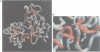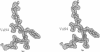Abstract
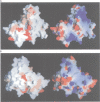
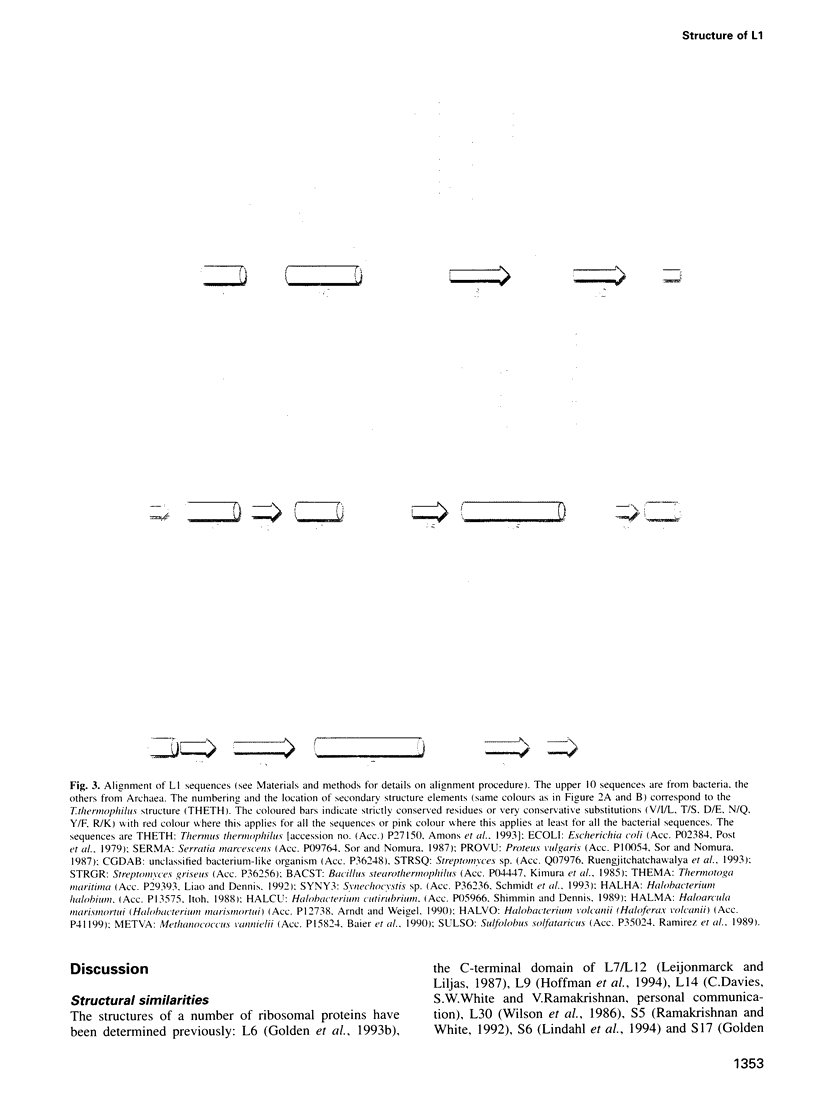
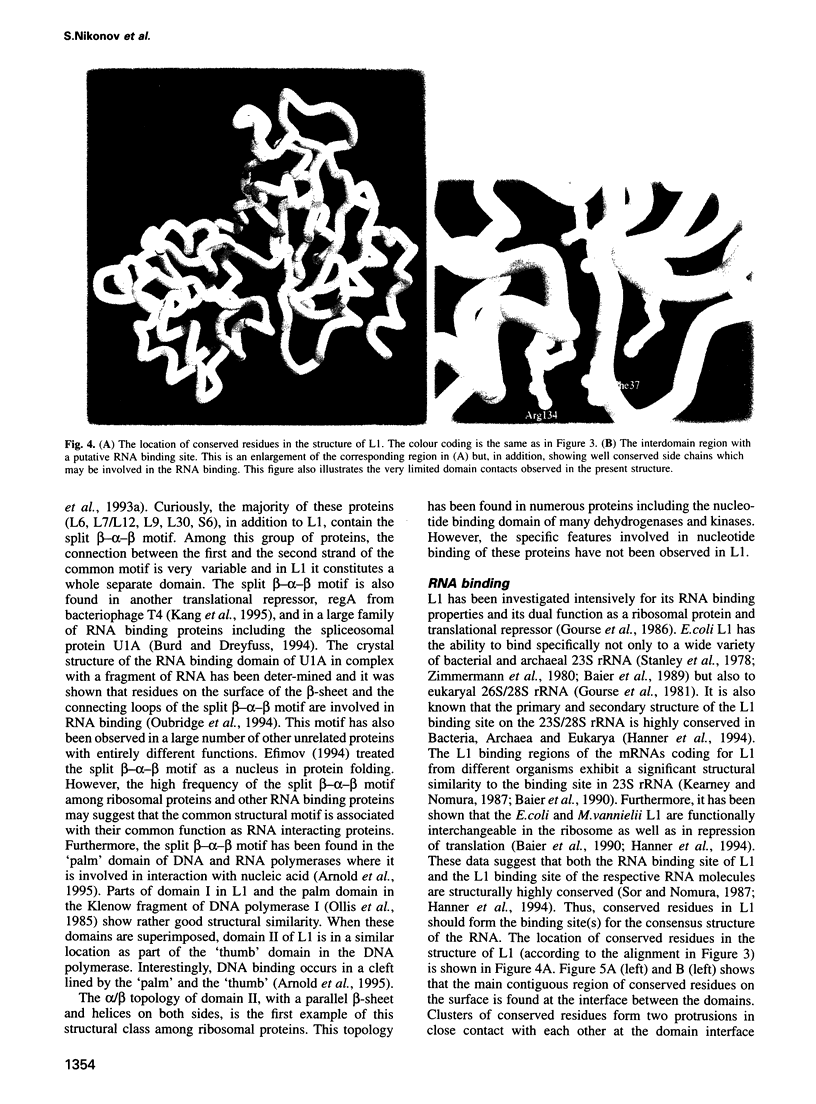
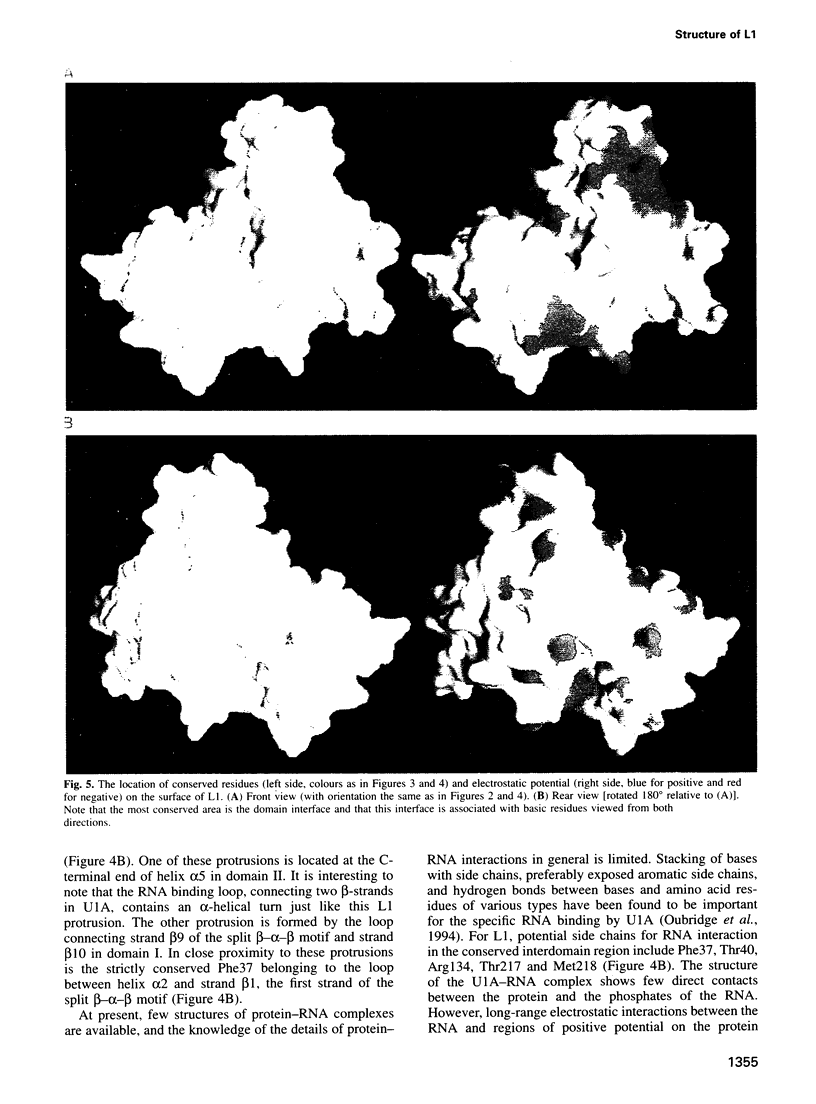
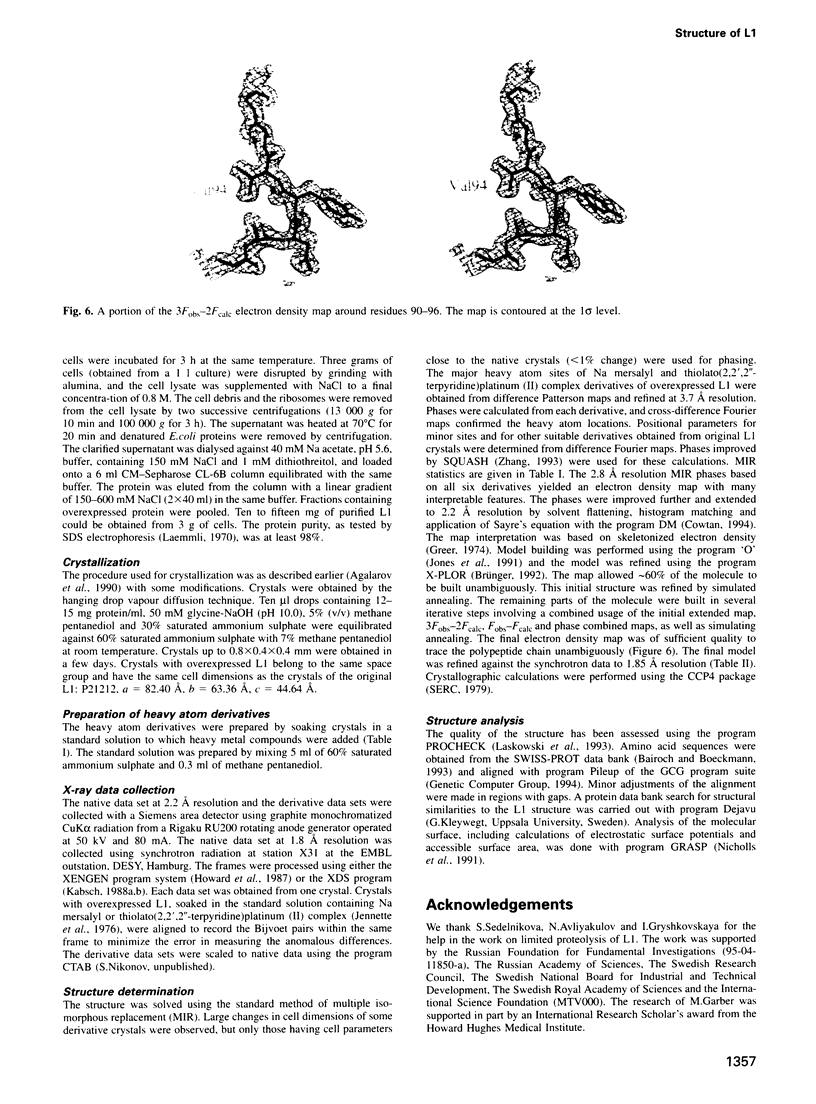
L1 has a dual function as a ribosomal protein binding rRNA and as a translational repressor binding mRNA. The crystal structure of L1 from Thermus thermophilus has been determined at 1.85 angstroms resolution. The protein is composed of two domains with the N- and C-termini in domain I. The eight N-terminal residues are very flexible, as the quality of electron density map shows. Proteolysis experiments have shown that the N-terminal tail is accessible and important for 23S rRNA binding. Most of the conserved amino acids are situated at the interface between the two domains. They probably form the specific RNA binding site of L1. Limited non-covalent contacts between the domains indicate an unstable domain interaction in the present conformation. Domain flexibility and RNA binding by induced fit seems plausible.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agalarov S. C., Eliseikina I. A., Sedelnikova S. E., Fomenkova N. P., Nikonov S. V., Garber M. B. Crystals of protein L1 from the 50 S ribosomal subunit of Thermus thermophilus. Preliminary crystallographic data. J Mol Biol. 1990 Dec 5;216(3):501–502. doi: 10.1016/0022-2836(90)90375-V. [DOI] [PubMed] [Google Scholar]
- Amons R., Muranova T. A., Rykunova A. I., Eliseikina I. A., Sedelnikova S. E. The complete primary structure of ribosomal protein L1 from Thermus thermophilus. J Protein Chem. 1993 Dec;12(6):725–734. doi: 10.1007/BF01024930. [DOI] [PubMed] [Google Scholar]
- Argos P. An investigation of protein subunit and domain interfaces. Protein Eng. 1988 Jul;2(2):101–113. doi: 10.1093/protein/2.2.101. [DOI] [PubMed] [Google Scholar]
- Arndt E., Weigel C. Nucleotide sequence of the genes encoding the L11, L1, L10 and L12 equivalent ribosomal proteins from the archaebacterium Halobacterium marismortui. Nucleic Acids Res. 1990 Mar 11;18(5):1285–1285. doi: 10.1093/nar/18.5.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E., Ding J., Hughes S. H., Hostomsky Z. Structures of DNA and RNA polymerases and their interactions with nucleic acid substrates. Curr Opin Struct Biol. 1995 Feb;5(1):27–38. doi: 10.1016/0959-440x(95)80006-m. [DOI] [PubMed] [Google Scholar]
- Baier G., Piendl W., Redl B., Stöffler G. Structure, organization and evolution of the L1 equivalent ribosomal protein gene of the archaebacterium Methanococcus vannielii. Nucleic Acids Res. 1990 Feb 25;18(4):719–724. doi: 10.1093/nar/18.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank, recent developments. Nucleic Acids Res. 1993 Jul 1;21(13):3093–3096. doi: 10.1093/nar/21.13.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Draper D. E. How do proteins recognize specific RNA sites? New clues from autogenously regulated ribosomal proteins. Trends Biochem Sci. 1989 Aug;14(8):335–338. doi: 10.1016/0968-0004(89)90167-9. [DOI] [PubMed] [Google Scholar]
- Efimov A. V. Common structural motifs in small proteins and domains. FEBS Lett. 1994 Dec 5;355(3):213–219. doi: 10.1016/0014-5793(94)01194-x. [DOI] [PubMed] [Google Scholar]
- Frank J., Zhu J., Penczek P., Li Y., Srivastava S., Verschoor A., Radermacher M., Grassucci R., Lata R. K., Agrawal R. K. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 1995 Aug 3;376(6539):441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- Golden B. L., Hoffman D. W., Ramakrishnan V., White S. W. Ribosomal protein S17: characterization of the three-dimensional structure by 1H and 15N NMR. Biochemistry. 1993 Nov 30;32(47):12812–12820. doi: 10.1021/bi00210a033. [DOI] [PubMed] [Google Scholar]
- Golden B. L., Ramakrishnan V., White S. W. Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein. EMBO J. 1993 Dec 15;12(13):4901–4908. doi: 10.1002/j.1460-2075.1993.tb06184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer J. Three-dimensional pattern recognition: an approach to automated interpretation of electron density maps of proteins. J Mol Biol. 1974 Jan 25;82(3):279–301. doi: 10.1016/0022-2836(74)90591-9. [DOI] [PubMed] [Google Scholar]
- Görlach M., Wittekind M., Beckman R. A., Mueller L., Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992 Sep;11(9):3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanner M., Mayer C., Köhrer C., Golderer G., Gröbner P., Piendl W. Autogenous translational regulation of the ribosomal MvaL1 operon in the archaebacterium Methanococcus vannielii. J Bacteriol. 1994 Jan;176(2):409–418. doi: 10.1128/jb.176.2.409-418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D. W., Davies C., Gerchman S. E., Kycia J. H., Porter S. J., White S. W., Ramakrishnan V. Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein. EMBO J. 1994 Jan 1;13(1):205–212. doi: 10.1002/j.1460-2075.1994.tb06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kang C., Chan R., Berger I., Lockshin C., Green L., Gold L., Rich A. Crystal structure of the T4 regA translational regulator protein at 1.9 A resolution. Science. 1995 May 26;268(5214):1170–1173. doi: 10.1126/science.7761833. [DOI] [PubMed] [Google Scholar]
- Kearney K. R., Nomura M. Secondary structure of the autoregulatory mRNA binding site of ribosomal protein L1. Mol Gen Genet. 1987 Nov;210(1):60–68. doi: 10.1007/BF00337759. [DOI] [PubMed] [Google Scholar]
- Kimura M., Kimura J., Ashman K. The complete primary structure of ribosomal proteins L1, L14, L15, L23, L24 and L29 from Bacillus stearothermophilus. Eur J Biochem. 1985 Aug 1;150(3):491–497. doi: 10.1111/j.1432-1033.1985.tb09049.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leijonmarck M., Liljas A. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 A. J Mol Biol. 1987 Jun 5;195(3):555–579. doi: 10.1016/0022-2836(87)90183-5. [DOI] [PubMed] [Google Scholar]
- Lindahl M., Svensson L. A., Liljas A., Sedelnikova S. E., Eliseikina I. A., Fomenkova N. P., Nevskaya N., Nikonov S. V., Garber M. B., Muranova T. A. Crystal structure of the ribosomal protein S6 from Thermus thermophilus. EMBO J. 1994 Mar 15;13(6):1249–1254. doi: 10.2210/pdb1ris/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E. A., Murphy M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994 Nov 1;50(Pt 6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Ito N., Avis J., Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995 Jun;20(6):235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Thornton J. M. Alpha plus beta folds revisited: some favoured motifs. Structure. 1993 Oct 15;1(2):105–120. doi: 10.1016/0969-2126(93)90026-d. [DOI] [PubMed] [Google Scholar]
- Oubridge C., Ito N., Evans P. R., Teo C. H., Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994 Dec 1;372(6505):432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Post L. E., Strycharz G. D., Nomura M., Lewis H., Dennis P. P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989 Apr 7;57(1):89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Schmidt J., Bubunenko M., Subramanian A. R. A novel operon organization involving the genes for chorismate synthase (aromatic biosynthesis pathway) and ribosomal GTPase center proteins (L11, L1, L10, L12: rplKAJL) in cyanobacterium Synechocystis PCC 6803. J Biol Chem. 1993 Dec 25;268(36):27447–27457. [PubMed] [Google Scholar]
- Shimmin L. C., Dennis P. P. Characterization of the L11, L1, L10 and L12 equivalent ribosomal protein gene cluster of the halophilic archaebacterium Halobacterium cutirubrum. EMBO J. 1989 Apr;8(4):1225–1235. doi: 10.1002/j.1460-2075.1989.tb03496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Mueller F., Orlova E. V., Schatz M., Dube P., Erdemir T., Zemlin F., Brimacombe R., van Heel M. The 70S Escherichia coli ribosome at 23 A resolution: fitting the ribosomal RNA. Structure. 1995 Aug 15;3(8):815–821. doi: 10.1016/s0969-2126(01)00216-7. [DOI] [PubMed] [Google Scholar]
- Subramanian A. R., Dabbs E. R. Functional studies on ribosomes lacking protein L1 from mutant Escherichia coli. Eur J Biochem. 1980 Nov;112(2):425–430. doi: 10.1111/j.1432-1033.1980.tb07222.x. [DOI] [PubMed] [Google Scholar]
- Wilson K. S., Appelt K., Badger J., Tanaka I., White S. W. Crystal structure of a prokaryotic ribosomal protein. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7251–7255. doi: 10.1073/pnas.83.19.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath A. Approaching atomic resolution in crystallography of ribosomes. Annu Rev Biophys Biomol Struct. 1992;21:77–93. doi: 10.1146/annurev.bb.21.060192.000453. [DOI] [PubMed] [Google Scholar]