(9):2087–2094. doi: 10.1002/j.1460-2075.1984.tb02095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Gowen J W, Gay E H. Chromosome Constitution and Behavior in Eversporting and Mottling in Drosophila Melanogaster. Genetics. 1934 May;19(3):189–208. doi: 10.1093/genetics/19.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigliatti T. Position-effect variegation--an assay for nonhistone chromosomal proteins and chromatin assembly and modifying factors. Methods Cell Biol. 1991;35:587–627. [PubMed] [Google Scholar]
- Hartmann-Goldstein I. J. On the relationship between heterochromatization and variegation in Drosophila, with special reference to temperature-sensitive periods. Genet Res. 1967 Oct;10(2):143–159. doi: 10.1017/s0016672300010880. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation and chromosome structure of a heat shock puff in Drosophila. Chromosoma. 1981;83(3):381–393. doi: 10.1007/BF00327360. [DOI] [PubMed] [Google Scholar]
- James T. C., Eissenberg J. C., Craig C., Dietrich V., Hobson A., Elgin S. C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989 Oct;50(1):170–180. [PubMed] [Google Scholar]
- Karess R. E., Rubin G. M. Analysis of P transposable element functions in Drosophila. Cell. 1984 Aug;38(1):135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Karpen G. H. Position-effect variegation and the new biology of heterochromatin. Curr Opin Genet Dev. 1994 Apr;4(2):281–291. doi: 10.1016/s0959-437x(05)80055-3. [DOI] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990 Oct 5;63(1):97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991 Mar 8;64(5):941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Kornher J. S., Kauffman S. A. Variegated expression of the Sgs-4 locus in Drosophila melanogaster. Chromosoma. 1986;94(3):205–216. doi: 10.1007/BF00288495. [DOI] [PubMed] [Google Scholar]
- Laird C. D. Proposed mechanism of inheritance and expression of the human fragile-X syndrome of mental retardation. Genetics. 1987 Nov;117(3):587–599. doi: 10.1093/genetics/117.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson P., Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992 Dec;56(4):543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J., Kotarski M. A., Tartof K. D. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988 Sep;120(1):181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz A., Ostermann K., Fleck O., Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994 May 27;143(1):139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- Lyon M. F. Some milestones in the history of X-chromosome inactivation. Annu Rev Genet. 1992;26:16–28. doi: 10.1146/annurev.ge.26.120192.000313. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987 Sep 4;237(4819):1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- Paro R., Hogness D. S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990 Dec;6(12):416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Pillus L., Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989 Nov 17;59(4):637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S., Goday C. Unusual kinetochores and chromatin diminution in Parascaris. Trends Genet. 1989 Sep;5(9):310–315. doi: 10.1016/0168-9525(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Rastelli L. White gene expression, repressive chromatin domains and homeotic gene regulation in Drosophila. Bioessays. 1994 Aug;16(8):549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- Qian S., Varjavand B., Pirrotta V. Molecular analysis of the zeste-white interaction reveals a promoter-proximal element essential for distant enhancer-promoter communication. Genetics. 1992 May;131(1):79–90. doi: 10.1093/genetics/131.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992 Sep;14(9):605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Reuter G., Werner W., Hoffmann H. J. Mutants affecting position-effect heterochromatinization in Drosophila melanogaster. Chromosoma. 1982;85(4):539–551. doi: 10.1007/BF00327349. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., Engels W. R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988 Mar;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S., Chue C., Goebl M., Craig C., Clark R. F., Powers J. A., Eissenberg J. C., Elgin S. C., Rothfield N. F., Earnshaw W. C. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J Cell Sci. 1993 Feb;104(Pt 2):573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- Simon J. A., Sutton C. A., Lobell R. B., Glaser R. L., Lis J. T. Determinants of heat shock-induced chromosome puffing. Cell. 1985 Apr;40(4):805–817. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- Singh P. B., Miller J. R., Pearce J., Kothary R., Burton R. D., Paro R., James T. C., Gaunt S. J. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991 Feb 25;19(4):789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. B. Molecular mechanisms of cellular determination: their relation to chromatin structure and parental imprinting. J Cell Sci. 1994 Oct;107(Pt 10):2653–2668. doi: 10.1242/jcs.107.10.2653. [DOI] [PubMed] [Google Scholar]
- Spradling A. C. Position effect variegation and genomic instability. Cold Spring Harb Symp Quant Biol. 1993;58:585–596. doi: 10.1101/sqb.1993.058.01.065. [DOI] [PubMed] [Google Scholar]
- Steller H., Pirrotta V. Expression of the Drosophila white gene under the control of the hsp70 heat shock promoter. EMBO J. 1985 Dec 30;4(13B):3765–3772. doi: 10.1002/j.1460-2075.1985.tb04146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. P., Zhou S. X., Tan S. S. X-chromosome activity of the mouse primordial germ cells revealed by the expression of an X-linked lacZ transgene. Development. 1994 Oct;120(10):2925–2932. doi: 10.1242/dev.120.10.2925. [DOI] [PubMed] [Google Scholar]
- Tan S. S., Williams E. A., Tam P. P. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat Genet. 1993 Feb;3(2):170–174. doi: 10.1038/ng0293-170. [DOI] [PubMed] [Google Scholar]
- Tartof K. D., Bremer M. Mechanisms for the construction and developmental control of heterochromatin formation and imprinted chromosome domains. Dev Suppl. 1990:35–45. [PubMed] [Google Scholar]
- Tartof K. D., Hobbs C., Jones M. A structural basis for variegating position effects. Cell. 1984 Jul;37(3):869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]
- Thompson J. S., Hecht A., Grunstein M. Histones and the regulation of heterochromatin in yeast. Cold Spring Harb Symp Quant Biol. 1993;58:247–256. doi: 10.1101/sqb.1993.058.01.029. [DOI] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Sedat J., Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982 Sep 6;146(1):148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- Wallrath L. L., Elgin S. C. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995 May 15;9(10):1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Wareham K. A., Lyon M. F., Glenister P. H., Williams E. D. Age related reactivation of an X-linked gene. 1987 Jun 25-Jul 1Nature. 327(6124):725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- Wustmann G., Szidonya J., Taubert H., Reuter G. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol Gen Genet. 1989 Jun;217(2-3):520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]
- Zhimulev I. F., Belyaeva E. S., Bgatov A. V., Baricheva E. M., Vlassova I. E. Cytogenetic and molecular aspects of position effect variegation in Drosophila melanogaster. II. Peculiarities of morphology and genetic activity of the 2B region in the T(1;2)dorvar7 chromosome in males. Chromosoma. 1988;96(3):255–261. doi: 10.1007/BF00302365. [DOI] [PubMed] [Google Scholar]